Abstract
The oxidative decarboxylation of the iron(II) α-hydroxy acid (mandelic acid) complex model, biomimetic of Rieske dioxygenase, has been investigated at the density functional level. The explored mechanism sheds light on the role of the α-hydroxyl group on the dioxygen activation. The potential energy surfaces have been explored in different electronic spin states. The rate-determining step of the process is the proton transfer. The oxidative decarboxylation preferentially takes place on the quintet state.
1. Introduction
In recent years, the employment of biomimetic catalysts considerably increased not only for their related high selectivity, but because they are allowed to operate in very mild conditions and they adapt to different types of substrates [1,2].
Encouraged by the new reactions catalyzed by Rieske dioxygenases [1,3,4], sustainable mononuclear nonheme iron complexes have been proposed for bioinspired oxidation catalysis. Dioxygenases are involved in the degradation of a wide range of natural and synthetic compounds by incorporating both atoms of O2 into substrates.
Bioinspired catalysis using peroxides or peracids for the oxidation of hydrocarbon substrates is more common and better characterized, than that based on dioxygen in catalytic oxidations, which has not been fully explored yet and the relative reaction mechanism is often unknown. This last reaction path requires both electron and proton sources [5,6,7,8,9,10,11,12,13] and cosubstrate-assisted O2 molecular activation by transition metal containing complexes can be an alternative pathway [14,15,16,17,18,19] resorting to the sacrificial reductants represented a good solution [12,20]. Paine’s and other groups have investigated a family of iron complexes supported by the monoanionic facial trinitrogen donor ligand, hydrotris(3,5-diphenylpyrazole-1-yl)borate (TpPh2 ligand) that activates O2 by 2 electrons reduction in the oxidative decarboxylation of the α-hydroxy acids as sacrificial reductants [3,21,22,23]. These studies provide evidence that the the hydroxyl group in the dioxygen activation is important. The iron complexes mentioned above reacting with O2 are able to carry out the stoichiometric cis-dihydroxylation of olefins producing the diol product [3], as in that with H2O2 [4,24,25]. The mononuclear non-heme iron(II) complex shown in Scheme 1, the object of the present investigation, represents the first biomimetic complex reproducing the dioxygenase activity of α-ketoglutarate-dependent enzymes.

Scheme 1.
Structure of hydrotris(3,5-diphenylpyrazole-1-yl)borate FeII (mandelate).
Since, as occurred in similar systems [26] no intermediate was detected in the course of the reaction, the steps up to the oxidative decarboxylation of the keto acid play a key role in the control of the rate of the reaction.
To provide deeper insights on the reaction catalyzed by the biomimetic complex of Scheme 1, we have undertaken a theoretical investigation taking into account the spin state of the analyzed species and building up the potential energy surfaces for the reaction path leading to the formation of an iron−oxygen oxidant upon oxidative decarboxylation of α-hydroxy acid.
2. Results and Discussion
As a first step of work, we have optimized the naked biomimetic iron(II) α‑hydroxy acid complex (represented by model a in Figure S1 of the Supplementary Information) in septet, quintet, triplet and singlet spin states.
The coordination bond distances, which we have obtained together with X-ray counterpart [3] and relative electronic energies are collected in Table 1. Results show that the complex assumes the most stable configuration in the high-spin quintet state in well agreement with the available data for similar systems [3,27,28]. The singlet, triplet and septet spin states lie at 17.2, 13.1, and 65.1 kcal/mol above ground one, respectively.

Table 1.
Main geometrical parameters for naked complex a at all the spin states. Relative energies provided at M06L level of theory are kcal/mol and compared with the experimental X-ray counterpart [3]. Distances are in Å. See Scheme 1 for labels.
Comparison with the crystallographic structure (Table 1) shows that the geometrical parameters in the quintet state are reproduced fairly faithfully. Furthermore, the calculated Fe-N bond distances well match with other high-spin (TpPh2) FeII complexes [23,27].
The maximum deviation results to be less than 0.06 Å. From the superposition of the optimized structures at the considered spin states, it emerges that the mandelate ring shows major deviations whereas the coordination around the iron does not suffer drastic changes (Figure 1). This suggests that in the quintet, the carboxylic group of the mandelic acid acts as monodentate ligand.

Figure 1.
Superposition of the M06L optimized structures of model a at the different spin states considered.
For the O2 addition (a-O2 complex), two different binding modes, namely end-on (eo) and side-on (so), have been taken into account. From computations, the eo binding mode appears to be the preferred one in all the considered spin states. Looking at the Figure 2 collecting the optimized structures of the eo obtained adducts, the triplet a-O2 complex represents the lowest energy configuration followed by the septet, quintet and singlet. A support to this finding arises from the analysis of the triplet spin state HOMO frontier orbital (see Figure 3) that mainly involves the metal center and its surrounding with the mandelate ligand less interested in the occurring electron redistribution.
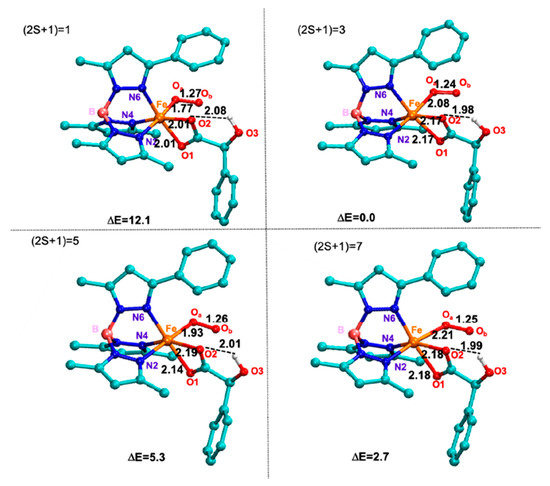
Figure 2.
M06L optimized structures and main geometrical parameters of a-O2 adducts. Selected bonds are in Å. Relative electronic energies are in kcal/mol.
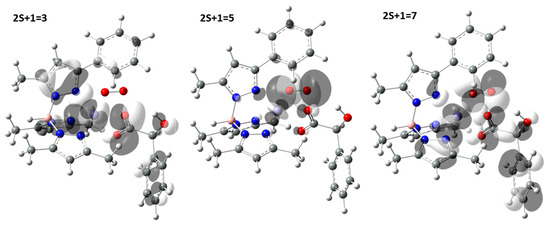
Figure 3.
Highest occupied molecular orbital (HOMO) composition of a-O2 adduct, in the triplet, quintet and septet states.
As concerns the geometrical structure, the entry of dioxygen induces a reorganization around the metal center that now appears to be hexacoordinated (octahedral geometry). The O2 occupies the apical position, while the carboxylate moiety (lying on the equatorial plane) acts as bidentate ligand with both the Fe-O bond lengths of 2.17 Å. These values well compare with those present in dioxygenase enzymes [20,26].
The Fe-Oa bond assumes the value of 2.08, 1.93, and 2.21 Å in the triplet, quintet and septet electronic states, respectively. The O-O bond distance in the ground state (1.24 Å) reveals a significant deviation from the corresponding value in the enzymatic environment (1.31 Å). This behavior can be ascribable to the weaker electro-donating character of TpPh2 ligand towards the iron(III), present in the a-O2 complex, relative to the usual biological ligands in the enzymes [26].
The considered reaction mechanism (Scheme 2) arises from experimental indications. In fact, biomimetic and enzymatic studies suggested that the iron(III) superoxide species, obtained upon addition of O2 at the iron(II) center, has the ability to abstract hydrogen atom from the O-H group of α-hydroxy acid.
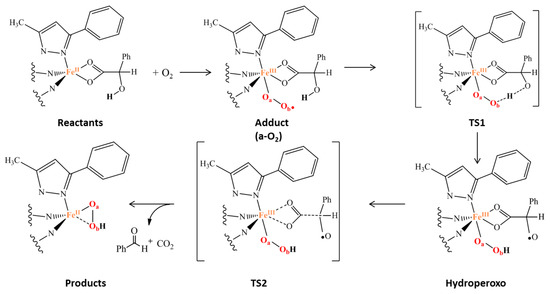
Scheme 2.
Proposed mechanism for the oxidative decarboxylation promoted by the non-heme biomimetic complex model (a).
Since the singlet electronic spin state of iron-dioxygen complexes lies at very high energy, only the septet, quintet and triplet potential energy surfaces (PESs) have been considered (see Figure 4).
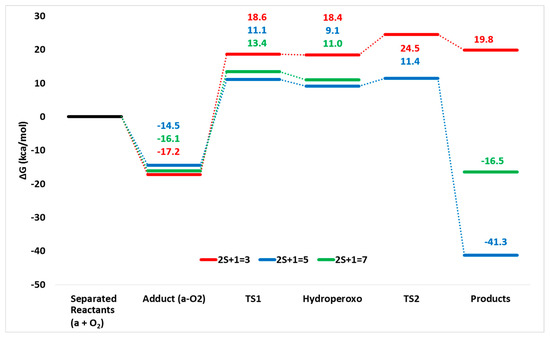
Figure 4.
M06L potential energy surfaces (PESs) for oxidative decarboxylation operated by non-heme biomimetic model (a) at different spin states (triplet, quintet and septet).
As far as the PES for the septet is concerned, no transition state connecting the hydroperoxide with the final oxidant agent (TS2 of Scheme 2) has been intercepted along the surface. A similar finding has been previously obtained for two enzymatic systems [26,29]. Instead, on the quintet and triplet PESs, both TS1 and TS2 have been characterized confirming that the oxidative decarboxylation is a multistep process.
The addition of O2 molecule starting to the complex (a) leads to the formation of adducts that in all the three considered spin states lie below the reactants’ asymptote suggesting as their formation is an exergonic process (see Figure 4).
On the three computed potential energy surfaces, the first transition state (TS1) is located at 18.6 (triplet), 11.1 (quintet) and 13.4 (septet) kcal/mol and their corresponding optimized structures are shown in Figure 5.
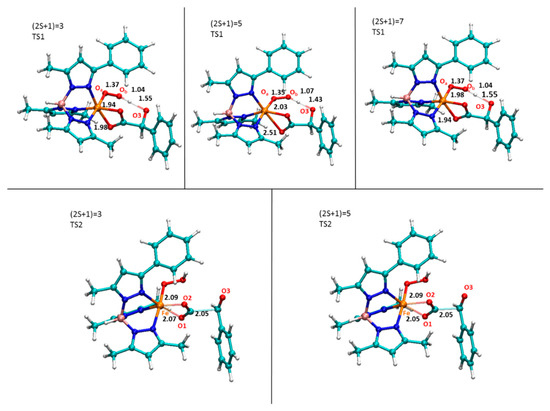
Figure 5.
M06L optimized structures of TS1 and TS2 intercepted along the three explored PESs. Selected bonds are in Å.
For the TS1 in the triplet state, the barrier is 7.5 kcal/mol higher than that of quintet and a spin crossing from these two PESs occurs. This is due to the fact that the resulting iron(III) hydroperoxo species prefers to be a quintet (9.1 kcal/mol), while the corresponding in the triplet state is 9.3 higher in energy. The TS1 describes the proton transfer from the α-OH group of the mandelate to the Ob atom of the molecular oxygen, as confirmed by the imaginary frequency (638.1i cm−1). This result enhances the role played by the α-hydroxy acid ligand in this process as proposed by Paine et al. [3].
The next formed species, containing the Fe(III)-OOH moiety, is found at 9.1 kcal/mol and 11.0 kcal/mol in the quintet and septet state, respectively. In contrast, the iron(III) hydroperoxo in the triplet state lies at higher energy. This finding reflects the relative geometrical differences (see Figure 6a) In fact, in the quintet state, the carboxylate moiety is monocoordinated to iron (O1-Fe: 1.96 Å, O2-Fe: 3.18 Å) and the C-C bond results to be more elongated (1.71 Å) with respect to the previous steps. The benzaldehyde is prone to be released but it still retained by an H bond between its carbonyl (2.07 Å) and the OOH moiety. From the iron(III) hydroperoxo, the reaction proceeds with the definitive C-C bond breaking (2.05 Å) via TS2 that is characterized by an imaginary frequency related to the C-C stretching mode (211.8i cm−1) as shown in Figure 4. In addition, this bond’s elongation generates the loss of the H bond since the OH-O=C is now 3.35 Å.
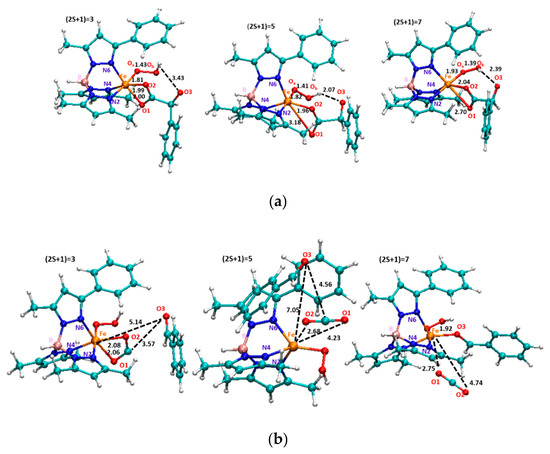
Figure 6.
M06L optimized structures and main geometrical parameters of hyproperoxo (a) and prod species (b) in all the three electronic states considered. Selected bonds are in Å.
The product in its quintet spin state, at 41.3 kcal/mol below the separated reactants, evidences as the decarboxylation is a highly exergonic process. In the product, the CO2 is already formed (O-C-O angle is 180°) and it lies at 2.67 Å from the metallic center engaged in a sandwich-like topology between two phenyl rings (ligand and benzaldehyde) (see Figure 6b). Some changes occur also in the metallic core due to the elongation of Oa-Ob bond (1.48 Å) and to the Fe-Oa distance of 1.91 Å. In this way, the iron(II) hydroperoxo oxidant agent is formed and ready to act as nucleophilic agent in the next oxidation reactions that are not object of the present study.
Paine et al. underline as “this iron(II) hydroperoxo species may also undergo O-O bond heterolysis to form an iron(IV)-oxo-hydroxo intermediate and both oxidant species can carry out the oxidation on different kind of substrates” [3]. To give a contribution to this aspect, geometry optimizations of the two forms have been performed and the resulting optimized geometries (shown in Figure S2) indicate that the quintet spin state proves to be the most stable one.
3. Conclusions
On the basis of our density functional theory investigation on the oxidative decarboxylation promoted by a biomimetic nonheme complex, the following conclusions can be drawn:
- The favored reaction path occurs in the quintet spin state as in the corresponding Rieske oxygenase enzymes;
- A spin cross takes place between the triplet and quintet electronic spin states after the adduct formation;
- The α-hydroxy group plays an important role in the oxidative decarboxylation;
- The rate determining step is the proton transfer;
- The overall reaction results to be more exergonic in the quintet PES.
4. Computational Details
All density functional theory (DFT) based calculations have been performed by using Gaussian09 code [30]. Following previous suggestions available in literature about the reliable results of the M06L [31,32] exchange-correlation functional in describing electronic structure and properties of metal containing systems also in various spin states [33,34,35,36], this functional has been adopted for the geometry optimizations.
The crystal structure (X ray) of the biomimetic catalyst [3] has been adopted as starting structure for our computations. In order to reduce the computational time, five phenyl rings of the TpPh2 ligand have been replaced by methyl groups (see a of Figure S1). The phenyl group of the TpPh2 ligand closely oriented to the iron center has been retained to better evaluate its role on the activated dioxygen species during the catalytic process.
Geometry optimizations and vibrational frequencies have been performed employing differentiated basis set. In particular, for O, N and H atoms in the active center the 6-31+G(d,p) set has been used while for the remaining ones the 6-31G(d,p) one has been chosen. The LANL2DZ effective core potential and the related basis set has been employed for iron.
In order to improve the energetic values, single point calculations with the larger basis set 6-311+G(d,p) for all atoms except for metal ion, has been performed on all the previously optimized structures. The solvent corrections were evaluated using the polarizable continuum model [37,38] as implemented in Gaussian09 taking into account the dielectric constant (ε = 24.93) that better reproduces the polarity of the solvents mixture used in the experimental work [3].
Owing to the unrestricted formalism of all calculations, the spin contamination and its effect on the calculated energetics was monitored as done for similar systems [35,39] and the <S2> values are collected in Supplementary Information section (Table S1).
Supplementary Materials
Supplementary materials can be found online.
Author Contributions
T.M. and N.R. conceived of the presented idea and supervised the project. A.P. and M.C.M. performed the computations. T.M. verified and analyzed the data. T.M. and N.R. wrote the manuscript with support from M.T., A.P. contributed to the final version of the manuscript, drafted the manuscript and designed the figures. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
The University of Calabria is gratefully acknowledged.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| TpPh2 | hydrotris (3,5-diphenylpyrazole-1-yl) borate |
| DFT | Density Functional Theory |
| PES | Potential Energy Surface |
| PCM | Polarizable Continuum Model |
| HOMO | High Occupied Molecular Orbital |
References
- Costas, M.; Mehn, M.P.; Jensen, M.P.; Que, L., Jr. Dioxygen activation at mononuclear nonheme iron active sites: Enzymes, models, and intermediates. Chem. Rev. 2004, 104, 939–986. [Google Scholar] [CrossRef] [PubMed]
- Talsi, E.P.; Bryliakov, K.P. Chemo- and stereoselective CH oxidations and epoxidations/cis-dihydroxylations with H2O2, catalyzed by non-heme iron and manganese complexes. Coord. Chem. Rev. 2012, 256, 1418–1434. [Google Scholar] [CrossRef]
- Paria, S.; Chatterjee, S.; Paine, T.K. Reactivity of an Iron− Oxygen Oxidant Generated upon Oxidative Decarboxylation of Biomimetic Iron(II) α-Hydroxy Acid Complexes. Inorg. Chem. 2014, 53, 2810–2821. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Costas, M.; Kim, J.; Tipton, A.K.; Que, L., Jr. Olefin Cis-Dihydroxylation versus Epoxidation by Non-Heme Iron Catalysts: Two Faces of an FeIII−OOH Coin. J. Am. Chem. Soc. 2002, 124, 3026–3035. [Google Scholar] [CrossRef] [PubMed]
- Thibon, A.; England, J.; Martinho, M.; Young, V.G., Jr.; Frisch, J.R.; Guillot, R.; Girerd, J.-J.; Münck, E.; Que, L., Jr.; Banse, F. Proton- and Reductant-Assisted Dioxygen Activation by a Nonheme Iron(II) Complex to Form an Oxoiron(IV) Intermediate. Angew. Chem. Int. Ed. Engl. 2008, 47, 7064–7067. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Lee, Y.-M.; Shin, W.; Fukuzumi, S.; Nam, W. Dioxygen Activation by Mononuclear Nonheme Iron(II) Complexes Generates Iron−Oxygen Intermediates in the Presence of an NADH Analogue and Proton. J. Am. Chem. Soc. 2009, 131, 13910–13911. [Google Scholar] [CrossRef]
- Mukherjee, A.; Martinho, M.; Bominaar, E.L.; Münck, E.; Que, L., Jr. Shape-Selective Interception by Hydrocarbons of the O2- Derived Oxidant of a Biomimetic Nonheme Iron Complex. Angew. Chem. Int. Ed. Engl. 2009, 48, 1780–1783. [Google Scholar] [CrossRef]
- Martinho, M.; Blain, G.; Banse, F. Activation of dioxygen by a mononuclear non-heme iron complex: Characterization of a Fe(III)-(OOH) intermediate. Dalton Trans. 2010, 39, 1630–1634. [Google Scholar] [CrossRef]
- Lee, Y.-M.; Hong, S.; Morimoto, Y.; Shin, W.; Fukuzumi, S.; Nam, W. Dioxygen Activation by a Non-Heme Iron(II) Complex: Formation of an Iron(IV)−Oxo Complex via C−H Activation by a Putative Iron(III)−Superoxo Species. J. Am. Chem. Soc. 2010, 132, 10668–10670. [Google Scholar] [CrossRef]
- Mandon, D.; Jaafar, H.; Thibon, A. Exploring the Oxygen sensitivity of FeCl2 complexes with tris(2-pyridylmethyl)amine-type ligands: O2 coordination and a quest for superoxide. N. J. Chem. 2011, 35, 1986–2000. [Google Scholar] [CrossRef]
- Badiei, Y.M.; Siegler, M.A.; Goldberg, D.P. O2 Activation by Bis(imino)pyridine Iron(II)-Thiolate Complexes. J. Am. Chem. Soc. 2011, 133, 1274–1277. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Goldsmith, R. Observation of a ferric hydroperoxide complex during the non-heme iron catalysed oxidation of alkenes and alkanes by O2. Chem. Commun. 2012, 48, 10532–10534. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Van Heuvelen, K.M.; Meier, K.K.; Münck, E.; Que, L., Jr. Sc3+-Triggered Oxoiron(IV) Formation from O2 and its Non-Heme Iron(II) Precursor via a Sc3+−Peroxo−Fe3+ Intermediate. J. Am. Chem. Soc. 2013, 135, 10198–10201. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.O.; Sastri, C.V.; Seo, M.S.; Kim, J.; Nam, W. Dioxygen Activation and Catalytic Aerobic Oxidation by a Mononuclear Nonheme Iron(II) Complex. J. Am. Chem. Soc. 2005, 127, 4178–4179. [Google Scholar] [CrossRef]
- Bruijnincx, P.C.A.; van Koten, G.; Gebbink, R.J.M.K. Mononuclear non-heme iron enzymes with the 2-His-1-carboxylate facial triad: Recent developments in enzymology and modeling studies. Chem. Soc. Rev. 2008, 37, 2716–2744. [Google Scholar] [CrossRef]
- Shook, R.L.; Peterson, S.M.; Greaves, J.; Moore, C.; Rheingold, A.L.; Borovik, A.S. Catalytic Reduction of Dioxygen to Water with a Monomeric Manganese Complex at Room Temperature. J. Am. Chem. Soc. 2011, 133, 5810–5817. [Google Scholar] [CrossRef]
- Paine, T.K.; Que, L., Jr. Dioxygen Activation by Biomimetic Iron(II) Complexes of α-Keto Acids and α-Hydroxy Acids. Struct. Bond. 2014, 160, 39–56. [Google Scholar]
- Sallmann, M.; Limberg, C. Utilizing the Trispyrazolyl Borate Ligand for the Mimicking of O2-Activating Mononuclear Nonheme Iron Enzymes. Acc. Chem. Res. 2015, 48, 2734–2743. [Google Scholar] [CrossRef]
- Sahu, S.; Goldberg, D.P. Activation of Dioxygen by Iron and Manganese Complexes: A Heme and Nonheme Perspective. J. Am. Chem. Soc. 2016, 138, 11410–11428. [Google Scholar] [CrossRef]
- Funabiki, T. Functional model oxygenations by nonheme iron complexes. In Advances in Catalytic Activation of Dioxygen by Metal Complexes; Simandi, L.I., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2003; pp. 157–226. [Google Scholar]
- Chatterjee, S.; Paine, T.K. Olefin cis-Dihydroxylation and Aliphatic C-H Bond Oxygenation by a Dioxygen-Derived Electrophilic Iron-Oxygen Oxidant. Angew. Chem. Int. Ed. Engl. 2015, 54, 9338–9342. [Google Scholar] [CrossRef]
- Chatterjee, S.; Paine, T.K. Hydroxylation versus Halogenation of Aliphatic C-H Bonds by a Dioxygen-Derived Iron−Oxygen Oxidant: Functional Mimicking of Iron Halogenases. Angew. Chem. Int. Ed. Engl. 2016, 55, 7717–7722. [Google Scholar] [CrossRef] [PubMed]
- Paria, S.; Que, L., Jr.; Paine, T.K. Oxidative Decarboxylation of Benzilic Acid by a Biomimetic Iron(II) Complex: Evidence for an Iron(IV)−Oxo−Hydroxo Oxidant from O2. Angew. Chem. Int. Ed. Engl. 2011, 50, 11129–11132. [Google Scholar] [CrossRef] [PubMed]
- Costas, M.; Que, L., Jr. Ligand Topology Tuning of Iron-Catalyzed Hydrocarbon Oxidations. Angew. Chem. Int. Ed. Engl. 2002, 41, 2179–2181. [Google Scholar] [CrossRef]
- Oldenburg, P.D.; Feng, Y.; Pryjomska-Ray, I.; Ness, D.; Que, L., Jr. Olefin cis-dihydroxylation with bio-inspired iron catalysts. evidence for an Fe(II)/Fe(IV) catalytic cycle. J. Am. Chem. Soc. 2010, 132, 17713–17723. [Google Scholar]
- Borowski, T.; Bassan, A.; Siegbahn, P.E.M. A Hybrid Density Functional Study of O−O Bond Cleavage and Phenyl Ring Hydroxylation for a Biomimetic Non-Heme Iron Complex. Inorg. Chem. 2004, 43, 3277–3291. [Google Scholar] [CrossRef]
- Mehn, M.P.; Fujisawa, K.; Hegg, E.L.; Que, L., Jr. Oxygen Activation by Nonheme Iron(II) Complexes: α-Keto Carboxylate versus Carboxylate. J. Am. Chem. Soc. 2003, 125, 7828–7842. [Google Scholar] [CrossRef]
- Siegbahn, P.E.M.; Borowski, T. Modeling Enzymatic Reactions Involving Transition Metals. Acc. Chem. Res. 2006, 39, 729–738. [Google Scholar] [CrossRef]
- Borowski, T.; Bassan, A.; Siegbahn, P.E.M. Mechanism of Dioxygen Activation in 2-Oxoglutarate-Dependent Enzymes: A Hybrid DFT Study. Chem. Eur. J. 2004, 10, 1031–1041. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Zakrzewski, V.G.; Montgomery, J.A., Jr.; Stratmann, R.E.; Burant, J.C.; et al. Gaussian 2009; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Zhao, Y.; Truhlar, D.G. A new local density functional for main-group thermochemistry, transition metal bonding, thermochemical kinetics, and noncovalent interactions. J. Chem. Phys. 2006, 125, 194101. [Google Scholar] [CrossRef]
- Valero, R.; Costa, R.; Moreira, I.D.P.R.; Truhlar, D.G.; Illas, F. Performance of the M06 family of exchange-correlation functionals for predicting magnetic coupling in organic and inorganic molecules. J. Chem. Phys. 2008, 128, 114103. [Google Scholar] [CrossRef]
- Marzouk, A.; Bolvin, H.; Reinhardt, P.; Manceron, L.; Perchard, J.P.; Tremblay, B.; Alikhani, M.E. A Combined Experimental and Theoretical Study of the Ti2 + N2O Reaction. J. Phys. Chem. A 2014, 118, 561–572. [Google Scholar] [CrossRef]
- Gusev, D.G. Assessing the Accuracy of M06-L Organometallic Thermochemistry. Organometallics 2013, 32, 4239–4243. [Google Scholar] [CrossRef]
- Marino, T.; Fortino, M.G.; Russo, N.; Toscano, M.; Alberto, M.E. Computational Mechanistic Insights on the NO Oxidation Reaction Catalyzed by Non-Heme Biomimetic Cr-N-Tetramethylated Cyclam Complexes. Int. J. Mol. Sci. 2019, 20, 3955. [Google Scholar] [CrossRef] [PubMed]
- Fortino, M.; Marino, T.; Russo, N. Theoretical Study of Silver-Ion-Mediated Base Pairs: The Case of C−Ag−C and C−Ag−A Systems. J. Phys. Chem. A 2015, 119, 5153–5157. [Google Scholar] [CrossRef] [PubMed]
- Boone, A.J.; Chang, C.H.; Greene, S.N.; Herz, T.; Richards, N.G.J. Modeling the spin-dependent properties of open-shell Fe(III)-containing systems: Towards a computational description of nitrile hydratase. Coord. Chem. Rev. 2003, 238–239, 291–314. [Google Scholar] [CrossRef]
- Barone, V.; Cossi, M. Quantum Calculation of Molecular Energies and Energy Gradients in Solution by a Conductor Solvent Model. J. Phys. Chem. A 1998, 102, 1995–2001. [Google Scholar] [CrossRef]
- Cossi, M.; Rega, N.; Scalmani, G.; Barone, V. Energies, Structures, and Electronic Properties of Molecules in Solution with the C-PCM Solvation Model. J. Comput. Chem. 2003, 24, 669–681. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).