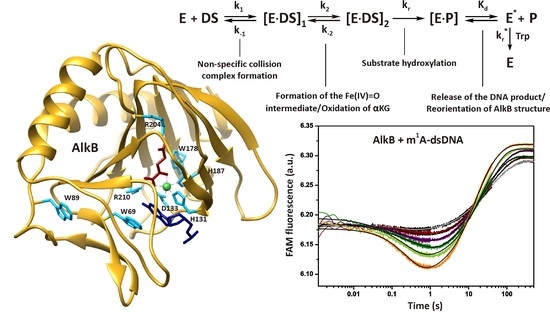
A Single-Turnover Kinetic Study of DNA Demethylation Catalyzed by Fe(II)/α-Ketoglutarate-Dependent Dioxygenase AlkB
Abstract
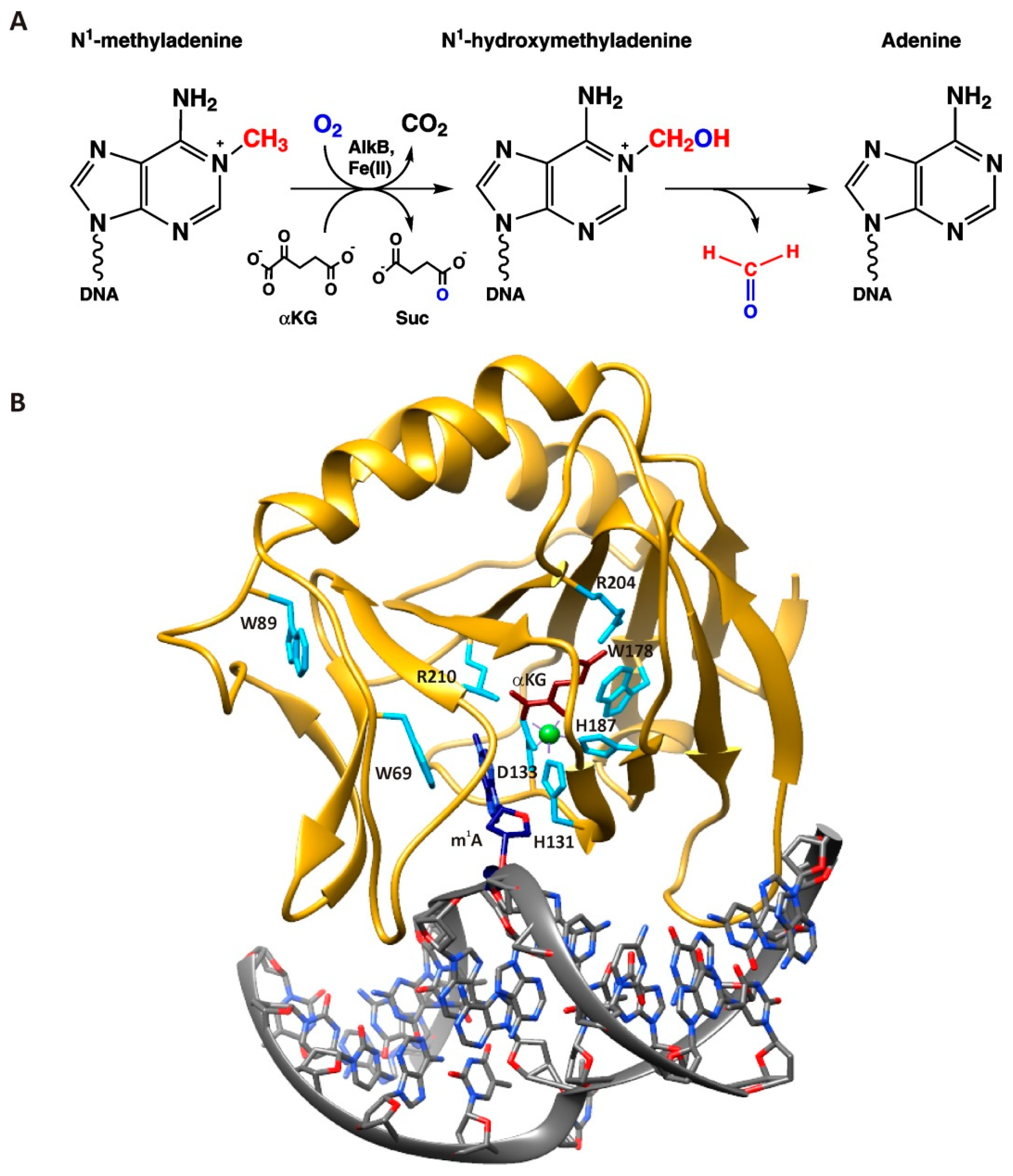
:1. Introduction
2. Results
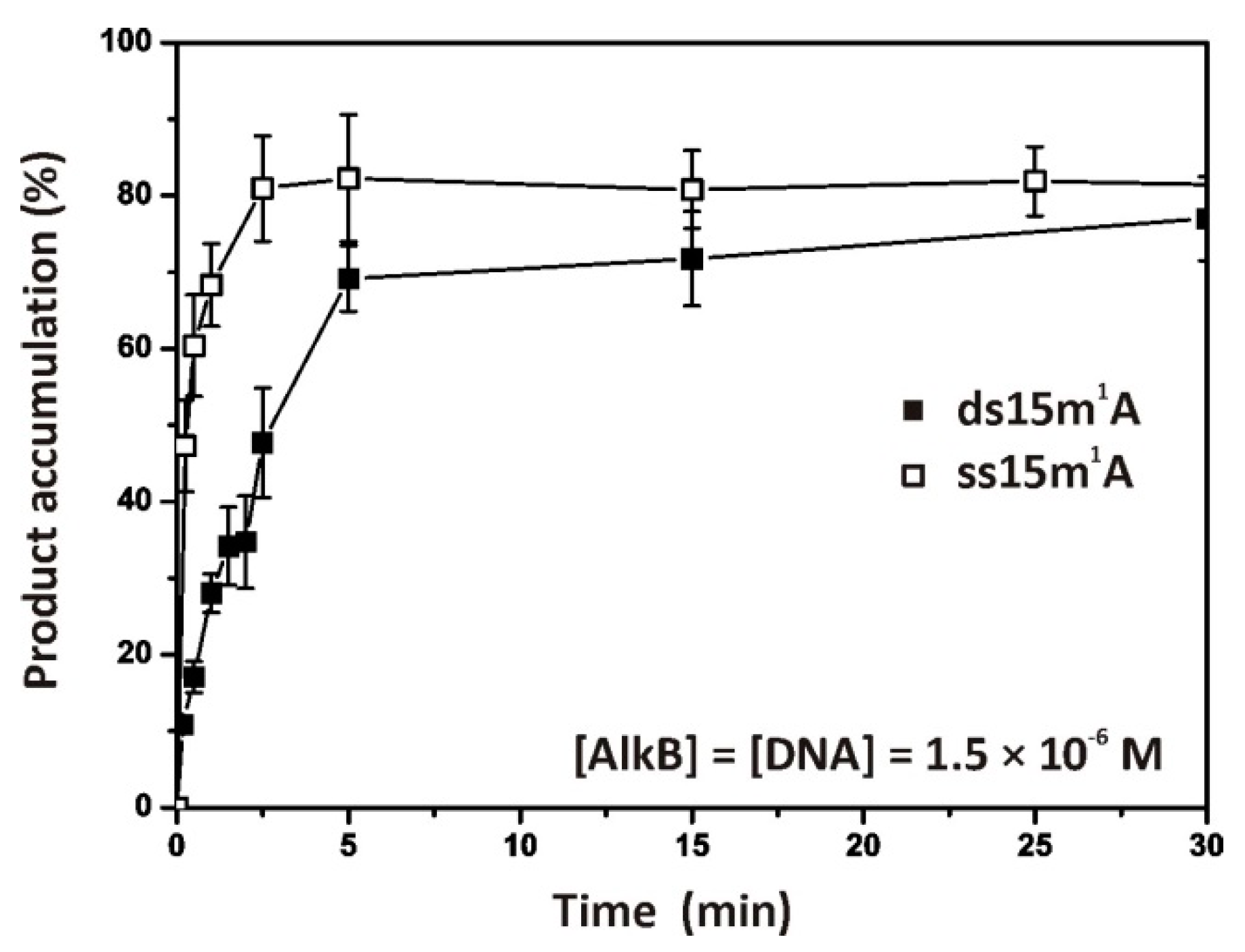
2.1. Repair of ssDNA and dsDNA Substrates Containing an m1A Lesion
2.2. Single-Turnover Kinetics of ssDNA Demethylation by AlkB
2.3. Single-Turnover Kinetics of dsDNA Demethylation by AlkB
2.4. Interaction of AlkB with Undamaged DNA Representing the Reaction Product
3. Discussion
4. Materials and Methods
4.1. ODNs
4.2. Protein Expression and Purification
4.3. A DNA Repair Assay
4.4. SF Fluorescence Measurements
4.5. Global Fitting of SF Data
4.6. Fluorescence Equilibrium Titration of AlkB
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Soll, J.M.; Sobol, R.W.; Mosammaparast, N. Regulation of DNA Alkylation Damage Repair: Lessons and Therapeutic Opportunities. Trends Biochem. Sci. 2017, 42, 206–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aravind, L.; Koonin, E.V. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001, 2, RESEARCH0007. [Google Scholar] [CrossRef] [PubMed]
- Falnes, P.O.; Johansen, R.F.; Seeberg, E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature 2002, 419, 178–182. [Google Scholar] [CrossRef] [PubMed]
- Trewick, S.C.; Henshaw, T.F.; Hausinger, R.P.; Lindahl, T.; Sedgwick, B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 2002, 419, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Falnes, P.O. Repair of 3-methylthymine and 1-methylguanine lesions by bacterial and human AlkB proteins. Nucleic Acids Res. 2004, 32, 6260–6267. [Google Scholar] [CrossRef] [Green Version]
- Delaney, J.C.; Smeester, L.; Wong, C.; Frick, L.E.; Taghizadeh, K.; Wishnok, J.S.; Drennan, C.L.; Samson, L.D.; Essigmann, J.M. AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat. Struct. Mol. Biol. 2005, 12, 855–860. [Google Scholar] [CrossRef]
- Aas, P.A.; Otterlei, M.; Falnes, P.O.; Vagbo, C.B.; Skorpen, F.; Akbari, M.; Sundheim, O.; Bjoras, M.; Slupphaug, G.; Seeberg, E.; et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 2003, 421, 859–863. [Google Scholar] [CrossRef]
- Falnes, P.O.; Bjoras, M.; Aas, P.A.; Sundheim, O.; Seeberg, E. Substrate specificities of bacterial and human AlkB proteins. Nucleic Acids Res. 2004, 32, 3456–3461. [Google Scholar] [CrossRef]
- Zhang, Z.; Ren, J.; Harlos, K.; McKinnon, C.H.; Clifton, I.J.; Schofield, C.J. Crystal structure of a clavaminate synthase-Fe(II)-2-oxoglutarate-substrate-NO complex: Evidence for metal centered rearrangements. Febs. Lett. 2002, 517, 7–12. [Google Scholar] [CrossRef]
- Elkins, J.M.; Ryle, M.J.; Clifton, I.J.; Dunning Hotopp, J.C.; Lloyd, J.S.; Burzlaff, N.I.; Baldwin, J.E.; Hausinger, R.P.; Roach, P.L. X-ray crystal structure of Escherichia coli taurine/alpha-ketoglutarate dioxygenase complexed to ferrous iron and substrates. Biochemistry 2002, 41, 5185–5192. [Google Scholar] [CrossRef]
- Yu, B.; Edstrom, W.C.; Benach, J.; Hamuro, Y.; Weber, P.C.; Gibney, B.R.; Hunt, J.F. Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB. Nature 2006, 439, 879–884. [Google Scholar] [CrossRef]
- Yu, B.; Hunt, J.F. Enzymological and structural studies of the mechanism of promiscuous substrate recognition by the oxidative DNA repair enzyme AlkB. Proc. Natl. Acad. Sci. USA 2009, 106, 14315–14320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.G.; Yi, C.; Duguid, E.M.; Sullivan, C.T.; Jian, X.; Rice, P.A.; He, C. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature 2008, 452, 961–965. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Ergel, B.; Gill, M.L.; Brown, L.; Yu, B.; Palmer, A.G., 3rd; Hunt, J.F. Protein dynamics control the progression and efficiency of the catalytic reaction cycle of the Escherichia coli DNA-repair enzyme AlkB. J. Biol. Chem. 2014, 289, 29584–29601. [Google Scholar] [CrossRef] [Green Version]
- Bleijlevens, B.; Shivarattan, T.; Flashman, E.; Yang, Y.; Simpson, P.J.; Koivisto, P.; Sedgwick, B.; Schofield, C.J.; Matthews, S.J. Dynamic states of the DNA repair enzyme AlkB regulate product release. Embo. Rep. 2008, 9, 872–877. [Google Scholar] [CrossRef] [Green Version]
- Bleijlevens, B.; Shivarattan, T.; van den Boom, K.S.; de Haan, A.; van der Zwan, G.; Simpson, P.J.; Matthews, S.J. Changes in protein dynamics of the DNA repair dioxygenase AlkB upon binding of Fe(2+) and 2-oxoglutarate. Biochemistry 2012, 51, 3334–3341. [Google Scholar] [CrossRef]
- Waheed, S.O.; Ramanan, R.; Chaturvedi, S.S.; Ainsley, J.; Evison, M.; Ames, J.M.; Schofield, C.J.; Christov, C.Z.; Karabencheva-Christova, T.G. Conformational flexibility influences structure-function relationships in nucleic acid N-methyl demethylases. Org. Biomol. Chem. 2019, 17, 2223–2231. [Google Scholar] [CrossRef]
- Kuznetsov, N.A.; Koval, V.V.; Nevinsky, G.A.; Douglas, K.T.; Zharkov, D.O.; Fedorova, O.S. Kinetic conformational analysis of human 8-oxoguanine-DNA glycosylase. J. Biol. Chem. 2007, 282, 1029–1038. [Google Scholar] [CrossRef] [Green Version]
- Kuznetsova, A.A.; Iakovlev, D.A.; Misovets, I.V.; Ishchenko, A.A.; Saparbaev, M.K.; Kuznetsov, N.A.; Fedorova, O.S. Pre-steady-state kinetic analysis of damage recognition by human single-strand selective monofunctional uracil-DNA glycosylase SMUG1. Mol. Biosyst. 2017, 13, 2638–2649. [Google Scholar] [CrossRef] [Green Version]
- Kanazhevskaya, L.Y.; Koval, V.V.; Zharkov, D.O.; Strauss, P.R.; Fedorova, O.S. Conformational transitions in human AP endonuclease 1 and its active site mutant during abasic site repair. Biochemistry 2010, 49, 6451–6461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanazhevskaya, L.Y.; Koval, V.V.; Vorobjev, Y.N.; Fedorova, O.S. Conformational dynamics of abasic DNA upon interactions with AP endonuclease 1 revealed by stopped-flow fluorescence analysis. Biochemistry 2012, 51, 1306–1321. [Google Scholar] [CrossRef] [PubMed]
- Henshaw, T.F.; Feig, M.; Hausinger, R.P. Aberrant activity of the DNA repair enzyme AlkB. J. Inorg. Biochem. 2004, 98, 856–861. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, N.A.; Vorobjev, Y.N.; Krasnoperov, L.N.; Fedorova, O.S. Thermodynamics of the multi-stage DNA lesion recognition and repair by formamidopyrimidine-DNA glycosylase using pyrrolocytosine fluorescence-stopped-flow pre-steady-state kinetics. Nucleic Acids Res. 2012, 40, 7384–7392. [Google Scholar] [CrossRef] [Green Version]
- Halford, S.E. An end to 40 years of mistakes in DNA-protein association kinetics? Biochem. Soc. Trans. 2009, 37, 343–348. [Google Scholar] [CrossRef] [Green Version]
- Jean, J.M.; Hall, K.B. 2-Aminopurine electronic structure and fluorescence properties in DNA. Biochemistry 2002, 41, 13152–13161. [Google Scholar] [CrossRef]
- Samson, L.; Cairns, J. A new pathway for DNA repair in Escherichia coli. Nature 1977, 267, 281–283. [Google Scholar] [CrossRef]
- Martinez, S.; Hausinger, R.P. Catalytic Mechanisms of Fe(II)- and 2-Oxoglutarate-dependent Oxygenases. J. Biol. Chem. 2015, 290, 20702–20711. [Google Scholar] [CrossRef] [Green Version]
- Price, J.C.; Barr, E.W.; Tirupati, B.; Bollinger, J.M., Jr.; Krebs, C. The first direct characterization of a high-valent iron intermediate in the reaction of an alpha-ketoglutarate-dependent dioxygenase: a high-spin FeIV complex in taurine/alpha-ketoglutarate dioxygenase (TauD) from Escherichia coli. Biochemistry 2003, 42, 7497–7508. [Google Scholar] [CrossRef]
- Huang, J.L.; Tang, Y.; Yu, C.P.; Sanyal, D.; Jia, X.; Liu, X.; Guo, Y.; Chang, W.C. Mechanistic Investigation of Oxidative Decarboxylation Catalyzed by Two Iron(II)- and 2-Oxoglutarate-Dependent Enzymes. Biochemistry 2018, 57, 1838–1841. [Google Scholar] [CrossRef]
- Tarhonskaya, H.; Szollossi, A.; Leung, I.K.; Bush, J.T.; Henry, L.; Chowdhury, R.; Iqbal, A.; Claridge, T.D.; Schofield, C.J.; Flashman, E. Studies on deacetoxycephalosporin C synthase support a consensus mechanism for 2-oxoglutarate dependent oxygenases. Biochemistry 2014, 53, 2483–2493. [Google Scholar] [CrossRef] [PubMed]
- Tarhonskaya, H.; Hardy, A.P.; Howe, E.A.; Loik, N.D.; Kramer, H.B.; McCullagh, J.S.; Schofield, C.J.; Flashman, E. Kinetic Investigations of the Role of Factor Inhibiting Hypoxia-inducible Factor (FIH) as an Oxygen Sensor. J. Biol. Chem. 2015, 290, 19726–19742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Giri, N.C.; Zhang, R.; Yamane, K.; Zhang, Y.; Maroney, M.; Costa, M. Nickel ions inhibit histone demethylase JMJD1A and DNA repair enzyme ABH2 by replacing the ferrous iron in the catalytic centers. J. Biol. Chem. 2010, 285, 7374–7383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishina, Y.; He, C. Probing the structure and function of the Escherichia coli DNA alkylation repair AlkB protein through chemical cross-linking. J. Am. Chem. Soc. 2003, 125, 8730–8731. [Google Scholar] [CrossRef]
- Flashman, E.; Hoffart, L.M.; Hamed, R.B.; Bollinger, J.M., Jr.; Krebs, C.; Schofield, C.J. Evidence for the slow reaction of hypoxia-inducible factor prolyl hydroxylase 2 with oxygen. Febs. J. 2010, 277, 4089–4099. [Google Scholar] [CrossRef] [Green Version]
- Grzyska, P.K.; Ryle, M.J.; Monterosso, G.R.; Liu, J.; Ballou, D.P.; Hausinger, R.P. Steady-state and transient kinetic analyses of taurine/alpha-ketoglutarate dioxygenase: effects of oxygen concentration, alternative sulfonates, and active-site variants on the FeIV-oxo intermediate. Biochemistry 2005, 44, 3845–3855. [Google Scholar] [CrossRef]
- Lee, D.H.; Jin, S.G.; Cai, S.; Chen, Y.; Pfeifer, G.P.; O’Connor, T.R. Repair of methylation damage in DNA and RNA by mammalian AlkB homologues. J. Biol. Chem. 2005, 280, 39448–39459. [Google Scholar] [CrossRef] [Green Version]
- Kuzmic, P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal. Biochem. 1996, 237, 260–273. [Google Scholar] [CrossRef]
- Kuznetsov, N.A.; Koval, V.V.; Zharkov, D.O.; Vorobjev, Y.N.; Nevinsky, G.A.; Douglas, K.T.; Fedorova, O.S. Pre-steady-state kinetic study of substrate specificity of Escherichia coli formamidopyrimidine-DNA glycosylase. Biochemistry 2007, 46, 424–435. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |





| Name | Sequence 1 |
|---|---|
| 15A | 5′-ACAGGATCCGGCATA-3′ 2 |
| 15m1A | 5′-ACAGGm1ATCCGGCATA-3′ |
| 15m1A_2aPu | 5′-ACAGGm1A(2aPu)CCGGCATA-3′ |
| 15m1A_FRET | 5′-FAM-ACAGGm1ATCCGGCATA-BHQ1-3′ |
| 15T | 5′-TATGCCGGATCCTGT-3′ |
| 15TT | 5′-TATGCCGGTTCCTGT-3′ |
| Substrate | ss15m1A | ss15m1A_2aPu | ss15m1A_FRET | ds15m1A | ds15m1A_2aPu | ds15m1A_FRET |
|---|---|---|---|---|---|---|
| k1 × 10-6, M−1 s−1 | 455 ± 33 | 13 ± 1 | 19 ± 4 | 180 ± 80 | 6.6 ± 0.9 | 0.28 ± 0.03 |
| k−1, s−1 | 9.2 ± 5 | 8 ± 0.5 | 51 ± 8 | 34 ± 13 | 9.2 ± 1.5 | 3.8 ± 0.2 |
| k2, s−1 | 49 ± 4 | |||||
| k-2, s−1 | 35 ± 3 | |||||
| k3, s−1 | 3.2 ± 0.3 | 2.5 ± 0.4 | 4.3 ± 1.5 | 1.1 ± 0.2 | 2.5 ± 0.8 | 1.1 ± 0.2 |
| k-3, s−1 | 1.9 ± 0.2 | 1.5 ± 0.1 | 3.8 ± 0.6 | 1.1 ± 0.1 | 2.9 ± 0.3 | 0.78 ± 0.07 |
| k4, s−1 | 0.43 ± 0.12 | |||||
| k-4, s−1 | 0.063 ± 0.015 | |||||
| kr, s−1 | 0.47 ± 0.04 | 0.19 ± 0.03 | 0.45 ± 0.11 | 0.21 ± 0.04 | ||
| Kd, M | (2.2 ± 0.3) × 10−6 | (3.1 ± 0.8) × 10−7 | (1.8 ± 0.2) × 10−6 | (4.4 ± 0.2) × 10−6 | ||
| kr*, s−1 | 0.024 ± 0.004 | 0.014 ± 0.001 | ||||
| Fluorescent probe | Trp | 2aPu | FRET | Trp | 2aPu | FRET |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanazhevskaya, L.Y.; Alekseeva, I.V.; Fedorova, O.S. A Single-Turnover Kinetic Study of DNA Demethylation Catalyzed by Fe(II)/α-Ketoglutarate-Dependent Dioxygenase AlkB. Molecules 2019, 24, 4576. https://doi.org/10.3390/molecules24244576
Kanazhevskaya LY, Alekseeva IV, Fedorova OS. A Single-Turnover Kinetic Study of DNA Demethylation Catalyzed by Fe(II)/α-Ketoglutarate-Dependent Dioxygenase AlkB. Molecules. 2019; 24(24):4576. https://doi.org/10.3390/molecules24244576
Chicago/Turabian StyleKanazhevskaya, Lyubov Yu., Irina V. Alekseeva, and Olga S. Fedorova. 2019. "A Single-Turnover Kinetic Study of DNA Demethylation Catalyzed by Fe(II)/α-Ketoglutarate-Dependent Dioxygenase AlkB" Molecules 24, no. 24: 4576. https://doi.org/10.3390/molecules24244576
APA StyleKanazhevskaya, L. Y., Alekseeva, I. V., & Fedorova, O. S. (2019). A Single-Turnover Kinetic Study of DNA Demethylation Catalyzed by Fe(II)/α-Ketoglutarate-Dependent Dioxygenase AlkB. Molecules, 24(24), 4576. https://doi.org/10.3390/molecules24244576