Real-Time 3D Single Particle Tracking: Towards Active Feedback Single Molecule Spectroscopy in Live Cells
Abstract
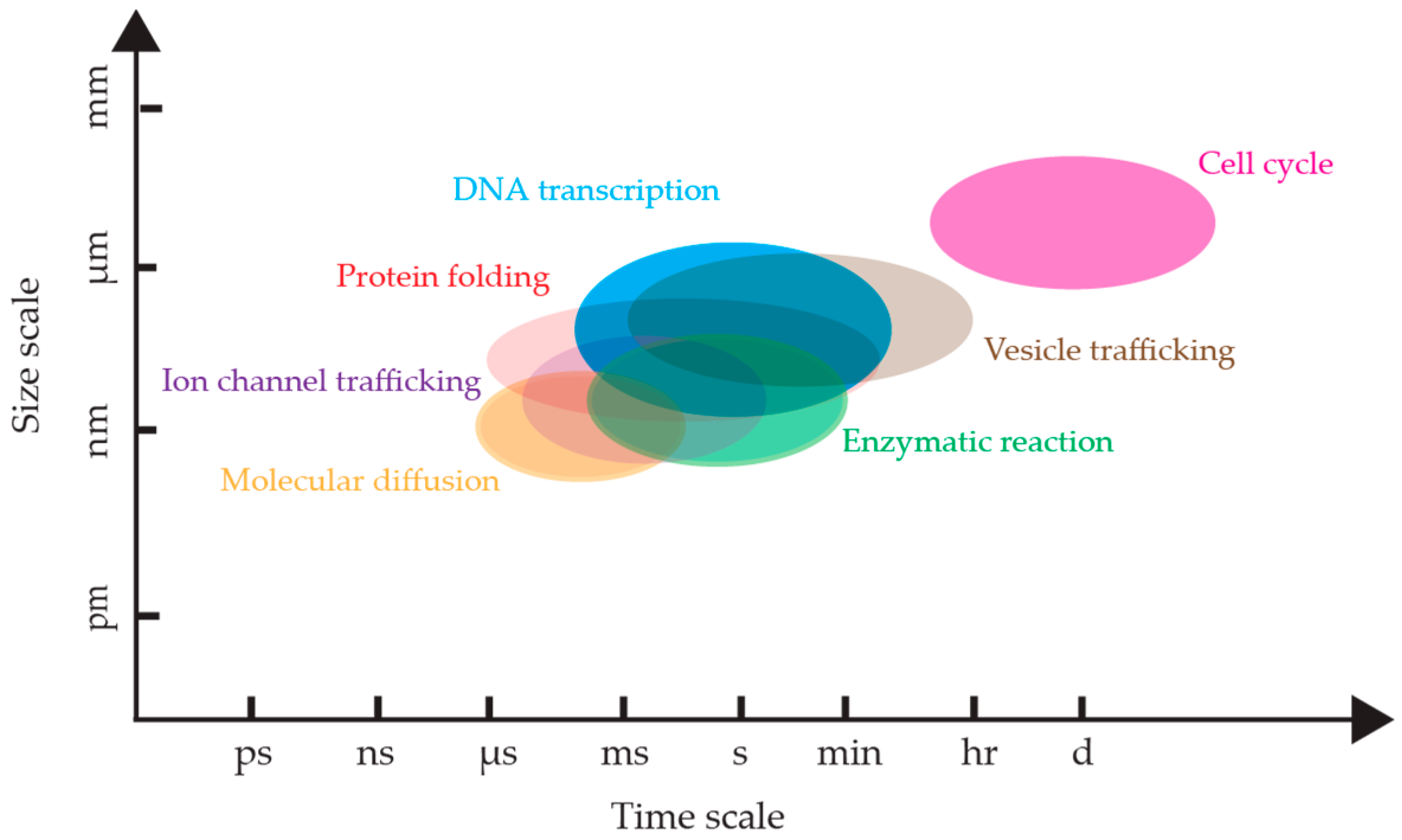
1. Introduction
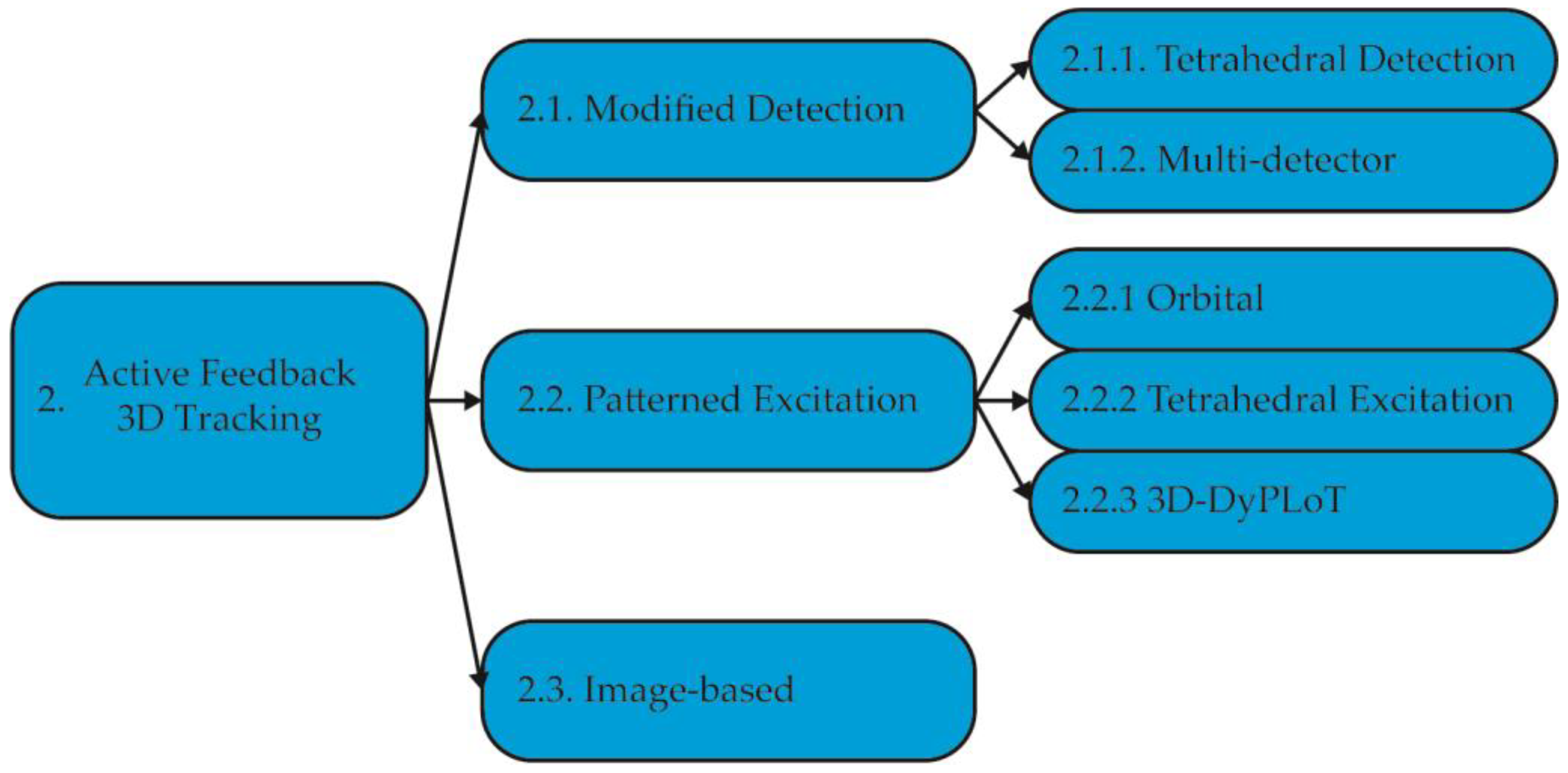
2. Active Feedback 3D Tracking Methods
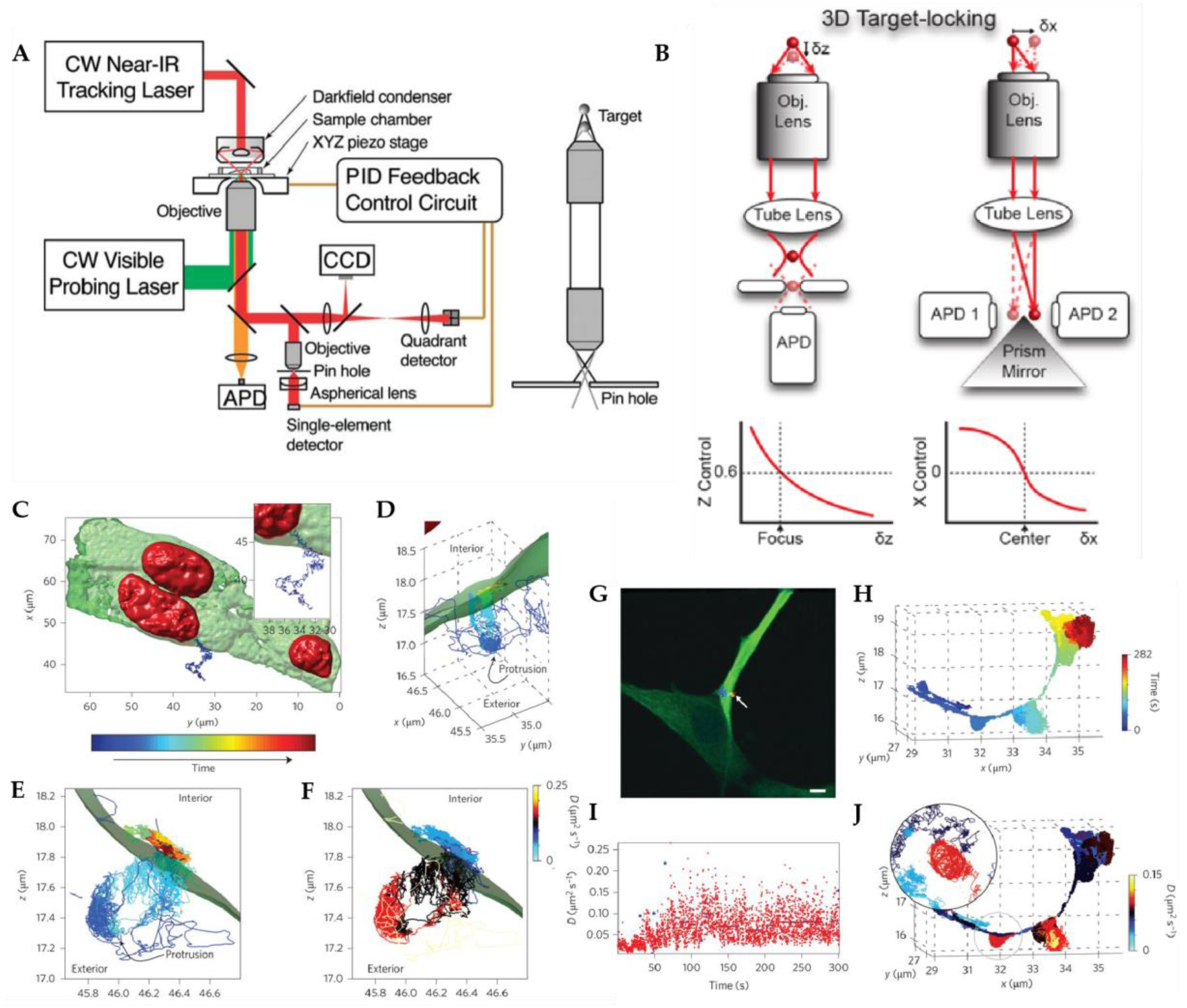
2.1. Active Feedback 3D Single Particle Tracking with Modified Detection
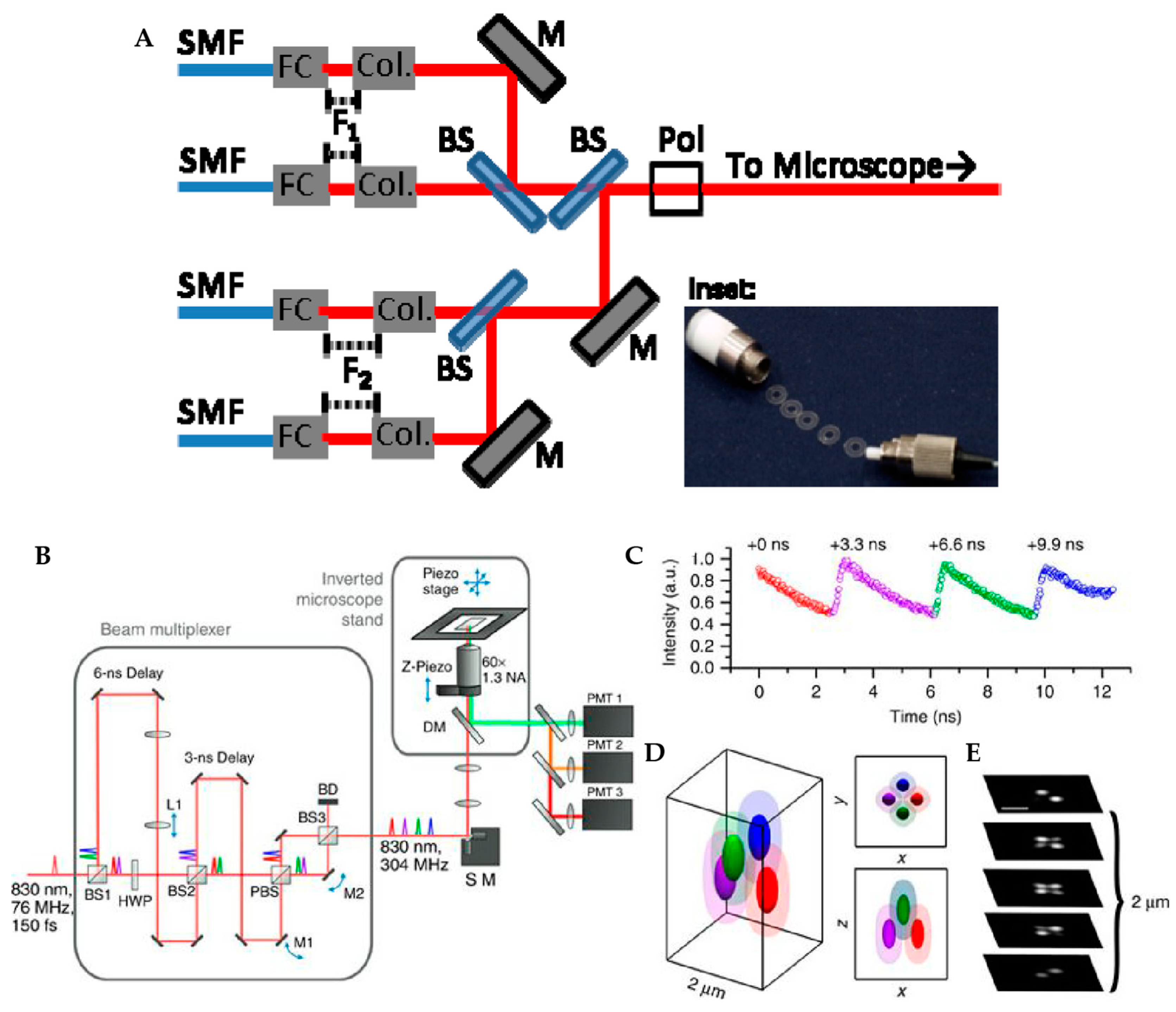
2.1.1. Tetrahedral Detection Active Feedback Tracking
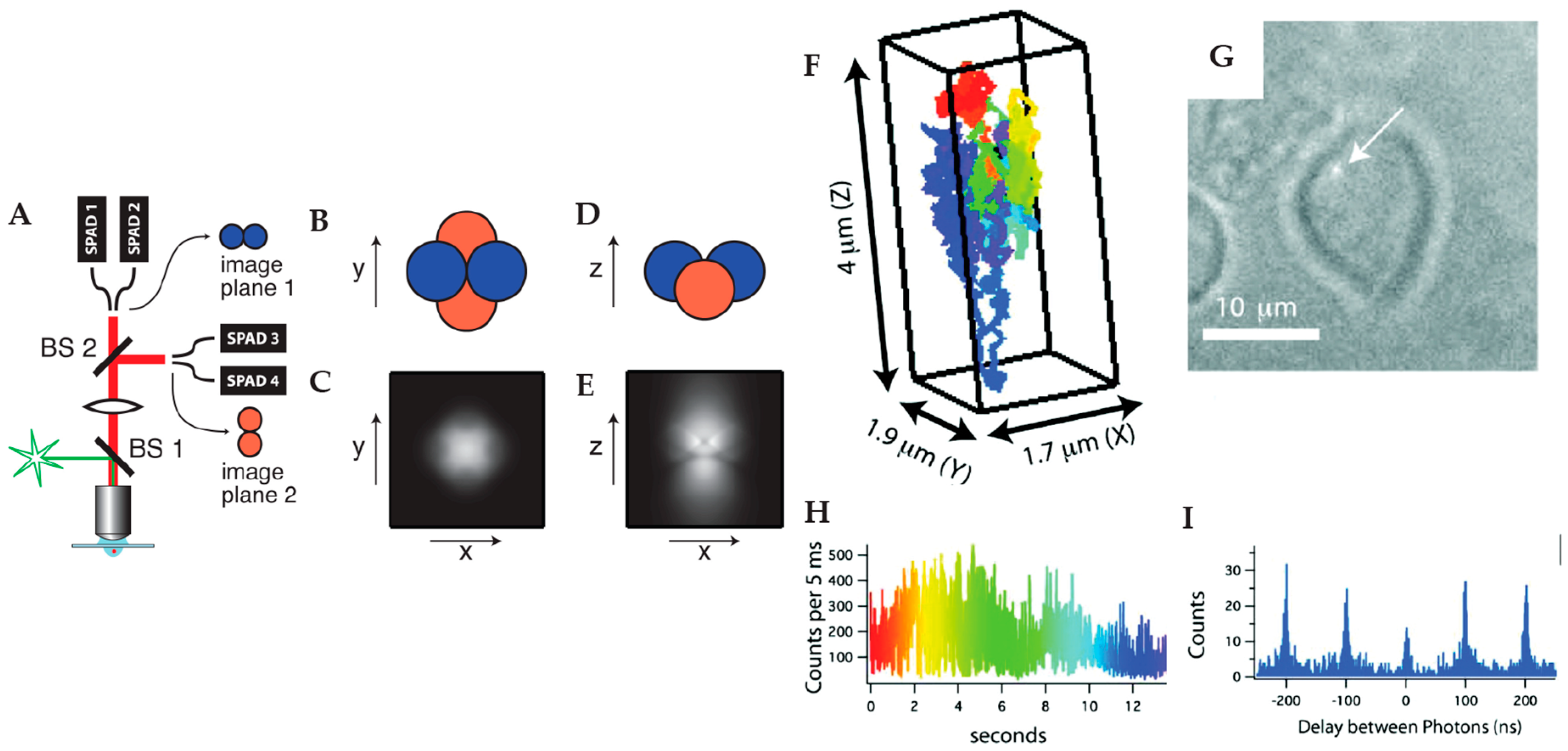
2.1.2. Multi-Detector Active Feedback Tracking
2.2. Active Feedback 3D Single Particle Tracking with Patterned Excitation
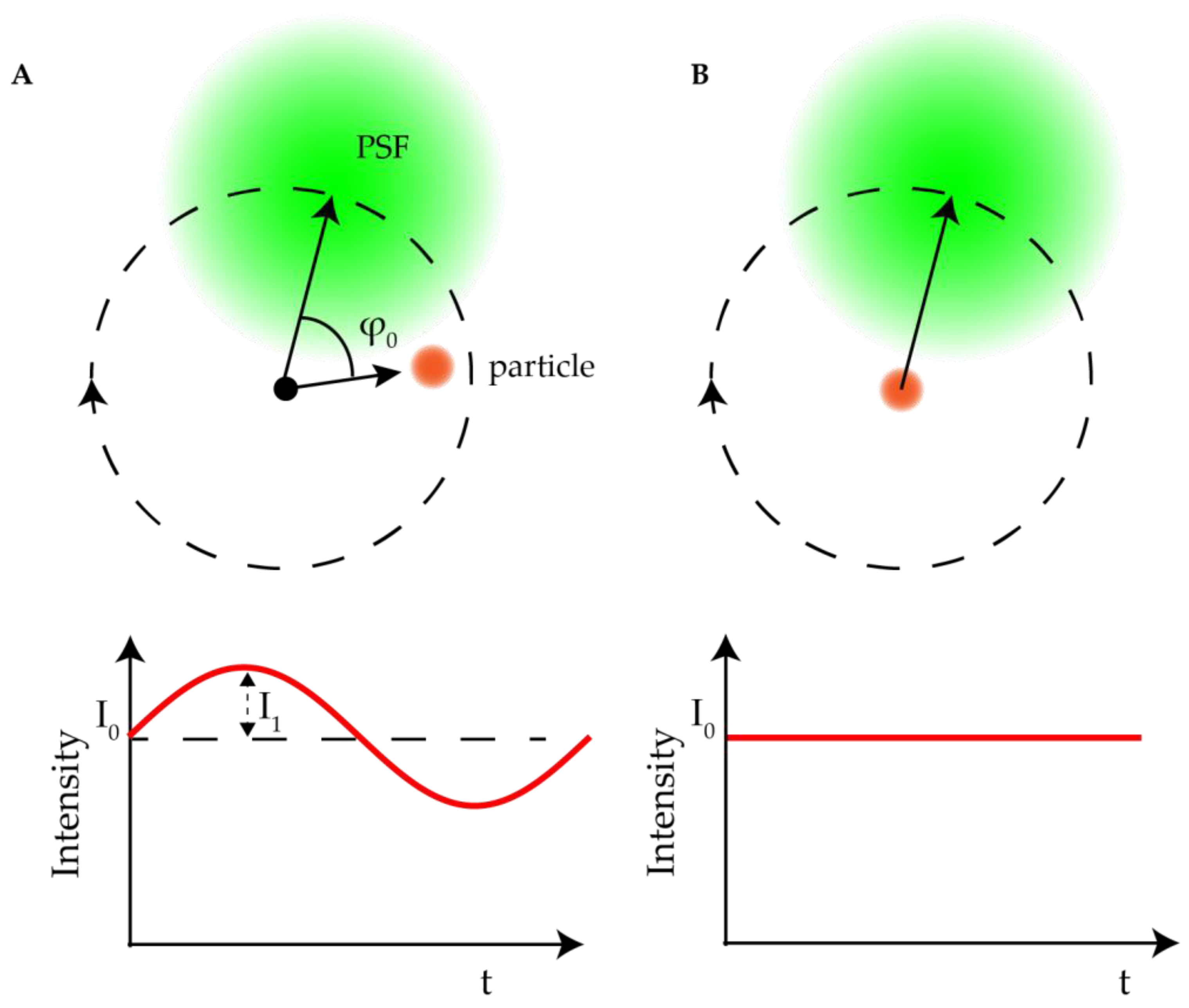
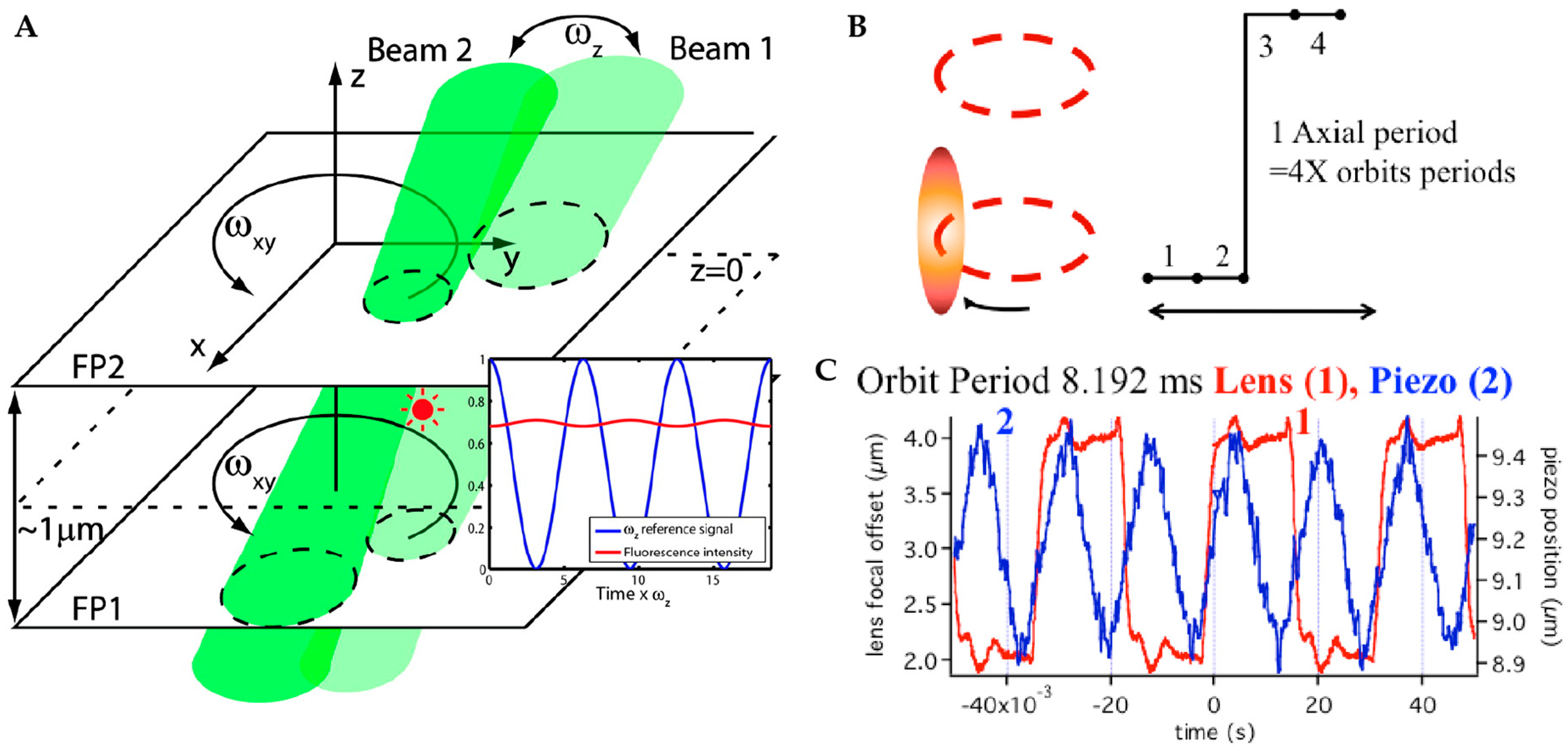
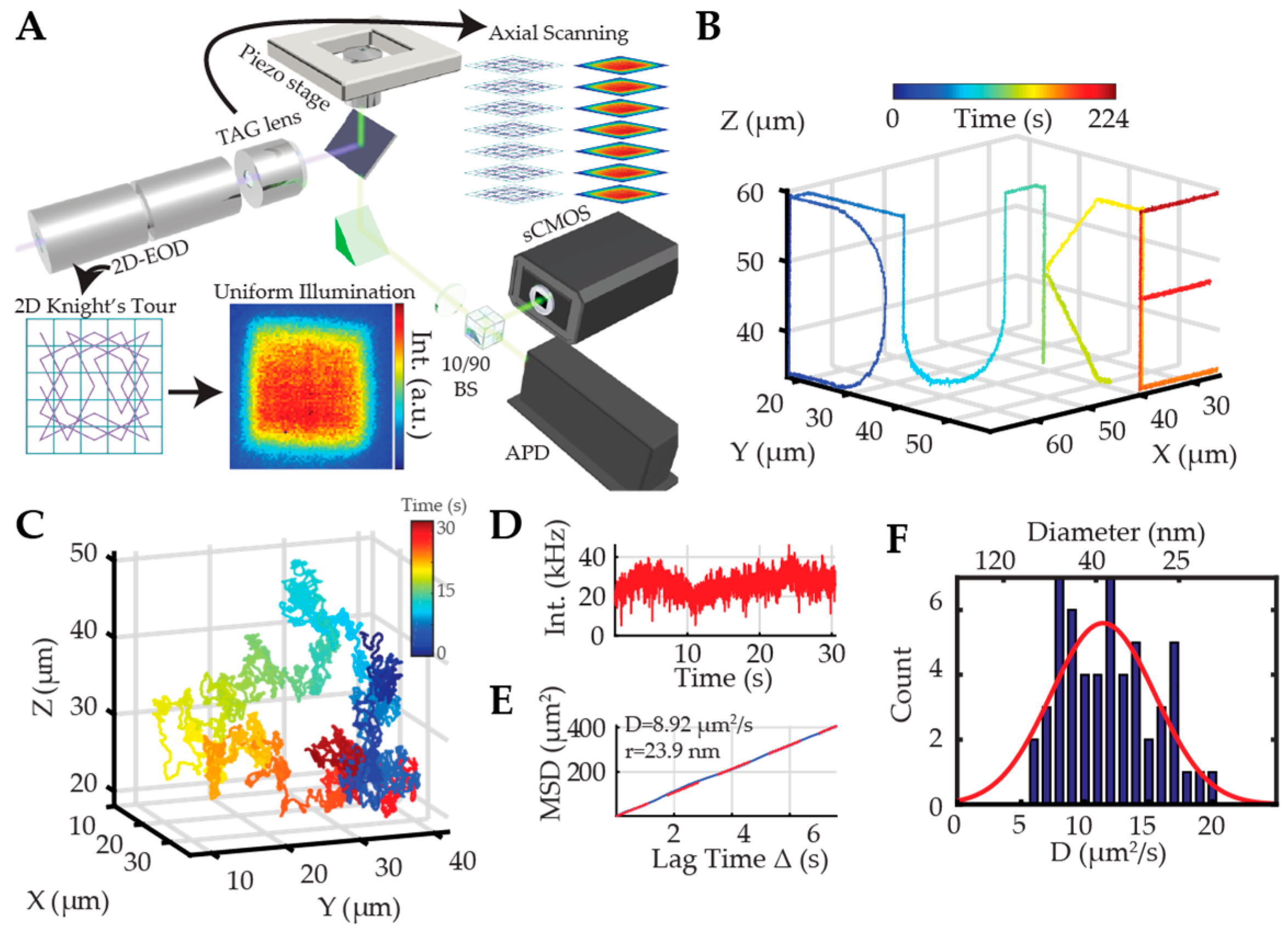
2.2.1. Orbital 3D Tracking Methods
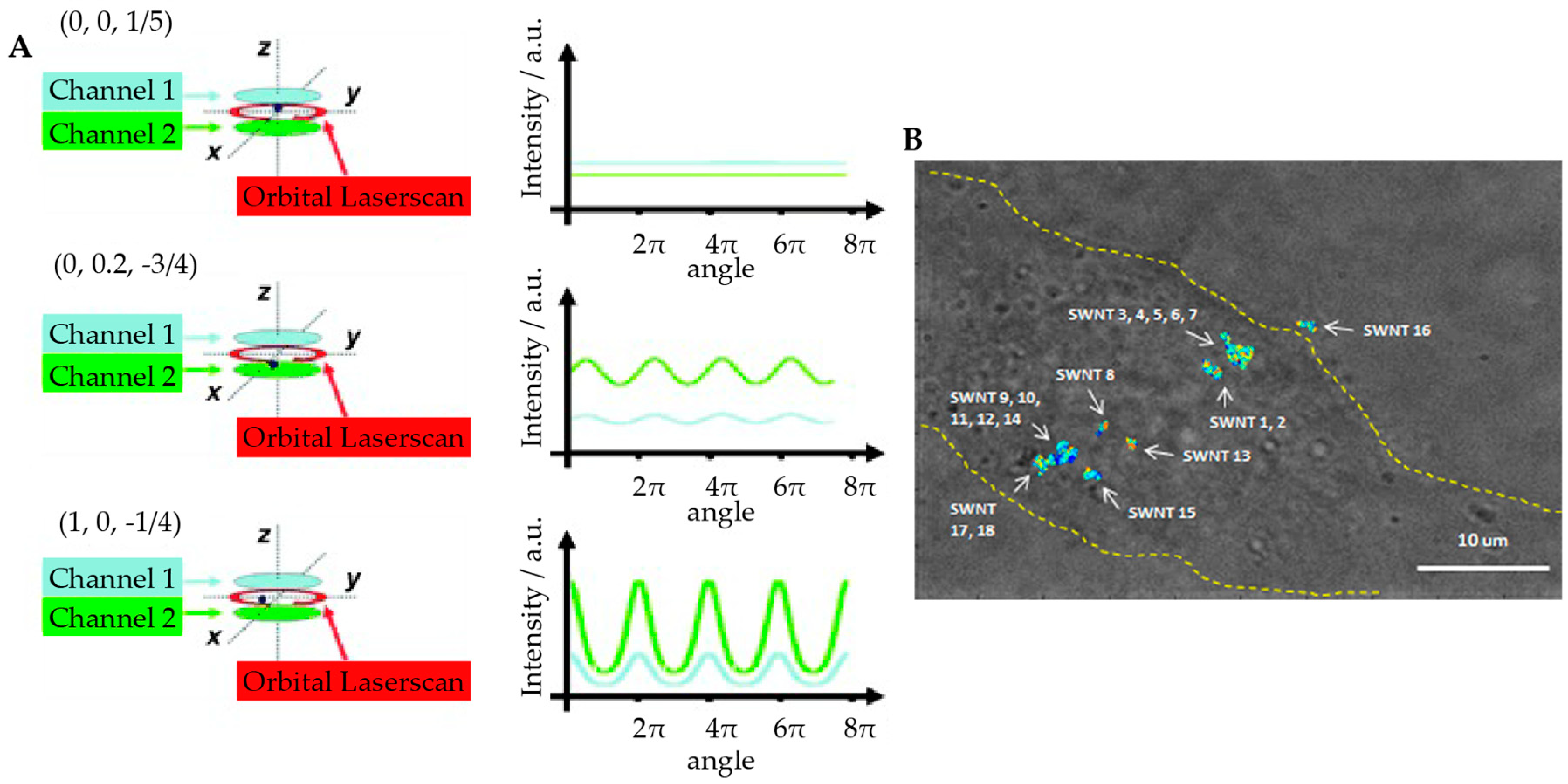
2.2.2. Tetrahedral Excitation 3D Tracking
2.2.3. Dynamic Photon Localization Tracking (3D-DyPLoT)
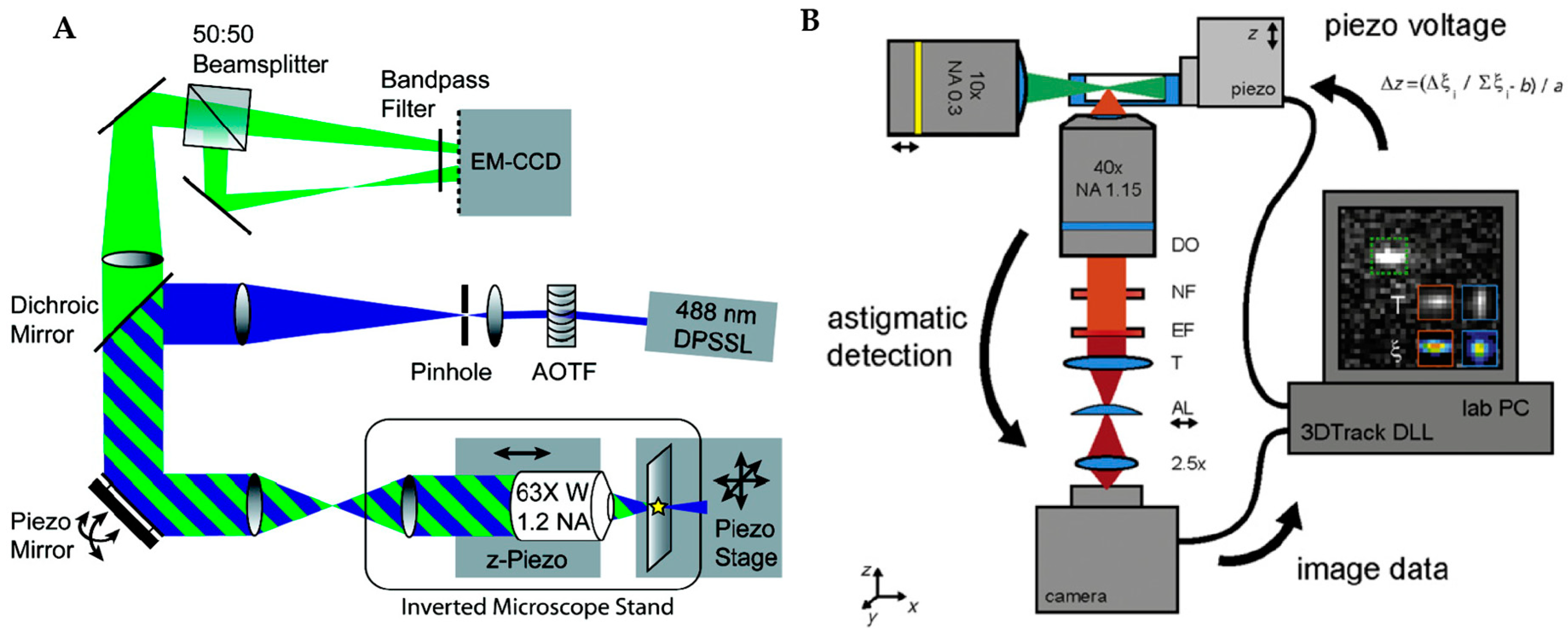
2.3. Image-Based Active Feedback Tracking
3. Method Comparison
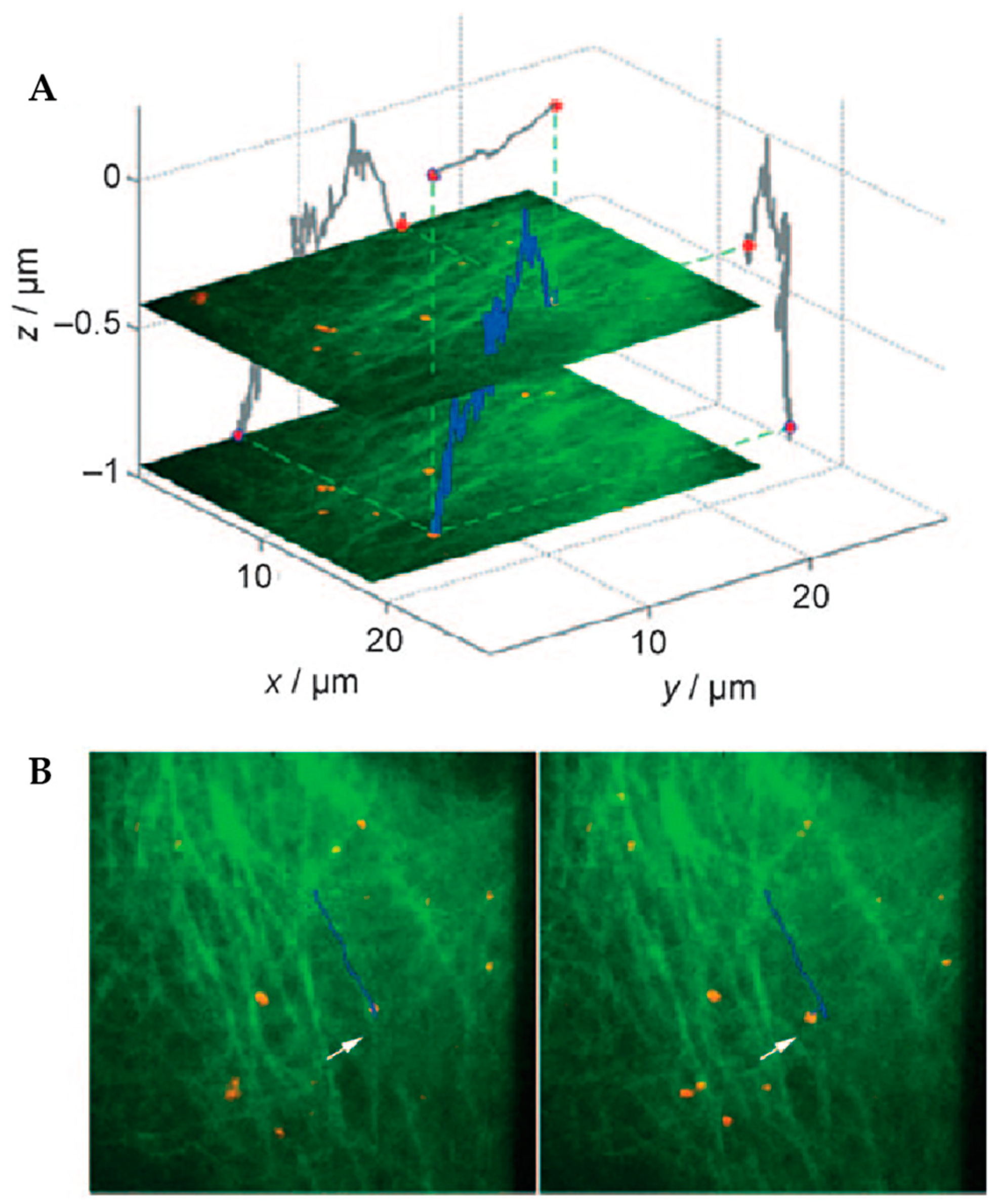
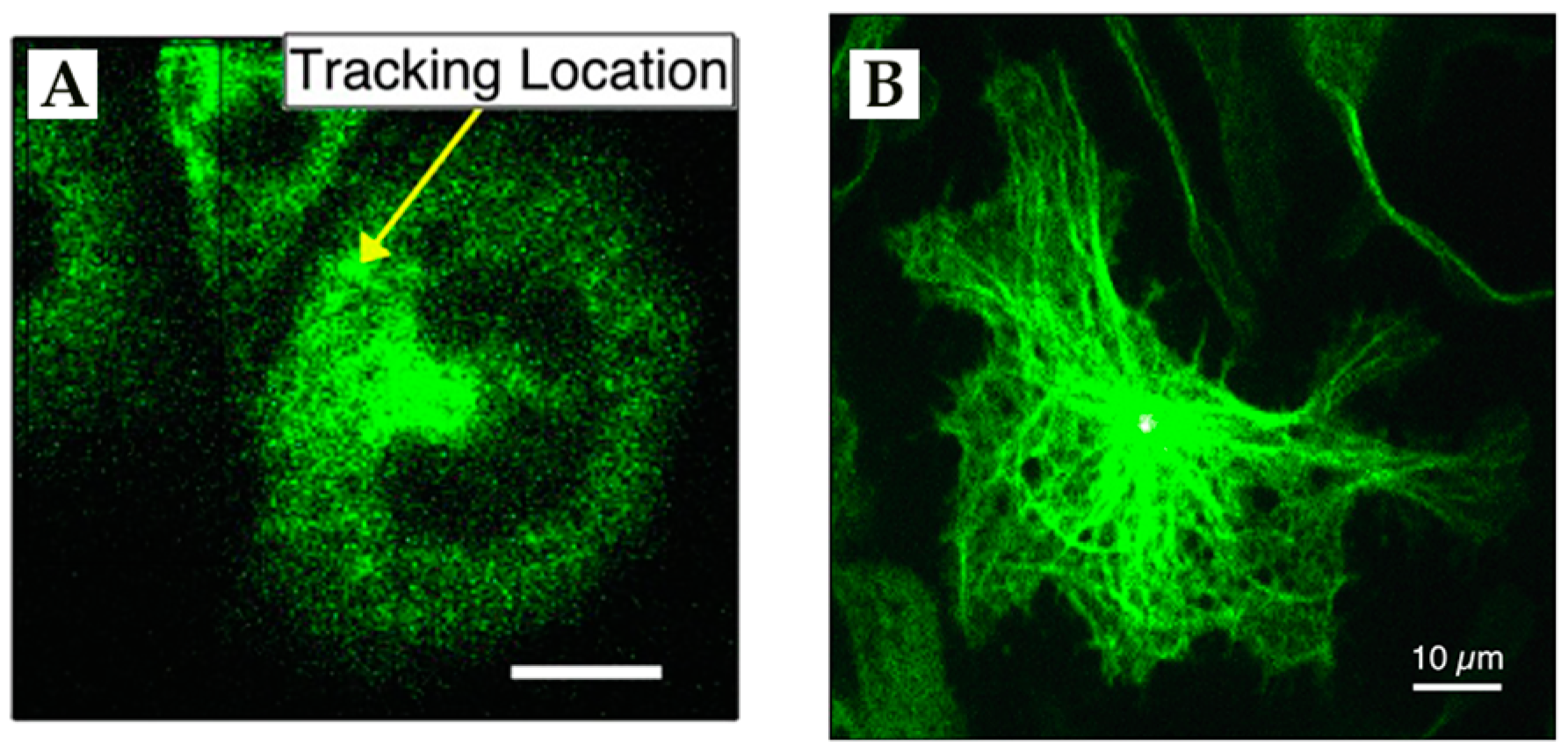
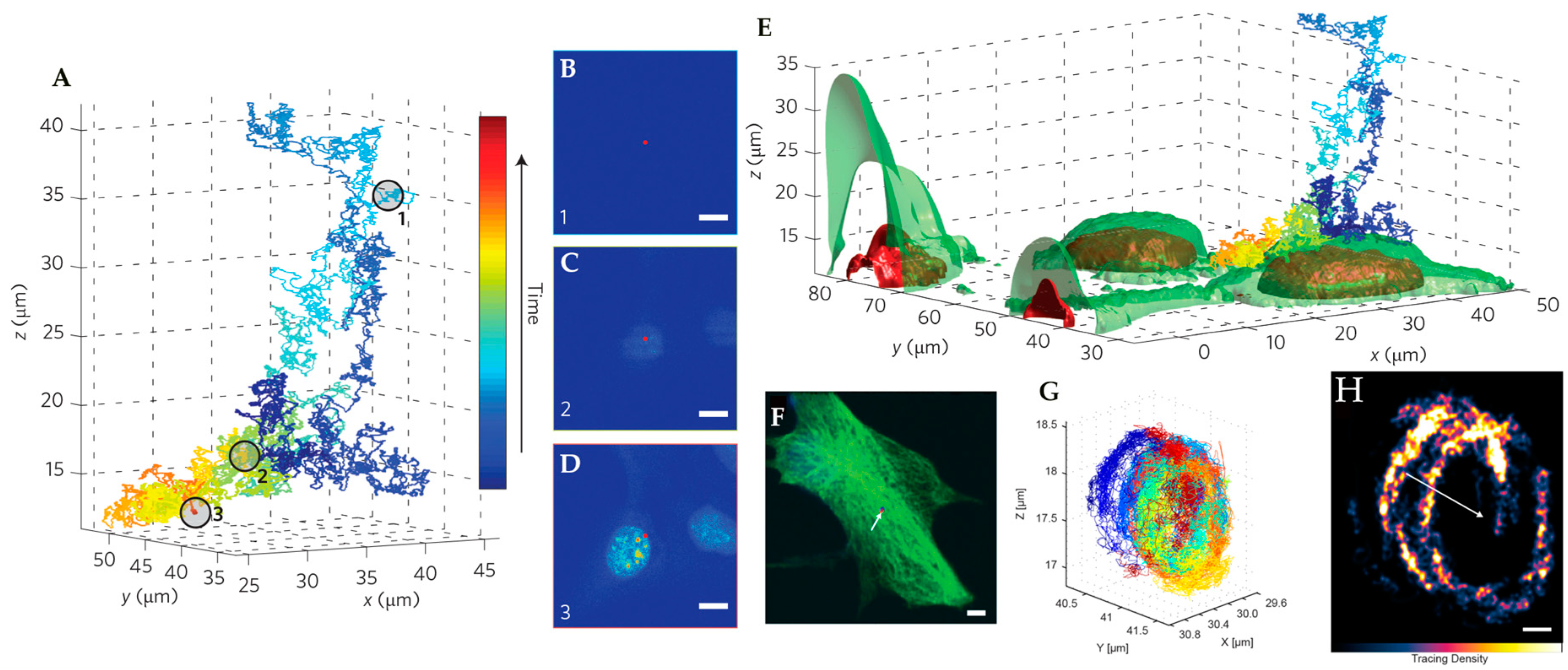
4. Environmental Contextualization in Active Feedback-Based 3D-SPT Methods
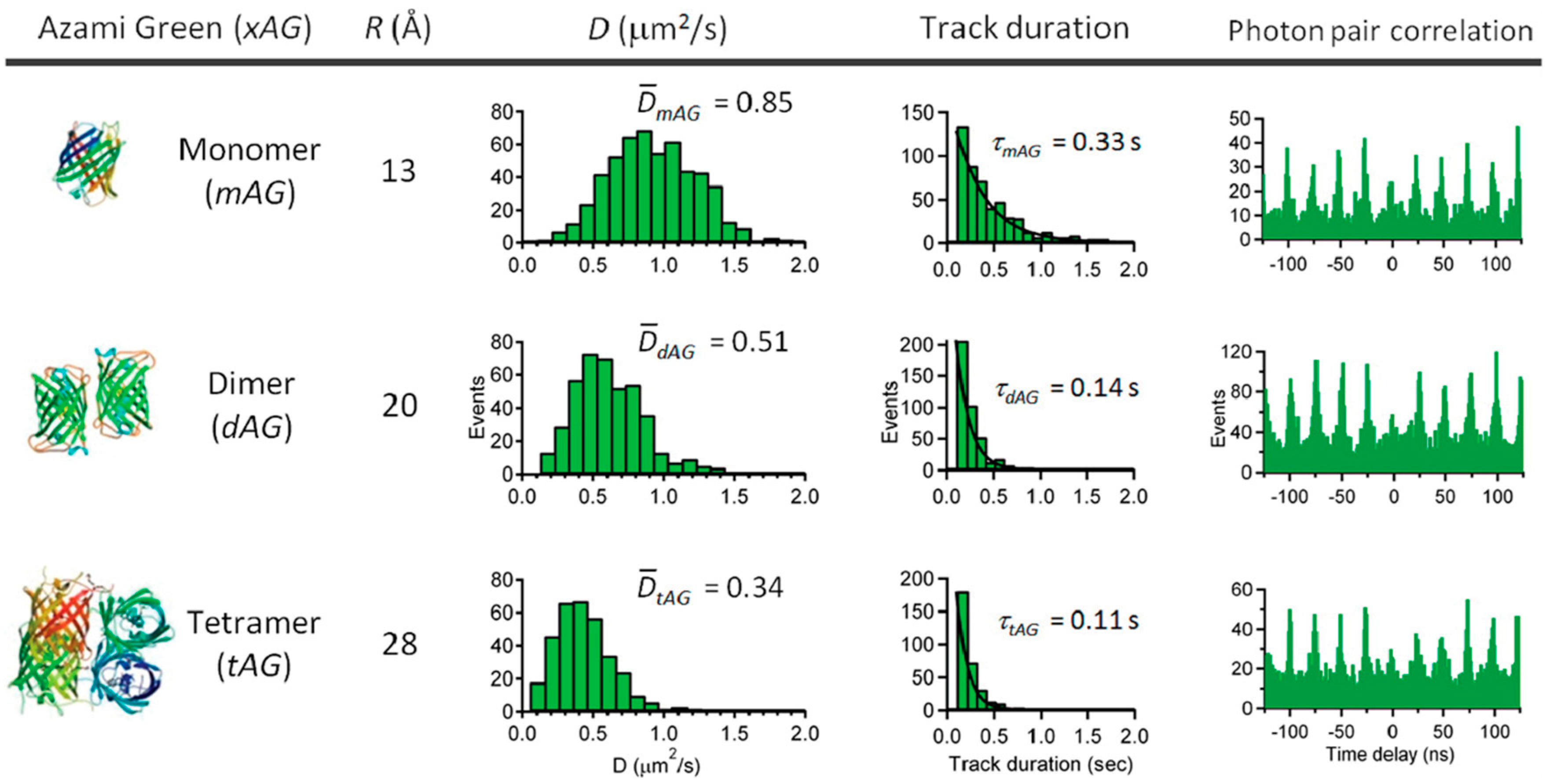
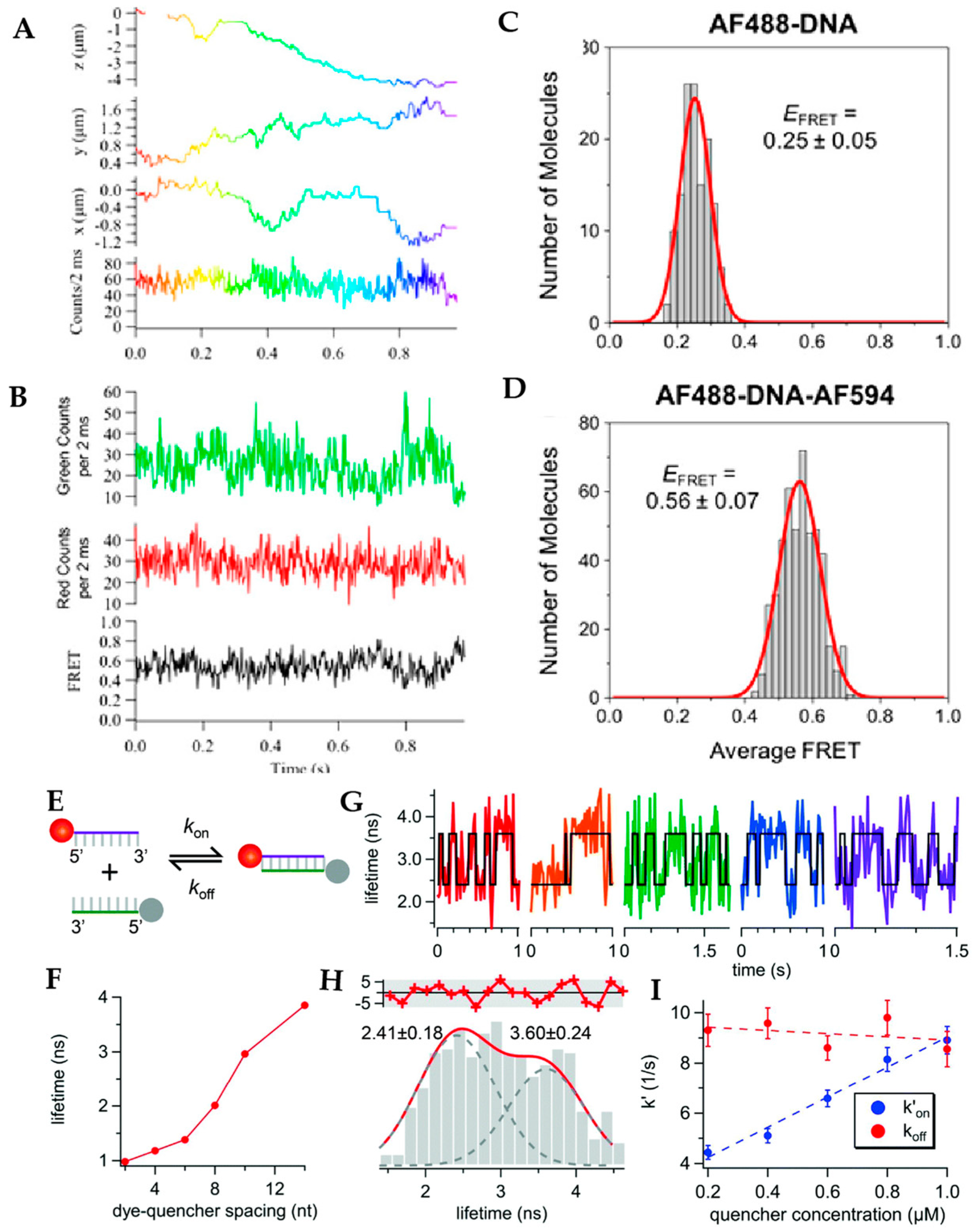
5. Active Feedback 3D Single Molecule Tracking: The Final Frontier
6. Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Moerner, W.E.; Orrit, M. Illuminating Single Molecules in Condensed Matter. Science 1999, 283, 1670–1676. [Google Scholar] [CrossRef]
- Ha, T.; Enderle, T.; Ogletree, D.F.; Chemla, D.S.; Selvin, P.R.; Weiss, S. Probing the interaction between two single molecules: Fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl. Acad. Sci. 1996, 93, 6264. [Google Scholar] [CrossRef]
- Lu, H.P.; Xun, L.; Xie, X.S. Single-Molecule Enzymatic Dynamics. Science 1998, 282, 1877–1882. [Google Scholar] [CrossRef]
- Shashkova, S.; Leake, M.C. Single-molecule fluorescence microscopy review: Shedding new light on old problems. Biosci. Rep. 2017, 37, BSR20170031. [Google Scholar] [CrossRef]
- Stracy, M.; Uphoff, S.; de Leon, F.G.; Kapanidis, A.N. In vivo single-molecule imaging of bacterial DNA replication, transcription, and repair. FEBS Lett. 2014, 588, 3585–3594. [Google Scholar] [CrossRef]
- Robinson, A.; van Oijen, A.M. Bacterial replication, transcription and translation: Mechanistic insights from single-molecule biochemical studies. Nat. Rev. Microbiol. 2013, 11, 303. [Google Scholar] [CrossRef]
- Alhadid, Y.; Chung, S.; Lerner, E.; Taatjes, D.J.; Borukhov, S.; Weiss, S. Studying transcription initiation by RNA polymerase with diffusion-based single-molecule fluorescence. Protein Sci. 2017, 26, 1278–1290. [Google Scholar] [CrossRef]
- Kapanidis, A.N.; Margeat, E.; Ho, S.O.; Kortkhonjia, E.; Weiss, S.; Ebright, R.H. Initial Transcription by RNA Polymerase Proceeds Through a DNA-Scrunching Mechanism. Science 2006, 314, 1144–1147. [Google Scholar] [CrossRef]
- Blanchard, S.C. Single-molecule observations of ribosome function. Curr. Opin. Struct. Biol. 2009, 19, 103–109. [Google Scholar] [CrossRef]
- Zhuang, X. Single-Molecule RNA Science. Annu. Rev. Biophys. Biomol. Struct. 2005, 34, 399–414. [Google Scholar] [CrossRef]
- Helm, M.; Kobitski, A.Y.; Nienhaus, G.U. Single-molecule Förster resonance energy transfer studies of RNA structure, dynamics and function. Biophys. Rev. 2009, 1, 161. [Google Scholar] [CrossRef]
- Ha, T.; Ting, A.Y.; Liang, J.; Caldwell, W.B.; Deniz, A.A.; Chemla, D.S.; Schultz, P.G.; Weiss, S. Single-molecule fluorescence spectroscopy of enzyme conformational dynamics and cleavage mechanism. Proc. Natl. Acad. Sci. USA 1999, 96, 893–898. [Google Scholar] [CrossRef]
- Klostermeier, D. Single-molecule FRET reveals nucleotide-driven conformational changes in molecular machines and their link to RNA unwinding and DNA supercoiling. Biochem. Soc. Trans. 2011, 39, 611–616. [Google Scholar] [CrossRef]
- Bisaria, N.; Herschlag, D. Probing the kinetic and thermodynamic consequences of the tetraloop/tetraloop receptor monovalent ion-binding site in P4–P6 RNA by smFRET. Biochem. Soc. Trans. 2015, 43, 172–178. [Google Scholar] [CrossRef]
- Henzler-Wildman, K.A.; Thai, V.; Lei, M.; Ott, M.; Wolf-Watz, M.; Fenn, T.; Pozharski, E.; Wilson, M.A.; Petsko, G.A.; Karplus, M.; et al. Intrinsic motions along an enzymatic reaction trajectory. Nature 2007, 450, 838. [Google Scholar] [CrossRef]
- Hanson, J.A.; Duderstadt, K.; Watkins, L.P.; Bhattacharyya, S.; Brokaw, J.; Chu, J.-W.; Yang, H. Illuminating the mechanistic roles of enzyme conformational dynamics. Proc. Natl. Acad. Sci. USA 2007, 104, 18055–18060. [Google Scholar] [CrossRef]
- Ferreon, A.C.M.; Gambin, Y.; Lemke, E.A.; Deniz, A.A. Interplay of α-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc. Natl. Acad. Sci. USA 2009, 106, 5645–5650. [Google Scholar] [CrossRef]
- Schuler, B.; Hofmann, H. Single-molecule spectroscopy of protein folding dynamics—expanding scope and timescales. Curr. Opin. Struct. Biol. 2013, 23, 36–47. [Google Scholar] [CrossRef]
- Maglione, M.; Sigrist, S.J. Seeing the forest tree by tree: Super-resolution light microscopy meets the neurosciences. Nat. Neurosci. 2013, 16, 790–797. [Google Scholar] [CrossRef]
- Fornasiero, E.F.; Opazo, F. Super-resolution imaging for cell biologists Concepts, applications, current challenges and developments. Bioessays 2015, 37, 436–451. [Google Scholar] [CrossRef]
- Sengupta, P.; van Engelenburg, S.B.; Lippincott-Schwartz, J. Superresolution Imaging of Biological Systems Using Photoactivated Localization Microscopy. Chem. Rev. 2014, 114, 3189–3202. [Google Scholar] [CrossRef]
- Liu, Z.; Lavis, L.D.; Betzig, E. Imaging Live-Cell Dynamics and Structure at the Single-Molecule Level. Mol. Cell 2015, 58, 644–659. [Google Scholar] [CrossRef]
- Kusumi, A.; Tsunoyama, T.A.; Hirosawa, K.M.; Kasai, R.S.; Fujiwara, T.K. Tracking single molecules at work in living cells. Nat. Chem. Biol. 2014, 10, 524–532. [Google Scholar] [CrossRef]
- Eggeling, C.; Willig, K.I.; Sahl, S.J.; Hell, S.W. Lens-based fluorescence nanoscopy. Q. Rev. Biophys. 2015, 48, 178–243. [Google Scholar] [CrossRef]
- Hell, S.W.; Sahl, S.J.; Bates, M.; Zhuang, X.W.; Heintzmann, R.; Booth, M.J.; Bewersdorf, J.; Shtengel, G.; Hess, H.; Tinnefeld, P.; et al. The 2015 super-resolution microscopy roadmap. J. Phys. D Appl. Phys. 2015, 48, 443001. [Google Scholar] [CrossRef]
- Ntziachristos, V. Going deeper than microscopy: The optical imaging frontier in biology. Nat. Methods 2010, 7, 603–614. [Google Scholar] [CrossRef]
- Ji, N. Adaptive optical fluorescence microscopy. Nat. Methods 2017, 14, 374–380. [Google Scholar] [CrossRef]
- Chen, T.; Dong, B.; Chen, K.C.; Zhao, F.; Cheng, X.D.; Ma, C.B.; Lee, S.; Zhang, P.; Kang, S.H.; Ha, J.W.; et al. Optical Super-Resolution Imaging of Surface Reactions. Chem. Rev. 2017, 117, 7510–7537. [Google Scholar] [CrossRef]
- Garcia, H.G.; Tikhonov, M.; Lin, A.; Gregor, T. Quantitative Imaging of Transcription in Living Drosophila Embryos Links Polymerase Activity to Patterning. Curr. Biol. 2013, 23, 2140–2145. [Google Scholar] [CrossRef]
- Bieri, O.; Wirz, J.; Hellrung, B.; Schutkowski, M.; Drewello, M.; Kiefhaber, T. The speed limit for protein folding measured by triplet-triplet energy transfer. Proc. Natl. Acad. Sci. USA 1999, 96, 9597–9601. [Google Scholar] [CrossRef]
- Kubelka, J.; Hofrichter, J.; Eaton, W.A. The protein folding ‘speed limit’. Curr. Opin. Struct. Biol. 2004, 14, 76–88. [Google Scholar] [CrossRef]
- Naganathan, A.N.; Munoz, V. Scaling of folding times with protein size. J. Am. Chem. Soc. 2005, 127, 480–481. [Google Scholar] [CrossRef]
- Kumar, M.; Mommer, M.S.; Sourjik, V. Mobility of Cytoplasmic, Membrane, and DNA-Binding Proteins in Escherichia coli. Biophys. J. 2010, 98, 552–559. [Google Scholar] [CrossRef]
- Williams, J.W.; Cady, L.C. Molecular diffusion in solution. Chem. Rev. 1934, 14, 171–217. [Google Scholar] [CrossRef]
- Verkman, A.S. Solute and macromolecule diffusion in cellular aqueous compartments. Trends Biochem. Sci. 2002, 27, 27–33. [Google Scholar] [CrossRef]
- Baserga, R. The Relationship of the Cell Cycle to Tumor Growth and Control of Cell Division: A Review. Cancer Res. 1965, 25, 581–595. [Google Scholar]
- Sudhof, T.C. The Synaptic Vesicle Cycle - a Cascade of Protein-Protein Interactions. Nature 1995, 375, 645–653. [Google Scholar] [CrossRef]
- Holt, M.; Cooke, A.; Neef, A.; Lagnado, L. High mobility of vesicles supports continuous exocytosis at a ribbon synapse. Curr. Biol. 2004, 14, 173–183. [Google Scholar] [CrossRef]
- Peng, A.; Rotman, Z.; Deng, P.Y.; Klyachko, V.A. Differential Motion Dynamics of Synaptic Vesicles Undergoing Spontaneous and Activity-Evoked Endocytosis. Neuron 2012, 73, 1108–1115. [Google Scholar] [CrossRef]
- Joensuu, M.; Padmanabhan, P.; Durisic, N.; Bademosi, A.T.D.; Cooper-Williams, E.; Morrow, I.C.; Harper, C.B.; Jung, W.; Parton, R.G.; Goodhil, G.J.; et al. Subdiffractional tracking of internalized molecules reveals heterogeneous motion states of synaptic vesicles. J. Cell Biol. 2016, 215, 277–292. [Google Scholar] [CrossRef]
- Syeda, R.; Qiu, Z.Z.; Dubin, A.E.; Murthy, S.E.; Florendo, M.N.; Mason, D.E.; Mathur, J.; Cahalan, S.M.; Peters, E.C.; Montal, M.; et al. LRRC8 Proteins Form Volume-Regulated Anion Channels that Sense Ionic Strength. Cell 2016, 164, 499–511. [Google Scholar] [CrossRef]
- Minn, A.J.; Velez, P.; Schendel, S.L.; Liang, H.; Muchmore, S.W.; Fesik, S.W.; Fill, M.; Thompson, C.B. Bcl-x(L) forms an ion channel in synthetic lipid membranes. Nature 1997, 385, 353–357. [Google Scholar] [CrossRef]
- Colquhoun, D.; Sakmann, B. Fluctuations in the Microsecond Time Range of the Current through Single Acetylcholine-Receptor Ion Channels. Nature 1981, 294, 464–466. [Google Scholar] [CrossRef]
- Kim, H.K.; Rasnik, I.; Liu, J.W.; Ha, T.J.; Lu, Y. Dissecting metal ion-dependent folding and catalysis of a single DNAzyme. Nat. Chem. Biol. 2007, 3, 763–768. [Google Scholar] [CrossRef]
- Deniz, A.A.; Dahan, M.; Grunwell, J.R.; Ha, T.; Faulhaber, A.E.; Chemla, D.S.; Weiss, S.; Schultz, P.G. Single-pair fluorescence resonance energy transfer on freely diffusing molecules: Observation of Förster distance dependence and subpopulations. Proc. Natl. Acad. Sci. USA 1999, 96, 3670–3675. [Google Scholar] [CrossRef]
- Deniz, A.A.; Laurence, T.A.; Beligere, G.S.; Dahan, M.; Martin, A.B.; Chemla, D.S.; Dawson, P.E.; Schultz, P.G.; Weiss, S. Single-molecule protein folding: Diffusion fluorescence resonance energy transfer studies of the denaturation of chymotrypsin inhibitor 2. Proc. Natl. Acad. Sci. USA 2000, 97, 5179–5184. [Google Scholar] [CrossRef]
- Kudalkar, E.M.; Davis, T.N.; Asbury, C.L. Single-Molecule Total Internal Reflection Fluorescence Microscopy. Cold Spring Harbor Protocols 2016, 2016, pdb.top077800. [Google Scholar] [CrossRef]
- Roy, R.; Hohng, S.; Ha, T. A practical guide to single-molecule FRET. Nat. Methods 2008, 5, 507. [Google Scholar] [CrossRef]
- Kim, J.-Y.; Kim, C.; Lee, N.K. Real-time submillisecond single-molecule FRET dynamics of freely diffusing molecules with liposome tethering. Nat. Commun. 2015, 6, 6992. [Google Scholar] [CrossRef]
- Leslie, S.R.; Fields, A.P.; Cohen, A.E. Convex Lens-Induced Confinement for Imaging Single Molecules. Anal. Chem. 2010, 82, 6224–6229. [Google Scholar] [CrossRef]
- Elting, M.W.; Leslie, S.R.; Churchman, L.S.; Korlach, J.; McFaul, C.M.J.; Leith, J.S.; Levene, M.J.; Cohen, A.E.; Spudich, J.A. Single-molecule fluorescence imaging of processive myosin with enhanced background suppression using linear zero-mode waveguides (ZMWs) and convex lens induced confinement (CLIC). Opt. Express 2013, 21, 1189–1202. [Google Scholar] [CrossRef][Green Version]
- Wang, Q.; Moerner, W.E. Optimal strategy for trapping single fluorescent molecules in solution using the ABEL trap. Appl. Phys. B 2010, 99, 23–30. [Google Scholar] [CrossRef]
- Schlau-Cohen, G.S.; Wang, Q.; Southall, J.; Cogdell, R.J.; Moerner, W.E. Single-molecule spectroscopy reveals photosynthetic LH2 complexes switch between emissive states. Proc. Natl. Acad. Sci. USA 2013, 110, 10899–10903. [Google Scholar] [CrossRef]
- Schlau-Cohen, G.S.; Yang, H.-Y.; Krüger, T.P.J.; Xu, P.; Gwizdala, M.; van Grondelle, R.; Croce, R.; Moerner, W.E. Single-Molecule Identification of Quenched and Unquenched States of LHCII. J. Phys. Chem. Lett. 2015, 6, 860–867. [Google Scholar] [CrossRef]
- Cohen, A.E.; Moerner, W.E. Controlling Brownian motion of single protein molecules and single fluorophores in aqueous buffer. Opt. Express 2008, 16, 6941–6956. [Google Scholar] [CrossRef]
- Goldsmith, R.H.; Moerner, W.E. Watching conformational- and photodynamics of single fluorescent proteins in solution. Nat. Chem. 2010, 2, 179–186. [Google Scholar] [CrossRef]
- Shen, H.; Tauzin, L.J.; Baiyasi, R.; Wang, W.X.; Moringo, N.; Shuang, B.; Landes, C.F. Single Particle Tracking: From Theory to Biophysical Applications. Chem. Rev. 2017, 117, 7331–7376. [Google Scholar] [CrossRef]
- Von Diezmann, A.; Shechtman, Y.; Moerner, W.E. Three-Dimensional Localization of Single Molecules for Super Resolution Imaging and Single-Particle Tracking. Chem. Rev. 2017, 117, 7244–7275. [Google Scholar] [CrossRef]
- Dupont, A.; Gorelashvili, M.; Schuller, V.; Wehnekamp, F.; Arcizet, D.; Katayama, Y.; Lamb, D.C.; Heinrich, D. Three-dimensional single-particle tracking in live cells: News from the third dimension. NJPh 2013, 15. [Google Scholar] [CrossRef]
- Toprak, E.; Balci, H.; Blehm, B.H.; Selvin, P.R. Three-dimensional particle tracking via bifocal imaging. Nano Lett. 2007, 7, 2043–2045. [Google Scholar] [CrossRef]
- Kao, H.P.; Verkman, A.S. Tracking of Single Fluorescent Particles in 3 Dimensions - Use of Cylindrical Optics to Encode Particle Position. Biophys. J. 1994, 67, 1291–1300. [Google Scholar] [CrossRef]
- Holtzer, L.; Meckel, T.; Schmidt, T. Nanometric three-dimensional tracking of individual quantum dots in cells. Appl. Phys. Lett. 2007, 90, Artn 053902. [Google Scholar] [CrossRef]
- Pavani, S.R.P.; Thompson, M.A.; Biteen, J.S.; Lord, S.J.; Liu, N.; Twieg, R.J.; Piestun, R.; Moerner, W.E. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc. Natl. Acad. Sci. USA 2009, 106, 2995–2999. [Google Scholar] [CrossRef]
- Lew, M.D.; Lee, S.F.; Badieirostami, M.; Moerner, W.E. Corkscrew point spread function for far-field three-dimensional nanoscale localization of pointlike objects. Opt. Lett. 2011, 36, 202–204. [Google Scholar] [CrossRef]
- Shechtman, Y.; Weiss, L.E.; Backer, A.S.; Sahl, S.J.; Moemer, W.E. Precise Three-Dimensional Scan-Free Multiple-Particle Tracking over Large Axial Ranges with Tetrapod Point Spread Functions. Nano Lett. 2015, 15, 4194–4199. [Google Scholar] [CrossRef]
- Berg, H.C. How to Track Bacteria. Rev. Sci. Instrum. 1971, 42, 868–871. [Google Scholar] [CrossRef]
- Lessard, G.A.; Goodwin, P.M.; Werner, J.H. Three-dimensional tracking of individual quantum dots. Appl. Phys. Lett. 2007, 91, Artn 224106. [Google Scholar] [CrossRef]
- Lessard, G.A.; Goodwin, P.M.; Werner, J.H. Three-dimensional tracking of fluorescent particles. Proc. SPIE 2006, 6092, Artn 609205. [Google Scholar] [CrossRef]
- Wells, N.P.; Lessard, G.A.; Werner, J.H. Confocal, Three-Dimensional Tracking of Individual Quantum Dots in High-Background Environments. Anal. Chem. 2008, 80, 9830–9834. [Google Scholar] [CrossRef]
- Wells, N.P.; Lessard, G.A.; Goodwin, P.M.; Phipps, M.E.; Cutler, P.J.; Lidke, D.S.; Wilson, B.S.; Werner, J.H. Time-Resolved Three-Dimensional Molecular Tracking in Live Cells. Nano Lett. 2010, 10, 4732–4737. [Google Scholar] [CrossRef]
- Keller, A.M.; DeVore, M.S.; Stich, D.G.; Vu, D.M.; Causgrove, T.; Werner, J.H. Multicolor Three-Dimensional Tracking for Single-Molecule Fluorescence Resonance Energy Transfer Measurements. Anal. Chem. 2018, 90, 6109–6115. [Google Scholar] [CrossRef]
- Cang, H.; Wong, C.M.; Xu, C.S.; Rizvi, A.H.; Yang, H. Confocal three dimensional tracking of a single nanoparticle with concurrent spectroscopic readouts. Appl. Phys. Lett. 2006, 88, Artn 223901. [Google Scholar] [CrossRef]
- Cang, H.; Xu, C.S.; Montiel, D.; Yang, H. Guiding a confocal microscope by single fluorescent nanoparticles. Opt Lett. 2007, 32, 2729–2731. [Google Scholar] [CrossRef]
- Cang, H.; Montiel, D.; Xu, C.S.; Yang, H. Observation of spectral anisotropy of gold nanoparticles. J. Chem. Phys. 2008, 129, Artn 044503. [Google Scholar] [CrossRef]
- Qian, B.; Montiel, D.; Bregulla, A.; Cichos, F.; Yang, H. Harnessing thermal fluctuations for purposeful activities: The manipulation of single micro-swimmers by adaptive photon nudging. Chem. Sci. 2013, 4, 1420–1429. [Google Scholar] [CrossRef]
- Welsher, K.; Yang, H. Multi-resolution 3D visualization of the early stages of cellular uptake of peptide-coated nanoparticles. Nat. Nanotechnol. 2014, 9, 198–203. [Google Scholar] [CrossRef]
- Enderlein, J. Tracking of fluorescent molecules diffusing within membranes. Appl. Phys. B-Lasers O 2000, 71, 773–777. [Google Scholar] [CrossRef]
- Levi, V.; Ruan, Q.; Kis-Petikova, K.; Gratton, E. Scanning FCS, a novel method for three-dimensional particle tracking. Biochem. Soc. Trans. 2003, 31, 997–1000. [Google Scholar] [CrossRef]
- Petersen, N.O.; Johnson, D.C.; Schlesinger, M.J. Scanning Fluorescence Correlation Spectroscopy .2. Application to Virus Glycoprotein Aggregation. Biophys. J. 1986, 49, 817–820. [Google Scholar] [CrossRef]
- Kis-Petikova, K.; Gratton, E. Distance measurement by circular scanning of the excitation beam in the two-photon microscope. Microsc. Res. Tech. 2004, 63, 34–49. [Google Scholar] [CrossRef]
- Levi, V.; Ruan, Q.Q.; Gratton, E. 3-D particle tracking in a two-photon microscope: Application to the study of molecular dynamics in cells. Biophys. J. 2005, 88, 2919–2928. [Google Scholar] [CrossRef]
- Lanzano, L.; Gratton, E. Orbital single particle tracking on a commercial confocal microscope using piezoelectric stage feedback. Methods Appl. Fluores 2014, 2, Artn 024010. [Google Scholar] [CrossRef]
- Annibale, P.; Dvornikov, A.; Gratton, E. Electrically tunable lens speeds up 3D orbital tracking. Biomed. Optics Express 2015, 6, 2181–2190. [Google Scholar] [CrossRef]
- Katayama, Y.; Burkacky, O.; Meyer, M.; Brauchle, C.; Gratton, E.; Lamb, D.C. Real-Time Nanomicroscopy via Three-Dimensional Single-Particle Tracking. Chemphyschem 2009, 10, 2458–2464. [Google Scholar] [CrossRef]
- Reuel, N.F.; Dupont, A.; Thouvenin, O.; Lamb, D.C.; Strano, M.S. Three-Dimensional Tracking of Carbon Nanotubes within Living Cells. ACS Nano 2012, 6, 5420–5428. [Google Scholar] [CrossRef]
- Welsher, K.; Liu, Z.; Sherlock, S.P.; Robinson, J.T.; Chen, Z.; Daranciang, D.; Dai, H. A route to brightly fluorescent carbon nanotubes for near-infrared imaging in mice. Nat. Nano 2009, 4, 773–780. [Google Scholar] [CrossRef]
- Welsher, K.; Sherlock, S.P.; Dai, H.J. Deep-tissue anatomical imaging of mice using carbon nanotube fluorophores in the second near-infrared window. Proc. Natl. Acad. Sci. USA 2011, 108, 8943–8948. [Google Scholar] [CrossRef]
- McHale, K.; Berglund, A.J.; Mabuchi, H. Quantum dot photon statistics measured by three-dimensional particle tracking. Nano Lett. 2007, 7, 3535–3539. [Google Scholar] [CrossRef]
- Du, K.; Liddle, J.A.; Berglund, A.J. Three-Dimensional Real-Time Tracking of Nanoparticles at an Oil-Water Interface. Langmuir 2012, 28, 9181–9188. [Google Scholar] [CrossRef]
- Du, K.; Ko, S.H.; Gallatin, G.M.; Yoon, H.P.; Liddle, J.A.; Berglund, A.J. Quantum dot-DNA origami binding: A single particle, 3D, real-time tracking study. Chem. Commun. 2013, 49, 907–909. [Google Scholar] [CrossRef]
- Germann, J.A.; Davis, L.M. Three-dimensional tracking of a single fluorescent nanoparticle using four-focus excitation in a confocal microscope. Opt. Express 2014, 22, 5641–5650. [Google Scholar] [CrossRef]
- Perillo, E.P.; Liu, Y.-L.; Huynh, K.; Liu, C.; Chou, C.-K.; Hung, M.-C.; Yeh, H.-C.; Dunn, A.K. Deep and high-resolution three-dimensional tracking of single particles using nonlinear and multiplexed illumination. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef]
- Hou, S.G.; Lang, X.Q.; Welsher, K. Robust real-time 3D single-particle tracking using a dynamically moving laser spot. Opt. Lett. 2017, 42, 2390–2393. [Google Scholar] [CrossRef]
- Hou, S.; Welsher, K. A Protocol for Real-time 3D Single Particle Tracking. JoVE 2018. [Google Scholar] [CrossRef]
- Fields, A.P.; Cohen, A.E. Optimal tracking of a Brownian particle. Opt. Express 2012, 20, 22585–22601. [Google Scholar] [CrossRef]
- Duocastella, M.; Sun, B.; Arnold, C.B. Simultaneous imaging of multiple focal planes for three-dimensional microscopy using ultra-high-speed adaptive optics. BIOMEDO 2012, 17, 050505. [Google Scholar] [CrossRef]
- Duocastella, M.; Theriault, C.; Arnold, C.B. Three-dimensional particle tracking via tunable color-encoded multiplexing. Opt. Lett. 2016, 41, 863–866. [Google Scholar] [CrossRef]
- Mermillod-Blondin, A.; McLeod, E.; Arnold, C.B. High-speed varifocal imaging with a tunable acoustic gradient index of refraction lens. Opt. Lett. 2008, 33, 2146–2148. [Google Scholar] [CrossRef]
- Karagyozov, D.; Skanata, M.M.; Lesar, A.; Gershow, M. Recording Neural Activity in Unrestrained Animals with Three-Dimensional Tracking Two-Photon Microscopy. Cell Rep. 2018, 25, 1371–1383. [Google Scholar] [CrossRef]
- Juette, M.F.; Bewersdorf, J. Three-Dimensional Tracking of Single Fluorescent Particles with Submillisecond Temporal Resolution. Nano Lett. 2010, 10, 4657–4663. [Google Scholar] [CrossRef]
- Spille, J.H.; Kaminski, T.P.; Scherer, K.; Rinne, J.S.; Heckel, A.; Kubitscheck, U. Direct observation of mobility state transitions in RNA trajectories by sensitive single molecule feedback tracking. Nucleic Acids Res. 2015, 43, ARTN e14. [Google Scholar] [CrossRef]
- DeVore, M.S.; Stich, D.G.; Keller, A.M.; Cleyrat, C.; Phipps, M.E.; Hollingsworth, J.A.; Lidke, D.S.; Wilson, B.S.; Goodwin, P.M.; Werner, J.H. Note: Time-gated 3D single quantum dot tracking with simultaneous spinning disk imaging. Rev. Sci. Instrum. 2015, 86, 126102. [Google Scholar] [CrossRef]
- McHale, K.; Mabuchi, H. Precise Characterization of the Conformation Fluctuations of Freely Diffusing DNA: Beyond Rouse and Zimm. J. Am. Chem. Soc. 2009, 131, 17901–17907. [Google Scholar] [CrossRef]
- Han, J.J.; Kiss, C.; Bradbury, A.R.M.; Werner, J.H. Time-Resolved, Confocal Single-Molecule Tracking of Individual Organic Dyes and Fluorescent Proteins in Three Dimensions. ACS Nano 2012, 6, 8922–8932. [Google Scholar] [CrossRef]
- Liu, C.; Obliosca, J.M.; Liu, Y.L.; Chen, Y.A.; Jiang, N.; Yeh, H.C. 3D single-molecule tracking enables direct hybridization kinetics measurement in solution. Nanoscale 2017, 9, 5664–5670. [Google Scholar] [CrossRef]
- Shain, W.J.; Vickers, N.A.; Goldberg, B.B.; Bifano, T.; Mertz, J. Extended depth-of-field microscopy with a high-speed deformable mirror. Opt. Lett. 2017, 42, 995–998. [Google Scholar] [CrossRef]

| Name | (a) Localization Precision X/Y/Z (nm) | (b) Precision Measurement Sample | (c) Temporal Resolution | (d) Fastest Tracked D (μm2/s) | (e) Count Rate (Fastest D, kHz) | (f) Tracking Duration (Fastest D) | (g) References |
|---|---|---|---|---|---|---|---|
| Tetrahedral detection | 70/70/110 | Sample: QD | Int.: 5 ms Piezo: ~1 ms | 3 | ~16 | 0.3 s | [69,70] |
| Rate: 40 kHz | |||||||
| D: 0.37 µm2/s | |||||||
| Multi-detector | 9.8/12.8/13.1 | Sample: FNP | Int.: 10 μs Piezo: ~1 ms | 4.81 | - | >8 s | [76] |
| Rate: - | |||||||
| D: Stationary | |||||||
| Orbital (piezo stage) | 2.4/2.8/2.4 | Sample: FNP | Piezo: 32 ms ETL: 8 ms | 0.04 | - | - | [81,82,83] |
| Rate: 28 kHz | |||||||
| D: Stationary | |||||||
| Orbital (laser modulation) | 352/352/272 | Sample: QD | Piezo: 1 ms | 20 | ~120 | 20 s | [88] |
| Rate: ~120 kHz | |||||||
| D: 20 µm2/s | |||||||
| Orbital (bi-plane detection) | 15/15/20 | Sample: FNP | Galvo: 32 ms | 2 | 50 | ~25 s | [84,90] |
| Rate: 10–40 kHz | |||||||
| D: Stationary | |||||||
| Tetrahedral excitation | 60/60/300 | Sample: FNP | Int. 1.86 ms | 12.2 | 473 | 0.078 s | [91] |
| Rate: ~5 kHz | |||||||
| D: Stationary | |||||||
| TSUNAMI | 16.2/16.7/35.1 | Sample: FNP | Piezo: ~1 ms Int.: 1 ms Offline: 50 μs | 7.5 | - | - | [92] |
| Rate: ~100 kHz | |||||||
| Sample speed: 6 µm2/s | |||||||
| 3D-DyPLoT | 6.6/8.7/15.6 | Sample: FNP | Int.: 20 μs Piezo: ~1 ms | 20 | 20 | 30 s for particle with 8.9 μm2/s dynamics | [93] |
| Rate: ~80 kHz | |||||||
| D: Stationary | |||||||
| Image-based (bi-plane) | 7/8/27 | Sample: FNP | Int.: 0.3 ms Piezo: 2 ms | 2.4 | 3-4 | 3 s | [100] |
| Rate: | |||||||
| D: Stationary | |||||||
| Image-based (light-sheet) | 10/10/40 | Sample: FNP | Int.: 1.1 ms | 9.4 | - | - | [101] |
| Rate: 750 photons | |||||||
| D: Stationary |
| Method | Tracking Temporal Resolution | Imaging Time | Simultaneous Imaging | Optical Sectioning | Reference |
|---|---|---|---|---|---|
| TSUNAMI | 50 µs | 0.7 s | NO | YES | [92] |
| Orbital | 32 ms | ~200 ms | YES | NO | [84] |
| Tetrahedral Detection | 5 ms | 40 ms | YES | YES | [102] |
| 3D-MM | 10 µs | 1.0 s | YES | YES | [76] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, S.; Johnson, C.; Welsher, K. Real-Time 3D Single Particle Tracking: Towards Active Feedback Single Molecule Spectroscopy in Live Cells. Molecules 2019, 24, 2826. https://doi.org/10.3390/molecules24152826
Hou S, Johnson C, Welsher K. Real-Time 3D Single Particle Tracking: Towards Active Feedback Single Molecule Spectroscopy in Live Cells. Molecules. 2019; 24(15):2826. https://doi.org/10.3390/molecules24152826
Chicago/Turabian StyleHou, Shangguo, Courtney Johnson, and Kevin Welsher. 2019. "Real-Time 3D Single Particle Tracking: Towards Active Feedback Single Molecule Spectroscopy in Live Cells" Molecules 24, no. 15: 2826. https://doi.org/10.3390/molecules24152826
APA StyleHou, S., Johnson, C., & Welsher, K. (2019). Real-Time 3D Single Particle Tracking: Towards Active Feedback Single Molecule Spectroscopy in Live Cells. Molecules, 24(15), 2826. https://doi.org/10.3390/molecules24152826





