The Oligomeric State of the Plasma Membrane H+-ATPase from Kluyveromyces lactis
Abstract
1. Introduction
2. Results and discussion
2.1. H+-ATPase Purification and Gel Filtration Chromatography
2.2. Blue Native Electrophoresis (BN-PAGE) of H+-ATPase Macromolecular Assembly States (MASs)
2.3. H+-ATPase Hexamer Identification by Western Blot (WB) in Native Plasma Membranes Treated with Dodecyl Maltoside (DDM)
2.4. H+-ATPase Kinetics
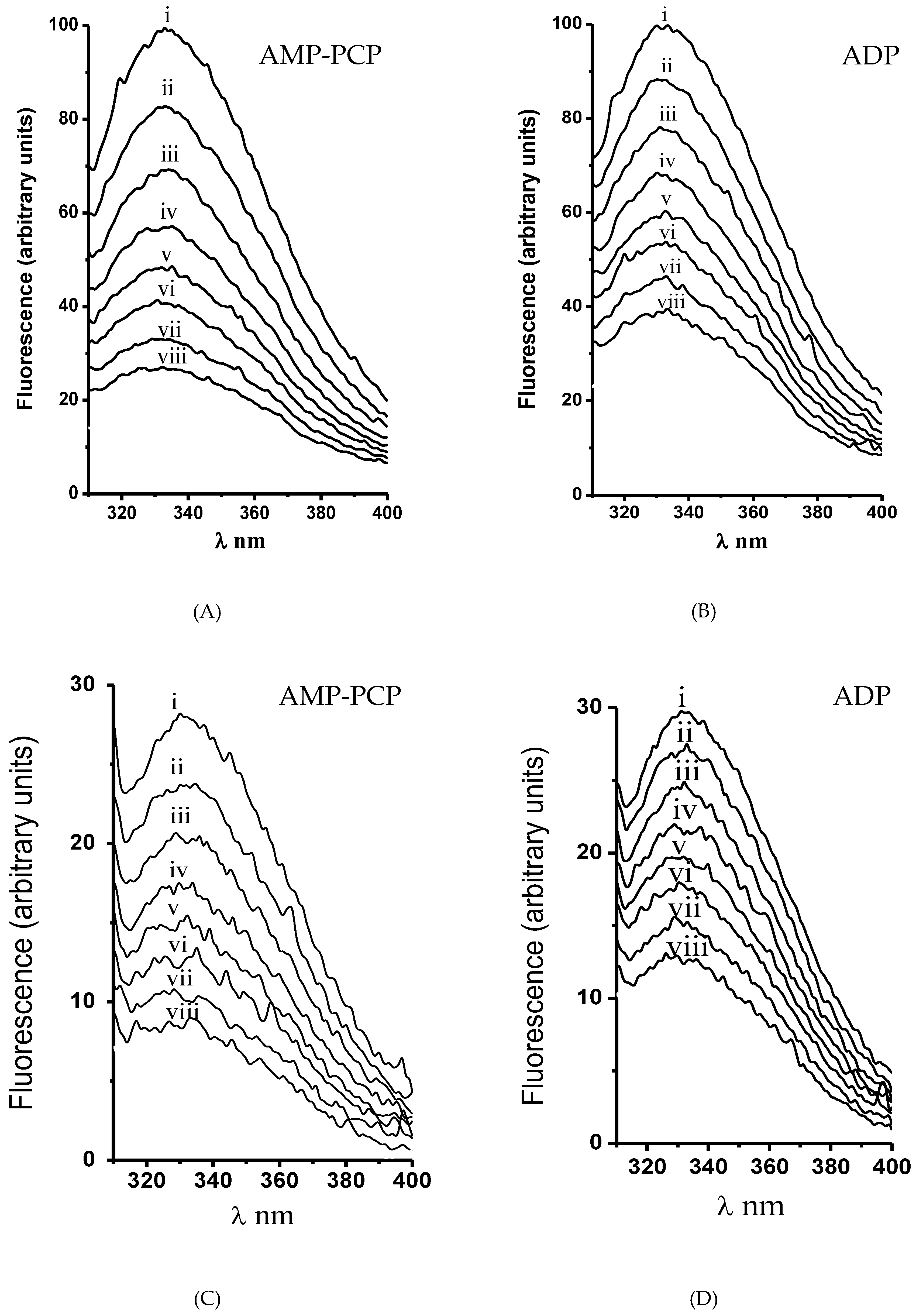
2.5. Intrinsic Fluorescence of H+-ATPase Hexamers
2.6. Nucleotide Affinity in H+-ATPase Hexamers
3. Materials and methods
3.1. Materials
3.2. Enzyme Purification
3.3. Size-Exclusion Chromatography
3.4. Blue Native Electrophoresis (BN-PAGE)
3.5. Western Blot (WB)
3.6. ATPase Activity Assay
3.7. H+-ATPase Intrinsic Fluorescence
3.8. Data Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Serrano, R.; Kielland-Brandt, M.C.; Fink, G.R. Yeast plasma membrane ATPase is essential for growth and has homology with (Na+ + K+), K+- and Ca2+-ATPases. Nature 1986, 319, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Serrano, R. In vivo glucose activation of the yeast plasma membrane ATPase. FEBS Lett. 1983, 156, 11–14. [Google Scholar] [CrossRef]
- Eraso, P.; Mazón, M.J.; Portillo, F. Yeast protein kinase Ptk2 localizes at the plasma membrane and phosphorylates in vitro the C-terminal peptide of the H+-ATPase. Biochim. Biophys. Acta (BBA) Biomembr. 2006, 1758, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Mazón, M.J.; Eraso, P.; Portillo, F. Specific phosphoantibodies reveal two phosphorylation sites in yeast Pma1 in response to glucose. FEMS Yeast Res. 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Portillo, F.; Eraso, P.; Serrano, R. Analysis of the regulatory domain of yeast plasma membrane H+-ATPase by directed mutagenesis and intragenic suppression. FEBS Lett. 1991, 287, 71–74. [Google Scholar] [CrossRef]
- Justesen, B.H.; Hansen, R.W.; Martens, H.J.; Theorin, L.; Palmgren, M.G.; Martinez, K.L.; Pomorski, T.G.; Fuglsang, A.T. Active plasma membrane P-type H+-ATPase reconstituted into nanodiscs is a monomer. J. Biol. Chem. 2013, 288, 26419–26429. [Google Scholar] [CrossRef] [PubMed]
- Goormaghtigh, E.; Chadwick, C.; Scarborough, G.A. Monomers of the Neurospora plasma membrane H+-ATPase catalyze efficient proton translocation. J. Biol. Chem. 1986, 261, 7466–7471. [Google Scholar]
- Dufour, J.-P.; Goffeau, A.A. Molecular and kinetic properties of the purified plasma membrane ATPase of the yeast Schizosaccharomyces pombe. Eur. J. Biochem. 1980, 105, 145–154. [Google Scholar] [CrossRef]
- Chadwick, C.C.; Goormaghtigh, E.; Scarborough, G.A. A hexameric form of the Neurospora crassa plasma membrane H+-ATPase. Arch. Biochem. Biophys. 1987, 252, 348–356. [Google Scholar] [CrossRef]
- Auer, M.; Scarborough, G.A.; Kühlbrandt, W. Surface crystallisation of the plasma membrane H+-ATPase on a carbon support film for electron crystallography. J. Mol. Biol. 1999, 287, 961–968. [Google Scholar] [CrossRef]
- Rhee, K.-H.; Scarborough, G.A.; Henderson, R. Domain movements of plasma membrane H(+)-ATPase: 3D structures of two states by electron cryo-microscopy. EMBO J. 2002, 21, 3582–3589. [Google Scholar] [CrossRef] [PubMed]
- Bowman, B.J.; Berenski, C.J.; Jung, C.Y. Size of the plasma membrane H+-ATPase from Neurospora crassa determined by radiation inactivation and comparison with the sarcoplasmic reticulum Ca2+-ATPase from skeletal muscle. J. Biol. Chem. 1985, 260, 8726–8730. [Google Scholar] [PubMed]
- Kanczewska, J.; Marco, S.; Vandermeeren, C.; Maudoux, O.; Rigaud, J.-L.; Boutry, M. Activation of the plant plasma membrane H+-ATPase by phosphorylation and binding of 14-3-3 proteins converts a dimer into a hexamer. Proc. Natl. Acad. Sci. USA 2005, 102, 11675–11680. [Google Scholar] [CrossRef] [PubMed]
- Ottmann, C.; Marco, S.; Jaspert, N.; Marcon, C.; Schauer, N.; Weyand, M.; Vandermeeren, C.; Duby, G.; Boutry, M.; Wittinghofer, A.; et al. Structure of a 14-3-3 coordinated hexamer of the plant plasma membrane H+-ATPase by combining X-ray crystallography and electron cryomicroscopy. Mol. Cell 2007, 25, 427–440. [Google Scholar] [CrossRef] [PubMed]
- Schuberth, C.; Wedlich-Söldner, R. Building a patchwork—The yeast plasma membrane as model to study lateral domain formation. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2015, 1853, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Spira, F.; Mueller, N.S.; Beck, G.; von Olshausen, P.; Beig, J.; Wedlich-Söldner, R. Patchwork organization of the yeast plasma membrane into numerous coexisting domains. Nat. Cell Biol. 2012, 14, 640–648. [Google Scholar] [CrossRef] [PubMed]
- Bagnat, M.; Chang, A.; Simons, K. Plasma membrane proton ATPase Pma1p requires raft association for surface delivery in yeast. Mol. Biol. Cell 2001, 12, 4129–4138. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Chang, A. Sphingoid base synthesis is required for oligomerization and cell surface stability of the yeast plasma membrane ATPase, Pma1. Proc. Natl. Acad. Sci. USA 2002, 99, 12853–12858. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.; Nakamoto, R.K.; Verjovski-Almeida, S.; Slayman, C.W. Structure and function of the yeast plasma-membrane H(+)-ATPase. Ann. N. Y. Acad. Sci. 1992, 671, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Hennessey, J.P.; Scarborough, G.A. Secondary structure of the Neurospora crassa plasma membrane H+-ATPase as estimated by circular dichroism. J. Biol. Chem. 1988, 263, 3123–3130. [Google Scholar] [PubMed]
- Bagnat, M.; Keranen, S.; Shevchenko, A.; Shevchenko, A.; Simons, K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA 2000, 97, 3254–3259. [Google Scholar] [CrossRef] [PubMed]
- Sampedro, J.G.; Ruiz-Granados, Y.G.; Nájera, H.; Téllez-Valencia, A.; Uribe, S. Fluorescence quenching by nucleotides of the plasma membrane H+-ATPase from Kluyveromyces lactis. Biochemistry 2007, 46, 5616–5622. [Google Scholar] [CrossRef] [PubMed]
- Sampedro, J.G.; Cortés, P.; Muñoz-Clares, R.A.; Fernández, A.; Uribe, S. Thermal inactivation of the plasma membrane H+-ATPase from Kluyveromyces lactis. Protection by trehalose. Biochim. Biophys. Acta (BBA) Protein Struct. Mol. Enzymol. 2001, 1544, 64–73. [Google Scholar] [CrossRef]
- Morsomme, P.; Chami, M.; Marco, S.; Nader, J.; Ketchum, K.A.; Goffeau, A.; Rigaud, J.-L. Characterization of a hyperthermophilic P-type ATPase from Methanococcus jannaschii expressed in yeast. J. Biol. Chem. 2002, 277, 29608–29616. [Google Scholar] [CrossRef] [PubMed]
- Gimpl, K.; Klement, J.; Keller, S. Characterising protein/detergent complexes by triple-detection size-exclusion chromatography. Biol. Proced. Online 2016, 18, 4. [Google Scholar] [CrossRef] [PubMed]
- Kunji, E.R.S.; Harding, M.; Butler, P.J.G.; Akamine, P. Determination of the molecular mass and dimensions of membrane proteins by size exclusion chromatography. Methods 2008, 46, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Schägger, H.; Cramer, W.A.; von Jagow, G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 1994, 217, 220–230. [Google Scholar] [CrossRef]
- Sampedro, J.G.; Guerra, G.; Pardo, J.P.; Uribe, S. Trehalose-mediated protection of the plasma membrane H+-ATPase from Kluyveromyces lactis during freeze-drying and rehydration. Cryobiology 1998, 37, 131–138. [Google Scholar] [CrossRef]
- Almeida, W.I.; Martins, O.B.; Carvalho-Alves, P.C. Self-association of isolated large cytoplasmic domain of plasma membrane H+-ATPase from Saccharomyces cerevisiae: Role of the phosphorylation domain in a general dimeric model for P-ATPases. Biochim. Biophys. Acta (BBA) Biomembr. 2006, 1758, 1768–1776. [Google Scholar] [CrossRef]
- Permyakov, S.; Suzina, N.; Valiakhmetov, A. Activation of H+-ATPase of the plasma membrane of Saccharomyces cerevisiae by glucose: The role of sphingolipid and lateral enzyme mobility. PLoS ONE 2012, 7, e30966. [Google Scholar] [CrossRef]
- Lee, M.C.S.; Hamamoto, S.; Schekman, R. Ceramide biosynthesis is required for the formation of the oligomeric H+-ATPase Pma1p in the yeast endoplasmic reticulum. J. Biol. Chem. 2002, 277, 22395–22401. [Google Scholar] [CrossRef]
- Chang, A.; Slayman, C.W. Maturation of the yeast plasma membrane [H+]ATPase involves phosphorylation during intracellular transport. J. Cell Biol. 1991, 115, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Gong, X.; Chang, A. An ER membrane protein, Sop4, facilitates ER export of the yeast plasma membrane [H+]ATPase, Pma1. Traffic 2002, 3, 730–739. [Google Scholar] [CrossRef] [PubMed]
- Pizzirusso, M.; Chang, A. Ubiquitin-mediated targeting of a mutant plasma membrane ATPase, Pma1-7, to the endosomal/vacuolar system in yeast. Mol. Biol. Cell 2004, 15, 2401–2409. [Google Scholar] [CrossRef] [PubMed]
- Cyrklaff, M.; Auer, M.; Kühlbrandt, W.; Scarborough, G.A. 2-D structure of the Neurospora crassa plasma membrane ATPase as determined by electron cryomicroscopy. EMBO J. 1995, 14, 1854–1857. [Google Scholar] [CrossRef]
- Liu, Y.; Sitaraman, S.; Chang, A. Multiple degradation pathways for misfolded mutants of the yeast plasma membrane ATPase, PMA1. J. Biol. Chem. 2006, 281, 31457–31466. [Google Scholar] [CrossRef] [PubMed]
- Eraso, P.; Portillo, F. Molecular mechanism of regulation of yeast plasma membrane H(+)-ATPase by glucose. Interaction between domains and identification of new regulatory sites. J. Biol. Chem. 1994, 269, 10393–10399. [Google Scholar]
- Gong, X.; Chang, A. A mutant plasma membrane ATPase, Pma1-10, is defective in stability at the yeast cell surface. Proc. Natl. Acad. Sci. USA 2001, 98, 9104–9109. [Google Scholar] [CrossRef]
- Duby, G.; Boutry, M. The plant plasma membrane proton pump ATPase: A highly regulated P-type ATPase with multiple physiological roles. Pflügers Arch. Eur. J. Physiol. 2009, 457, 645–655. [Google Scholar] [CrossRef]
- Piette, A.-S.; Derua, R.; Waelkens, E.; Boutry, M.; Duby, G. A phosphorylation in the C-terminal auto-inhibitory domain of the plant plasma membrane H+-ATPase activates the enzyme with no requirement for regulatory 14-3-3 proteins. J. Biol. Chem. 2011, 286, 18474–18482. [Google Scholar] [CrossRef]
- Callis, P.R. Binding phenomena and fluorescence quenching. II: Photophysics of aromatic residues and dependence of fluorescence spectra on protein conformation. J. Mol. Struct. 2014, 1077, 22–29. [Google Scholar] [CrossRef]
- Sampedro, J.G.; Nájera, H.; Uribe-Carvajal, S.; Ruiz-Granados, Y.G. Mapping the ATP binding site in the plasma membrane H+-ATPase from Kluyveromyces lactis. J. Fluoresc. 2014, 24, 1849–1859. [Google Scholar] [CrossRef] [PubMed]
- Callis, P.R.; Liu, T. Quantitative prediction of fluorescence quantum yields for tryptophan in proteins. J. Phys. Chem. B 2004, 108, 4248–4259. [Google Scholar] [CrossRef]
- Páez-Pérez, E.D.; De La Cruz-Torres, V.; Sampedro, J.G. Nucleotide binding in an engineered recombinant Ca2+-ATPase N-domain. Biochemistry 2016, 55, 6751–6765. [Google Scholar] [CrossRef]
- Petrushanko, I.Y.; Mitkevich, V.A.; Anashkina, A.A.; Klimanova, E.A.; Dergousova, E.A.; Lopina, O.D.; Makarov, A.A. Critical role of γ-phosphate in structural transition of Na,K-ATPase upon ATP binding. Sci. Rep. 2015, 4, 5165. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, J.T.; Kanashova, T.; Dittmar, G.; Palmgren, M. Isolation of native plasma membrane H+-ATPase (Pma1p) in both the active and basal activation states. FEBS Open Bio 2018, 8, 774–783. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar]
- Sampedro, J.G.; Muñoz-Clares, R.A.; Uribe, S. Trehalose-mediated inhibition of the plasma membrane H+-ATPase from Kluyveromyces lactis: Dependence on viscosity and temperature. J. Bacteriol. 2002, 184, 4384–4391. [Google Scholar] [CrossRef]
- Lakowicz, J.R. (Ed.) Principles of Fluorescence Spectroscopy; Springer US: Boston, MA, USA, 2006; ISBN 978-0-387-31278-1. [Google Scholar]
- Neet, K.E. Cooperativity in enzyme function: Equilibrium and kinetic aspects. Methods Enzymol. 1980, 64, 139–192. [Google Scholar]
- Luo, W.; Chang, A. An endosome-to-plasma membrane pathway involved in trafficking of a mutant plasma membrane ATPase in yeast. Mol. Biol. Cell 2000, 11, 579–592. [Google Scholar] [CrossRef]
- Patton, J.L.; Lester, R.L. Phosphatidylinositol phosphate, phosphatidylinositol bisphosphate, and the phosphoinositol sphingolipids are found in the plasma membrane and stimulate the plasma membrane H+-ATPase of Saccharomyces cerevisiae. Arch. Biochem. Biophys. 1992, 292, 70–76. [Google Scholar] [CrossRef]
Sample Availability: Samples of the chemical compounds, antibody, and the K. lactis yeast strain used are available from the authors. |



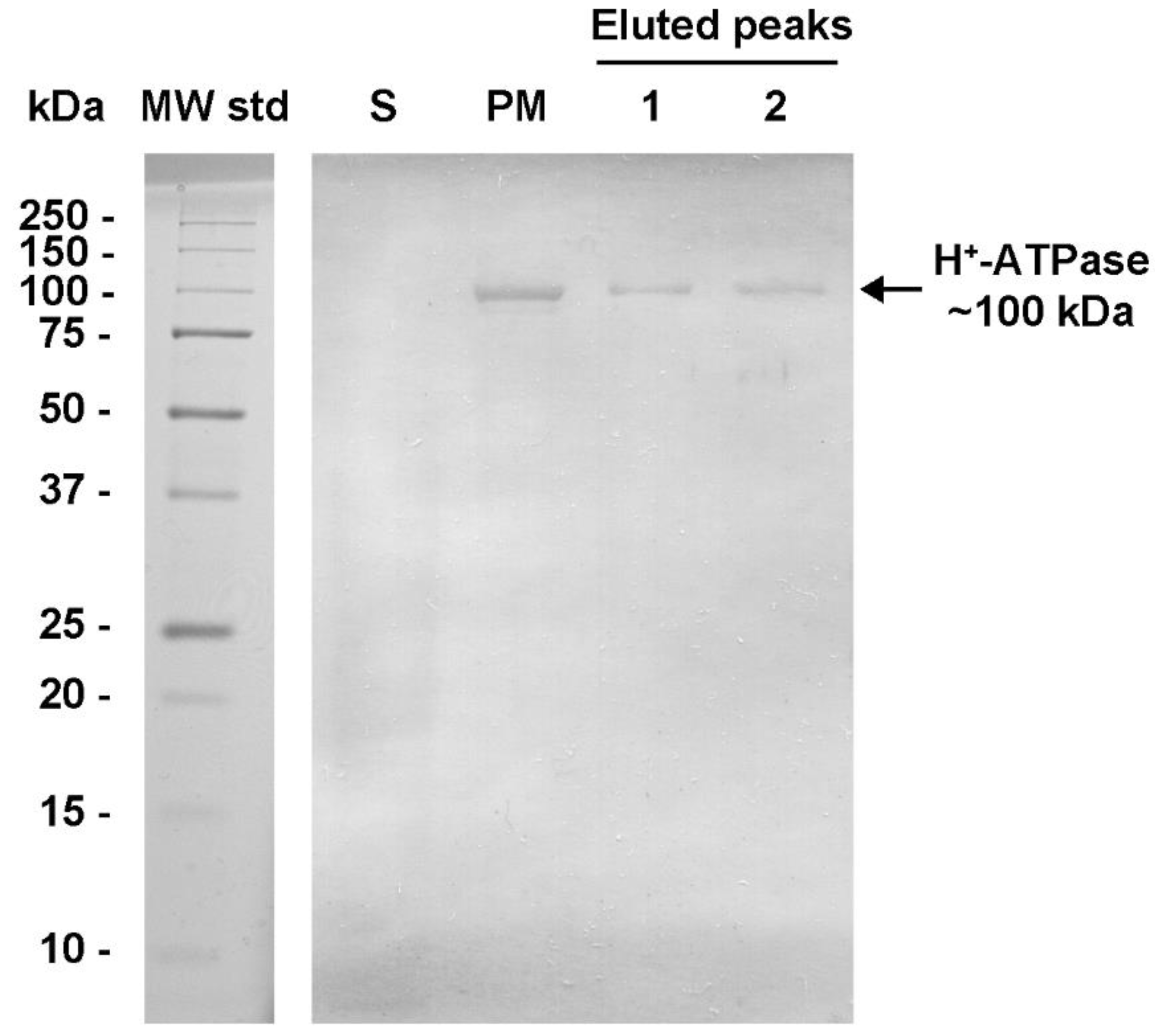




| H+-ATPase Hexamer | Vmax (μmols ATP mg prot.−1 min−1) | S0.5 (μM ATP) | Hill Number (n) |
|---|---|---|---|
| Elution Peak 1 | 17.68 ± 0.67 | 329 ± 19 | 1.98 ± 0.21 |
| Elution Peak 2 | 5.19 ± 0.26 | 364 ± 28 | 1.82 ± 0.22 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz-Granados, Y.G.; De La Cruz-Torres, V.; Sampedro, J.G. The Oligomeric State of the Plasma Membrane H+-ATPase from Kluyveromyces lactis. Molecules 2019, 24, 958. https://doi.org/10.3390/molecules24050958
Ruiz-Granados YG, De La Cruz-Torres V, Sampedro JG. The Oligomeric State of the Plasma Membrane H+-ATPase from Kluyveromyces lactis. Molecules. 2019; 24(5):958. https://doi.org/10.3390/molecules24050958
Chicago/Turabian StyleRuiz-Granados, Yadira G., Valentín De La Cruz-Torres, and José G. Sampedro. 2019. "The Oligomeric State of the Plasma Membrane H+-ATPase from Kluyveromyces lactis" Molecules 24, no. 5: 958. https://doi.org/10.3390/molecules24050958
APA StyleRuiz-Granados, Y. G., De La Cruz-Torres, V., & Sampedro, J. G. (2019). The Oligomeric State of the Plasma Membrane H+-ATPase from Kluyveromyces lactis. Molecules, 24(5), 958. https://doi.org/10.3390/molecules24050958







