Influence of Bacterial Physiology on Processing of Selenite, Biogenesis of Nanomaterials and Their Thermodynamic Stability
Abstract
1. Introduction
2. Results
2.1. SeO32− Bioprocessing by MPV1 Cultures
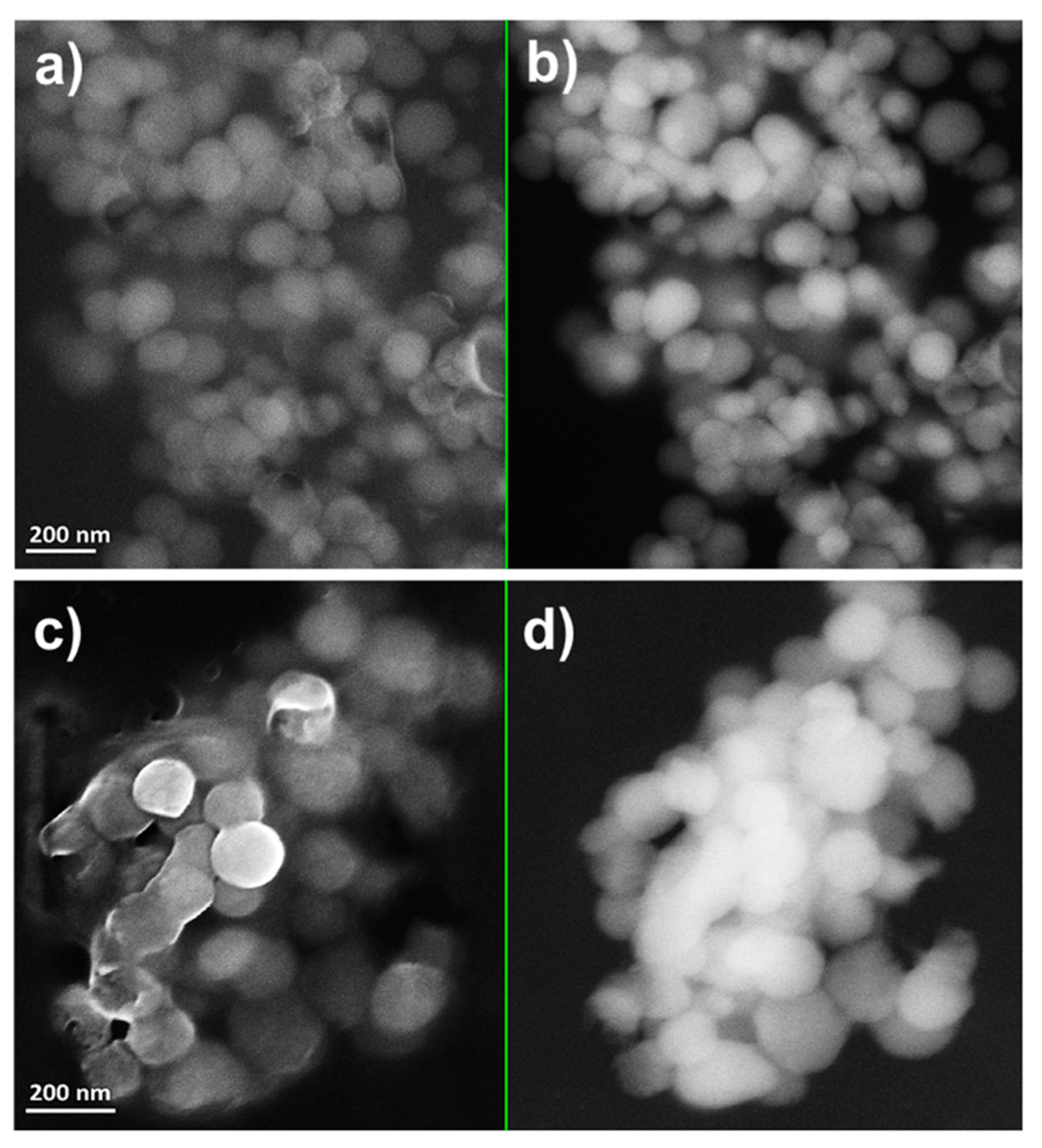
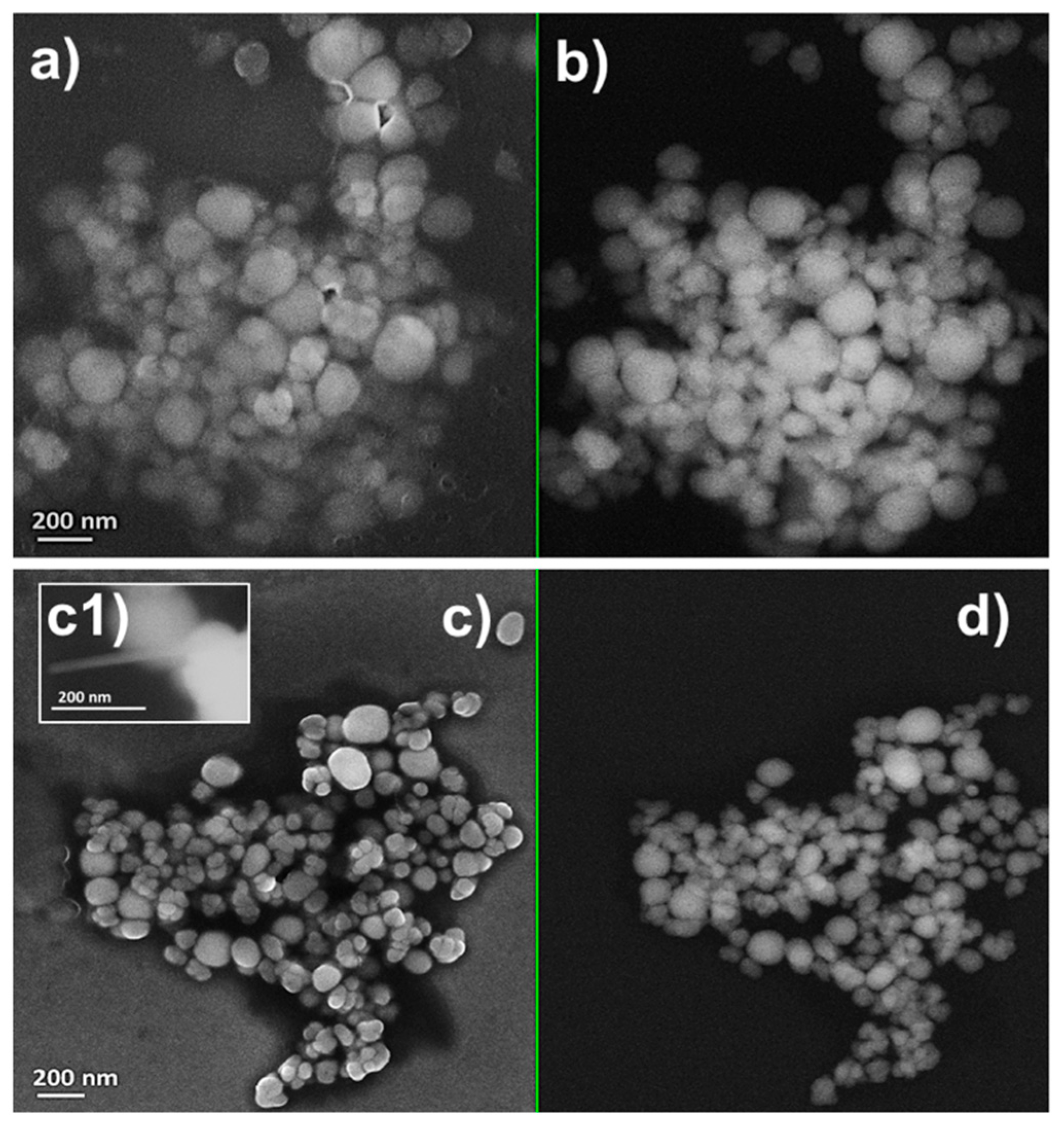
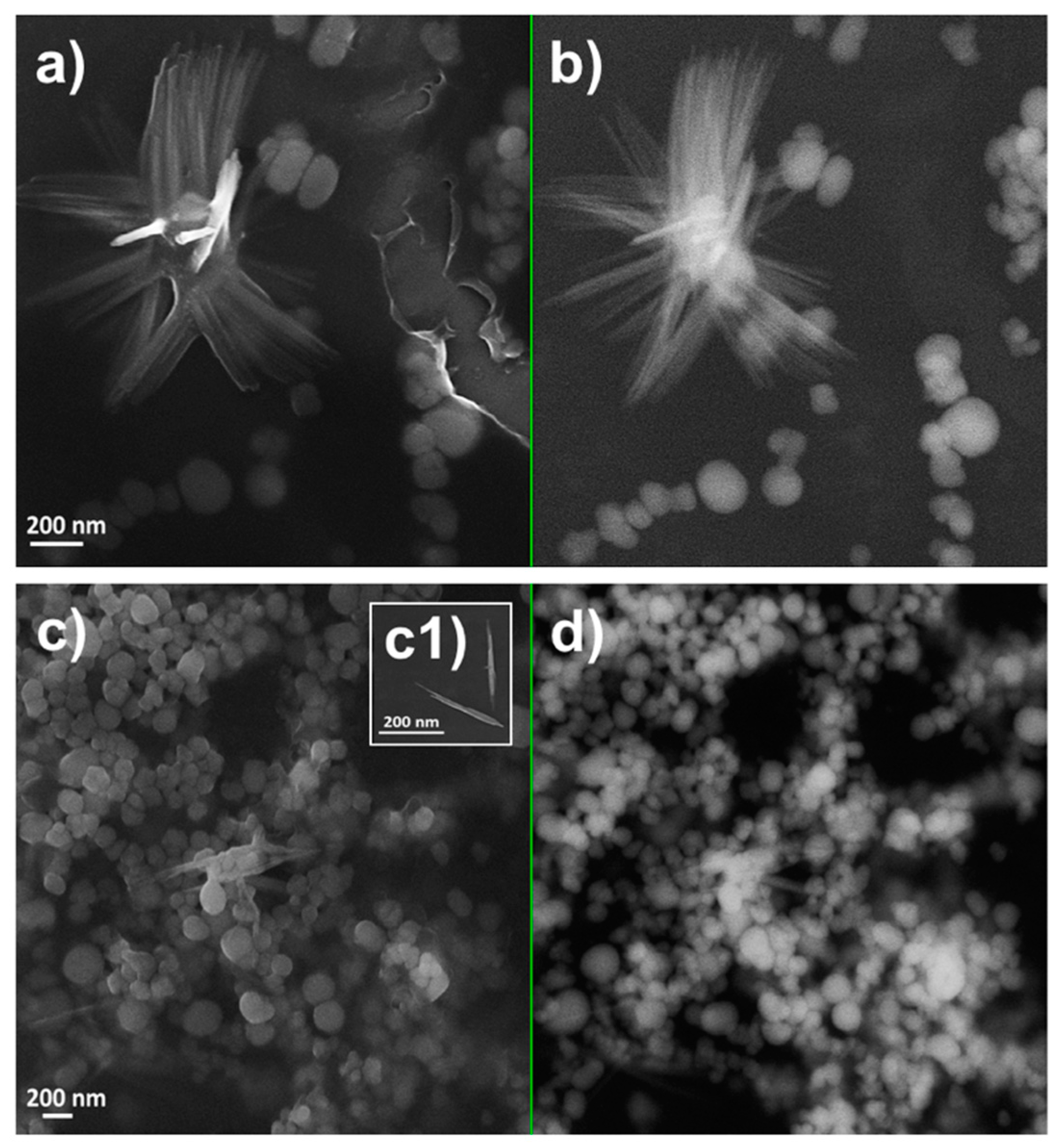
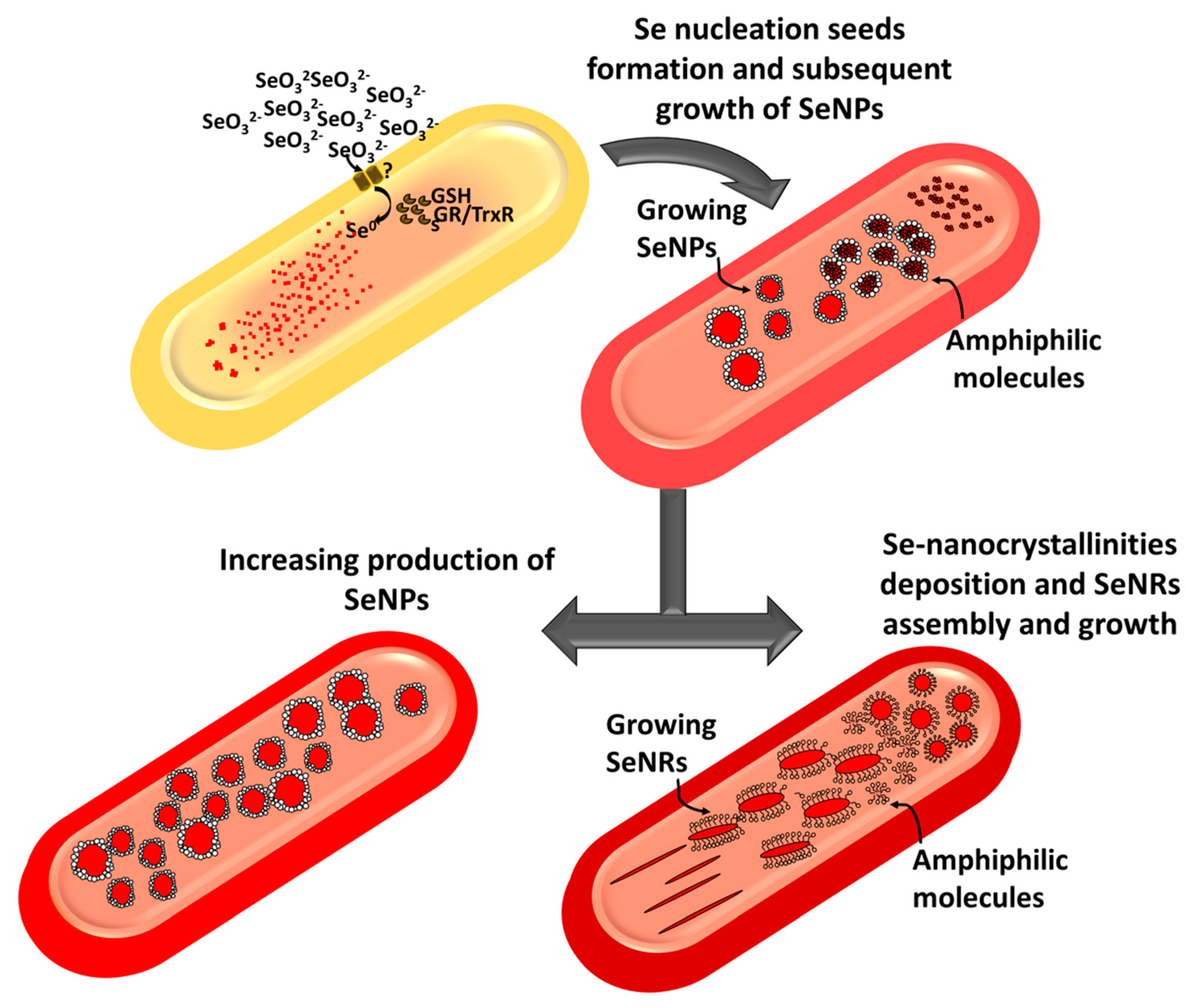
2.2. Characterization of Se Nanostructures Produced by MPV1 Cells
2.3. Tuning of Se Nanostructure Morphology by Varying MPV1 Physiological State
2.4. Physical-Chemical Characterization of the Biogenic Se Nanostructure Extracts
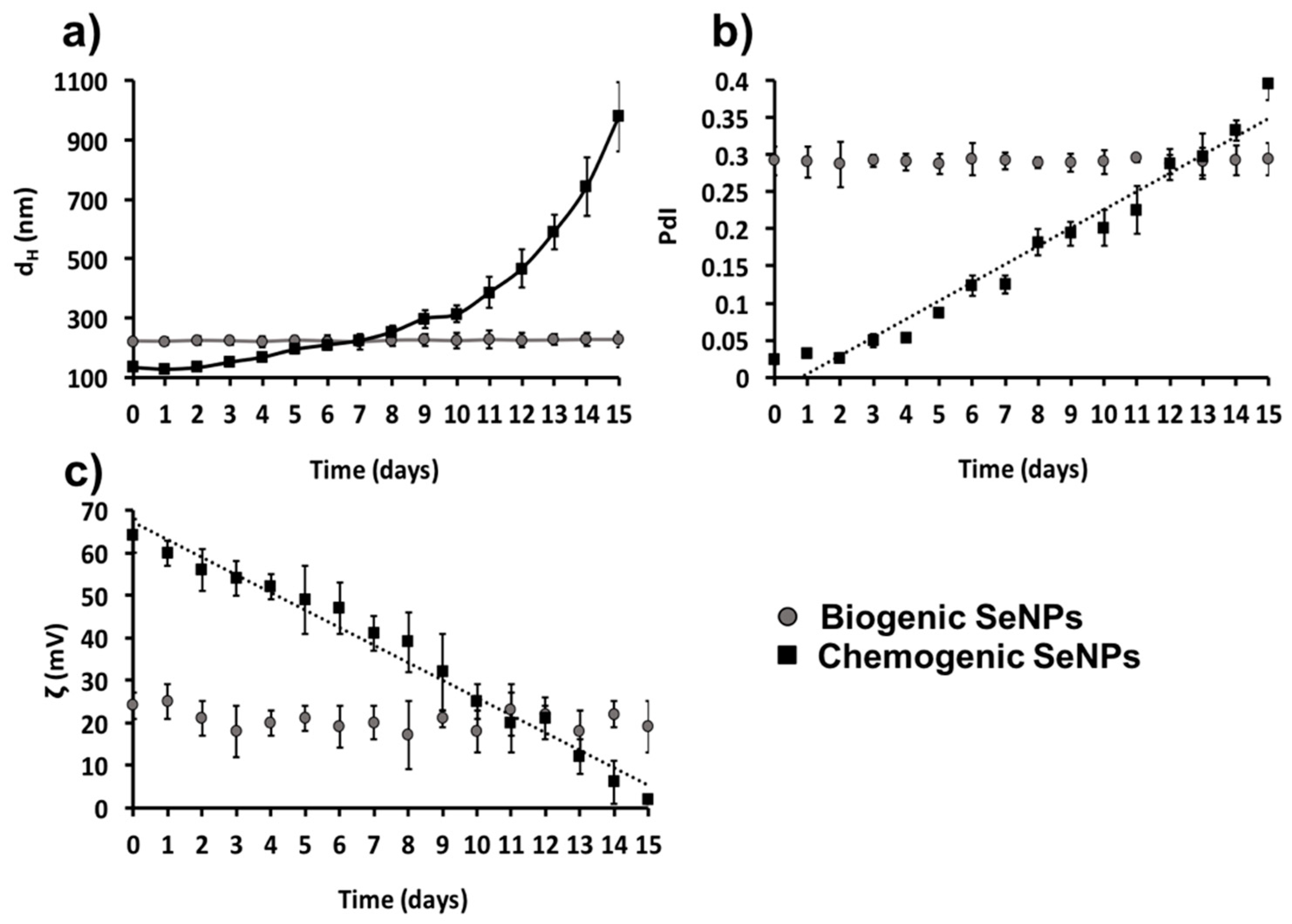
2.5. Role of Organic Material in Thermodynamic Stabilization of Biogenic Se Nanostructures
3. Discussion
4. Materials and Methods
4.1. Bacterial Culture Conditions
4.2. Biotic SeO32- Removal Efficiency
4.3. Measurement of Thiol Oxidation as Consequence of SeO32− Bioprocessing
4.4. Preparation and Recovery of Biogenic Se Nanomaterial Extracts and Their Supernatants
4.5. Physical-Chemical Characterization of Biogenic Se Nanomaterial Extracts
4.6. Monitoring Thermodynamic Stability of Biogenic Se Nanomaterial Extracts and Chemogenic Se Nanoparticles
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lenz, M.; Lens, P.N.L. The essential toxin: The changing perception of selenium in environmental sciences. Sci. Total. Environ. 2009, 407, 3620–3633. [Google Scholar] [CrossRef] [PubMed]
- Barceloux, D.G. Selenium. J. Toxicol. Clin. Toxicol. 1999, 37, 145–172. [Google Scholar] [CrossRef] [PubMed]
- Zannoni, D.; Borsetti, F.; Harrison, J.J.; Turner, R.J. The bacterial response to the chalcogen metalloids Se and Te. Adv. Microb. Physiol. 2008, 53, 1–71. [Google Scholar] [PubMed]
- Gerrard, T.; Telford, J.; Williams, H. Detection of selenium deposits in Escherichia coli by electron microscopy. J. Bacteriol. 1974, 119, 1057–1060. [Google Scholar] [PubMed]
- Horikoshi, S.; Serpone, N. Chapter 1, general introduction to nanoparticles. In Microwaves in Nanoparticle Synthesis: Fundamentals and Applications; Horikoshi, S., Serpone, N., Eds.; Wiley-VCH Verlag GmbH & Co: Weinheim, Germany, 2013; pp. 1–24. [Google Scholar]
- Wadhwani, S.A.; Shedbalkar, U.U.; Singh, R.; Chopade, B.A. Biogenic selenium nanoparticles: current status and future prospects. Appl. Microbiol. Biotechnol. 2016, 100, 2555–2566. [Google Scholar] [CrossRef] [PubMed]
- Painter, E.P. The chemistry and toxicity of selenium compounds with special reference to the selenium problem. Chem. Rev. 1941, 28, 179–213. [Google Scholar] [CrossRef]
- Ganther, H.E. Reduction of the selenotrisulfide derivative of glutathione to a persulfide analog by a gluthatione reductase. Biochemistry. 1971, 10, 4089–4098. [Google Scholar] [CrossRef]
- Kessi, J.; Hanselmann, K.W. Similarities between the abiotic reduction of selenite with glutathione and the dissimilatory reaction mediated by Rhodospirillum rubrum and Escherichia coli. J. Biol. Chem. 2004, 279, 50662–50669. [Google Scholar] [CrossRef]
- Harrison, G.; Curie, C.; Laishley, E.J. Purification and characterization of an inducible dissimilatory type sulphite reductase from Clostridium pasteurianium. Arch. Microbiol. 1984, 138, 72–78. [Google Scholar] [CrossRef]
- DeMoll-Decker, H.; Macy, J.M. The periplasmic nitrite reductase of Thauera selenatis may catalyze the reduction of selenite to elemental selenium. Arch. Microbiol. 1993, 160, 241–247. [Google Scholar]
- Avazeri, C.; Turner, R.J.; Pommier, J.; Weiner, J.H.; Giordano, G.; Vermeglio, A. Tellurite and selenate reductase activity of nitrate reductases from Escherichia coli: correlation with tellurite resistance. Microbiology 1997, 143, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Schroder, I.; Rech, S.; Krafft, T.; May, J.M. Purification and characterization of the selenate reductase from Thauera selenatis. J. Biol. Chem. 1997, 272, 23765–23768. [Google Scholar] [CrossRef] [PubMed]
- Kessi, J. Enzymatic systems proposed to be involved in the dissimilatory reduction of selenite in the purple non-sulfur bacteria Rhodospirillum rubrum and Rhodobacter capsulatus. Microbiology 2006, 152, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Basaglia, M.; Toffanin, A.; Baldan, E.; Bottegal, M.; Shapleigh, J.P.; Casella, S. Selenate-reducing capacity of the copper-containing nitrite reductase of Rhizobium sullae. FEMS Microbiol. Lett. 2007, 269, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Li, D.B.; Cheng, Y.Y.; Wu, C.; Li, W.W.; Li, N.; Yang, Z.C.; Tong, Z.H.; Yu, H.Q. Selenite reduction by Shewanella oneidensis MR-1 is mediated by fumarate reductase in periplasm. Sci. Rep. 2014, 4, 3735. [Google Scholar] [CrossRef]
- Hockin, S.L.; Gadd, L. Linked redox precipitation of sulfur and selenium under anaerobic conditions by sulfate-reducing bacterial biofilms. Appl. Environ. Microbiol. 2003, 69, 7063–7072. [Google Scholar] [CrossRef] [PubMed]
- Zawadzka, A.M.; Crawford, R.L.; Paszczynki, A.J. Pyridine-2,6-bis(thiocarboxylic acid) produced by Pseudomonas stutzeri KC reduces and precipitates selenium and tellurium oxyanions. Appl. Environ. Microbiol. 2006, 72, 3119–3129. [Google Scholar] [CrossRef]
- Tam, K.; Ho, C.T.; Lee, J.H.; Chang, C.H.; Rheem, Y.; Chen, W.; Hur, H.G.; Myung, N.V. Growth mechanism of amorphous selenium nanoparticles synthesized by Shewanella sp. HN-41. Biosci. Biotechnol. Biochem. 2010, 74, 696–700. [Google Scholar] [CrossRef]
- Che, L.; Dong, Y.; Wu, M.; Zhao, Y.; Liu, L.; Zhou, H. Characterization of Selenite Reduction by Lysinibacillus sp. ZYM-1 and Photocatalytic Performance of Biogenic Selenium Nanospheres. ACS Sustainable Chem. Eng. 2017, 5, 2535–2543. [Google Scholar] [CrossRef]
- Ruiz Fresneda, M.A.; Delgado Martin, J.; Gomez Bolivar, J.; Fernandez Cantos, M.V.; Bosch-Estevez, G.; Martinez Moreno, M.F.; Merroun, M.L. Green synthesis and biotransformation of amorphous Se nanospheres to trigonal 1D Se nanostructures: impact on Se mobility within the concept of radioactive waste disposal. Environ. Sci. Nano. 2018, 5, 2103–2116. [Google Scholar] [CrossRef]
- Pal, S.; Tak, Y.K.; Song, J.M. Does the antibacterial activity of silver nanoparticles depend on the shape of the nanoparticle? A study of the gram-negative bacterium Escherichia coli. Appl. Environ. Microbiol. 2007, 73, 1712–1720. [Google Scholar] [CrossRef] [PubMed]
- George, S.; Lin, S.; Ji, Z.; Thomas, C.R.; Li, L.; Mecklenburg, M.; Meng, H.; Wang, X.; Zhang, H.; Xia, T.; et al. Surface defects on plate-shaped silver nanoparticles contribute to its hazard potential in a fish gill cell line and zebrafish embryos. ACS Nano. 2012, 6, 3745–3759. [Google Scholar] [CrossRef] [PubMed]
- Rajput, S.; Werezuk, R.; Lange, R.M.; McDermott, M.T. Fungal Isolate Optimized for Biogenesis of Silver Nanoparticles with Enhanced Colloidal Stability. Langmuir 2016, 32, 8688–8697. [Google Scholar] [CrossRef] [PubMed]
- Piacenza, E.; Presentato, A.; Turner, R.J. Stability of biogenic metal(loid) nanomaterials related to the colloidal stabilization theory of chemical nanostructures. Crit. Rev. Biotechnol. 2018, 38, 1137–1156. [Google Scholar] [CrossRef] [PubMed]
- Zonaro, E.; Piacenza, E.; Presentato, A.; Monti, F.; Dell’Anna, R.; Lampis, S.; Vallini, G. Ochrobactrum sp. MPV1 from a dump of roasted pyrites can be exploited as bacterial catalyst for the biogenesis of selenium and tellurium nanoparticles. Microb. Cell Fact. 2017, 16, 215. [Google Scholar] [CrossRef] [PubMed]
- Piacenza, E.; Presentato, A.; Ambrosi, A.; Speghini, A.; Turner, R.J.; Vallini, G.; Lampis, S. Physical-chemical properties of biogenic selenium nanostructures produced by Stenotrophomonas maltophilia SeITE02 and Ochrobactrum sp. MPV1. Front. Microbiol. 2018, 9, 3178. [Google Scholar] [CrossRef] [PubMed]
- Presentato, A.; Piacenza, E.; Anikovkiy, M.; Cappelletti, M.; Zannoni, D.; Turner, R.J. Biosynthesis of selenium-nanoparticles and -nanorods as a product of selenite bioconversion by the aerobic bacterium Rhodococcus aetherivorans BCP1. N. Biotechnol. 2018, 41, 1–8. [Google Scholar] [CrossRef]
- Nancharaiah, Y.V.; Lens, P.N.L. Selenium biomineralization for biotechnological applications. Trends Biotechnol. 2015, 33, 323–330. [Google Scholar] [CrossRef]
- Song, D.; Li, X.; Cheng, Y.; Xiao, X.; Lu, Z.; Wang, L.; Wang, F. Aerobic biogenesis of selenium nanoparticles by Enterobacter cloacae Z0206 as a consequence of fumarate reductase mediated selenite reduction. Sci. Rep. 2017, 7, 3239. [Google Scholar] [CrossRef]
- Piacenza, E.; Presentato, A.; Zonaro, E.; Lampis, S.; Vallini, G.; Turner, R.J. Chapter 5: Microbial-Based Bioremediation of Selenium and Tellurium Compounds. In Biosorption; Derco, J., Vrana, B., Eds.; IntechOpen: London, UK, 2018; pp. 117–147. [Google Scholar]
- Wang, Y.; Shu, X.; Hou, J.; Lu, W.; Zhao, W.; Huang, S.; Wu, L. Selenium Nanoparticle Synthesized by Proteus mirabilis YC801: An Efficacious Pathway for Selenite Biotransformation and Detoxification. Int. J. Mol. Sci. 2018, 19, 3809. [Google Scholar] [CrossRef]
- Wang, Y.; Shu, X.; Zhou, Q.; Fan, T.; Wang, T.; Chen, X.; Li, M.; Ma, Y.; Ni, J.; Hou, J.; et al. Selenite Reduction and the Biogenesis of Selenium Nanoparticles by Alcaligenes faecalis Se03 Isolated from the Gut of Monochamus alternatus (Coleoptera: Cerambycidae). Int. J. Mol. Sci. 2018, 19, 2799. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Ren, B.; Li, H.; Lin, Z.; Banuelos, G.; Li, L.; Zhao, G.; Guo, Y. Biosynthesis of selenium nanoparticles and effects of selenite, selenate, and selenomethionine on cell growth and morphology in Rahnella aquatilis HX2. Appl. Microbiol. Biotechnol. 2018, 102, 6196–6205. [Google Scholar] [CrossRef] [PubMed]
- Che, L.; Xu, W.; Zhan, J.; Zhang, L.; Liu, L.; Zhou, H. Complete Genome Sequence of Bacillus cereus CC-1, A Novel Marine Selenate/Selenite Reducing Bacterium Producing Metallic Selenides Nanomaterials. Curr. Microbiol. 2019, 76, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Biswas, K.C.; Barton, L.L.; Tsui, W.L.; Shuman, K.; Gillespie, J.; Eze, C.S. A novel method for the measurement of elemental selenium produced by bacterial reduction of selenite. J. Microbiol. Meth. 2011, 86, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Wu, S.; Li, N.; Wang, D.; Zheng, S.; Wang, G. Novel bacterial selenite reductase CsrF responsible for Se(IV) and Cr(VI) reduction that produces nanoparticles in Alishewanella sp. WH16-1. J. Haz. Mat. 2018, 342, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Hunter, W.J.; Manter, D.K. Reduction of Selenite to Elemental Red Selenium by Pseudomonas sp. Strain CA5. Curr. Microbiol. 2009, 58, 493–498. [Google Scholar] [CrossRef]
- Wadhwani, S.A.; Shedbalkar, U.U.; Singh, R.; Chopade, B.A. Biosynthesis of gold and selenium nanoparticles by purified protein from Acinetobacter sp. SW 30. Enzyme Microb. Technol. 2018, 111, 81–86. [Google Scholar] [CrossRef]
- Jahan, M.I.; Tobe, R.; Mihara, H. Characterization of a Novel Porin-Like Protein, ExtI, from Geobacter sulfurreducens and Its Implication in the Reduction of Selenite and Tellurite. Int. J. Mol. Sci. 2018, 19, 809. [Google Scholar] [CrossRef]
- Gates, B.; Mayers, B.; Cattle, B.; Xia, Y. Synthesis and characterization of uniform nanowires of trigonal selenium. Adv. Funct. Mater. 2002, 12, 219–227. [Google Scholar] [CrossRef]
- Thanh, N.T.K.; Maclean, N.; Mahiddine, S. Mechanisms of nucleation and growth of nanoparticles in solution. Chem. Rev. 2014, 114, 7610–7630. [Google Scholar] [CrossRef]
- Vreeland, E.C.; Watt, J.; Schober, G.B.; Hance, B.G.; Austin, M.J.; Price, A.D.; Fellows, B.D.; Monson, T.C.; Hudak, N.S.; Maldonado-Camargo, L.; et al. Enhanced Nanoparticle Size Control by Extending LaMer’s Mechanism. Chem. Mater. 2015, 27, 6059–6066. [Google Scholar] [CrossRef]
- Shoeibi, S.; Mozdziak, P.; Golkar-Narenji, A. Biogenesis of Selenium Nanoparticles Using Green Chemistry. Top. Curr. Chem. (Cham.). 2017, 375, 88. [Google Scholar] [CrossRef] [PubMed]
- Research on Nanomaterials. Available online: https://www.epa.gov/chemical-research/research-nanomaterials (accessed on 8 July 2019).
- Potocnik, J. Commission recommendation of 18 October 2011 on the definition of nanomaterial. Off. J. Eur. Communities: Legis. 2011, L275, 38–40. [Google Scholar] [CrossRef]
- Wang, T.; Yang, L.; Zhang, B.; Liu, J. Extracellular biosynthesis and transformation of selenium nanoparticles and application in H2O2 biosensor. Colloids Surf. B. Bionterfaces. 2010, 80, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Debieux, C.M.; Dridge, E.J.; Mueller, C.M.; Splatt, P.; Paszkiewicz, K.; Knight, I.; Florance, H.; Love, J.; Titball, R.W.; Lewis, R.J.; et al. A bacterial process for selenium nanosphere assembly. PNAS 2011, 108, 13480–13485. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chen, T.; Yang, F.; Liu, J.; Zheng, W. Facile and controllable one-step fabrication of selenium nanoparticles assisted by L-cysteine. Mater. Lett. 2010, 64, 614–617. [Google Scholar] [CrossRef]
- Lin, Z.H.; Wang, C.R. Evidence on the size-dependent absorption spectral evolution of selenium nanoparticles. Mat. Chem. Phys. 2005, 92, 591–594. [Google Scholar] [CrossRef]
- Ahmad, M.S.; Yasser, M.M.; Sholkamy, E.N.; Ali, A.M.; Mehanni, M.M. Anticancer activity of biostabilized selenium nanorods synthesized by Streptomyces bikiniensis strain Ess_amA-1. Int. J. Nanomed. 2015, 10, 3389–3401. [Google Scholar]
- Jain, R.; Jordan, N.; Weiss, S.; Foerstendorf, H.; Heim, K.; Kacker, R.; Hubner, R.; Kramer, H.; van Hullebusch, E.D.; Farges, F.; et al. Extracellular polymeric substances govern the surface charge of biogenic elemental selenium nanoparticles. Environ. Sci. Technol. 2015, 49, 1713–1720. [Google Scholar] [CrossRef]
- Estevam, E.C.; Griffin, S.; Nasim, M.J.; Denezhkin, P.; Schneider, R.; Lilischkis, R.; Dominguez-Alvarez, E.; Witek, K.; Latacz, G.; Keck, C.; et al. Natural selenium particles from Staphylococcus carnosus: hazards or particles with particular promise? J. Hazard. Mater. 2017, 324, 22–30. [Google Scholar] [CrossRef]
- Lampis, S.; Zonaro, E.; Bertolini, C.; Cecconi, D.; Monti, F.; Micaroni, M.; Turner, R.J.; Butler, C.S.; Vallini, G. Selenite biotransformation and detoxification by Stenotrophomonas maltophilia SeITE02: Novel clues on the route to bacterial biogenesis of selenium nanoparticles. J. Haz. Mat. 2017, 324, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Tugarova, A.V.; Kamnev, A.A. Proteins in microbial synthesis of selenium nanoparticles. Talanta 2017, 174, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Tugarova, A.; Mamchenkova, P.V.; Dyatlova, Y.A.; Kamnev, A.A. FTIR and Raman spectroscopic studies of selenium nanoparticles synthesized by the bacterium Azospirillum thiophilum. Spectrochim. Acta, A. Mol. Biomol. Spectrosc. 2018, 192, 458–463. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, D.; Qian, H.; Liang, Y.; Pan, X.; Gadd, G.M. Interactions between biogenic selenium nanoparticles and goethite colloids and consequence for remediation of elemental mercury contaminated groundwater. Sci. Tot. Environ. 2018, 613, 672–678. [Google Scholar] [CrossRef] [PubMed]
- Vogel, M.; Fischer, S.; Maffert, A.; Hübner, R.; Scheinost, A.C.; Franzen, C.; Steudtner, R. Biotransformation and detoxification of selenite by microbial biogenesis of selenium-sulfur nanoparticles. J. Hazard. Mater. 2018, 344, 749–757. [Google Scholar] [CrossRef] [PubMed]
- Israelachvili, J.N. Chapter 20: Soft and Biological Structures. In Intermolecular and Surface Forces, 3rd ed.; Israelachvili, J.N., Ed.; Academic Press: Oxford, MS, USA, 2011; pp. 535–576. [Google Scholar]
- Stetefeld, J.; McKenna, S.A.; Patel, T.R. Dynamic light scattering: a practical guide and applications in biomedical sciences. Biophys. Rev. 2016, 8, 409–427. [Google Scholar] [CrossRef] [PubMed]
- Presentato, A.; Piacenza, E.; Anikovkiy, M.; Cappelletti, M.; Zannoni, D.; Turner, R.J. Rhodococcus aetherivorans BCP1 as cell factory for the production of intracellular tellurium nanorods under aerobic conditions. Microb. Cell Fact. 2016, 15, 204. [Google Scholar] [CrossRef]
- Gates, B.; Yin, Y.; Xia, Y. A solution-phase approach to the synthesis of uniform nanowires of crystalline selenium with lateral dimensions in the range of 10−30 nm. J. Am. Chem. Soc. 2000, 122, 12582. [Google Scholar] [CrossRef]
- Jeong, U.; Camargo, P.H.C.; Lee, Y.H.; Xia, Y. Chemical transformation: a powerful route to metal chalcogenide nanowires. J. Mater. Chem. 2006, 16, 3893–3897. [Google Scholar] [CrossRef]
- Piacenza, E.; Presentato, A.; Zonaro, E.; Lampis, S.; Vallini, G.; Turner, R.J. Selenium and Tellurium Nanomaterials. Phys. Sci. Rev. 2018, 3, 20170100. [Google Scholar]
- Kumar, C.; Sujhitha, P.; Mamidyala, S.; Usharani, P.; Das, B.; Reddy, C. Ochrosin, a new biosurfactant produced by halophilic Ochrobactrum sp. strain BS-206 (MTCC 5720): purification, characterization and its biological evaluation. Process. Biochem. 2014, 49, 1708–1717. [Google Scholar] [CrossRef]
- Frassinetti, S.; Setti, L.; Corti, A.; Farrinelli, P.; Montevecchi, P.; Vallini, G. Biodegradation of dibenzothiophene by a nodulating isolate of Rhizobium meliloti. Can. J. Microbiol. 1998, 44, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Kessi, J.; Ramuz, M.; Wehrli, E.; Spycher, M.; Bachofen, R. Reduction of selenite and detoxification of elemental selenium by the phototrophic bacterium Rhodospirillum rubrum. Appl. Environ. Microbiol. 1999, 65, 4734–4740. [Google Scholar] [PubMed]
- Turner, R.J.; Weiner, J.H.; Taylor, D.E. Tellurite-mediate thiol oxidation in Escherichia coli. Microbiology 1999, 145, 2549–2557. [Google Scholar] [CrossRef] [PubMed]
- Markwell, M.A.K.; Haas, S.M.; Bieber, L.L.; Tolbert, N.E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 1978, 87, 206–210. [Google Scholar] [CrossRef]
Sample Availability: Not available. |
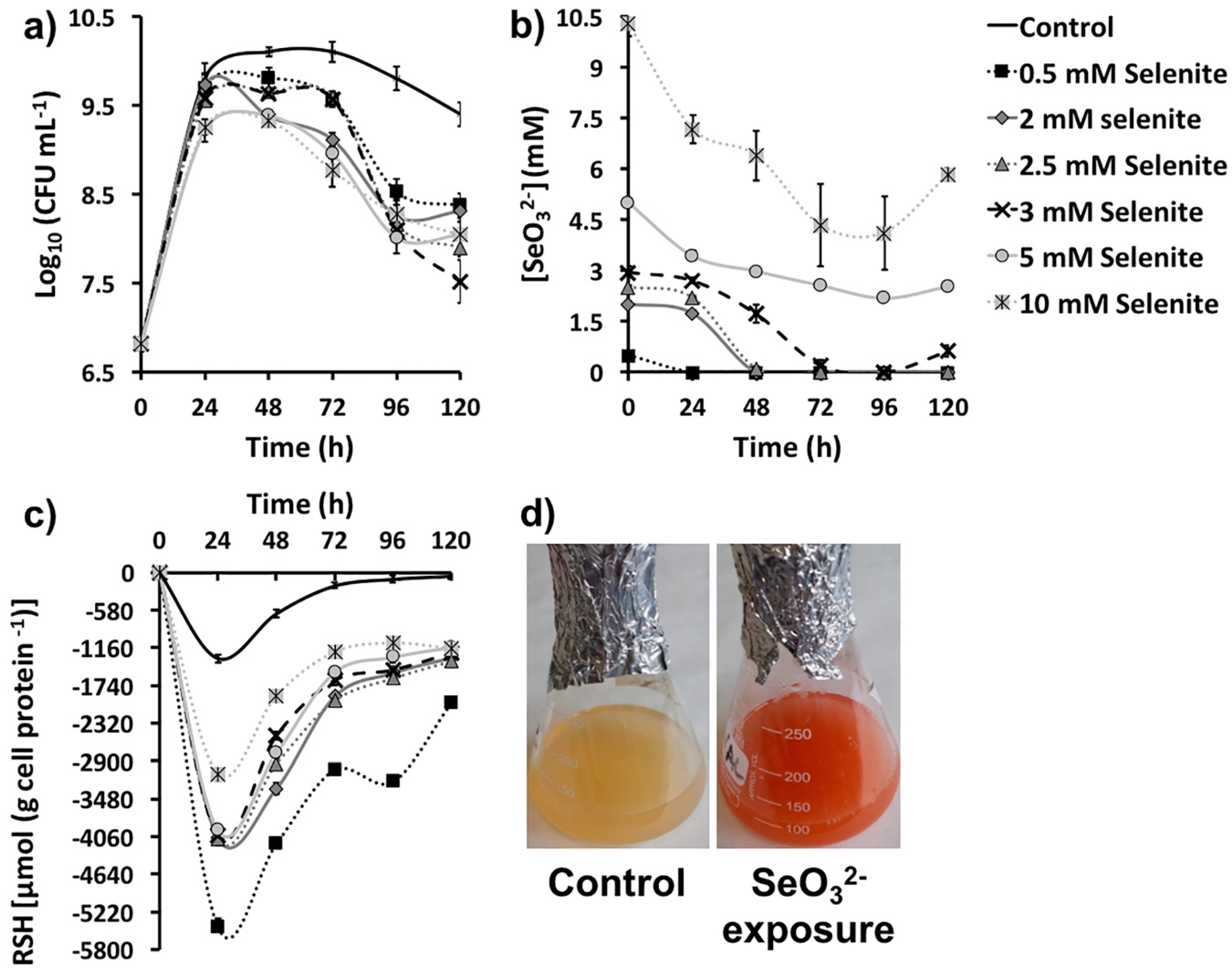

| SeO32- Removal (mM) as Function of Its Initial Concentration | ||||||
|---|---|---|---|---|---|---|
| Time (h) | 0.5 | 2 | 2.5 | 3 | 5 | 10 |
| 24 | 0.5 | 0.27 ± 0.09 | 0.31 ± 0.02 | 0.30 ± 0.08 | 1.56 ± 0.13 | 2.83 ± 0.12 |
| 48 | - | 2 | 2.39 ± 0.04 | 1.28 ± 0.11 | 2.02 ± 0.05 | 3.62 ± 0.09 |
| 72 | - | - | 2.5 | 2.79 ± 0.13 | 2.43 ± 0.04 | 5.67 ± 0.05 |
| 96 | - | - | - | 3 | 2.81 ± 0.03 | 5.89 ± 0.07 |
| 120 | - | - | - | 2.47 ± 0.10 | 2.46 ± 0.09 | 4.16 ± 0.04 |
| 144 | N.D. | N.D. | N.D. | N.D. | 2.51 ± 0.10 | 2.93 ± 0.09 |
| 168 | N.D. | N.D. | N.D. | N.D. | 2.54 ± 0.08 | 2.51 ± 0.11 |
| MPV1 Culture Conditions to Produce SeNSs | Acronym | Recovery Procedure |
|---|---|---|
| Growth for 24 h in the presence of 0.5 mM SeO32− | SeNPsMPV1-0.5_24_e | [27] |
| Growth for 120 h in the presence of 0.5 mM SeO32− | SeNPsMPV1-0.5_120_e | |
| Growth for 48 h in the presence of 2 mM SeO32− | SeNPsMPV1-2_48_e | |
| Growth for 120 h in the presence of 2 mM SeO32− | SeNPsMPV1-2_120_e | |
| Growth for 120 h in the presence of 5 mM SeO32− | SeNPsMPV1-5_120_e | |
| Growth for 120 h in the presence of 10 mM SeO32− | SeNSsMPV1-10_120_e | |
| Growth for 120 h in the presence of glucose and 0.5 mM SeO32− | SeNSsMPV1_G_e | [28] |
| Growth for 120 h in the presence of pyruvate and 0.5 mM SeO32− | SeNSsMPV1_P_e |
| Biogenic SeNS Extracts | Average NP Diameter (nm) | Average NR Length (nm) |
|---|---|---|
| SeNPsMPV1-0.5_24_e | 122 ± 40 | N.D. |
| SeNPsMPV1-0.5_120_e | 146 ± 25 | N.D. |
| SeNPsMPV1-2_48_e | 118 ± 36 | N.D. |
| SeNPsMPV1-2_120_e | 132 ± 21 | N.D. |
| SeNPsMPV1-5_120_e | 125 ± 32 | N.D. |
| SeNPsMPV1-10_120_e | 92 ± 26 | N.D. |
| SeNSsMPV1-G_e | 125 ± 37 | 513 ± 92 |
| SeNSsMPV1-P_e | 127 ± 52 | 418 ± 115 |
| Biogenic Extract | Se | C | O | N | S |
|---|---|---|---|---|---|
| SeNPsMPV1-0.5_120_e | ✓ | ✓ | ✓ | ✓ | ✓ |
| SeNPsMPV1-2_120_e | ✓ | ✓ | ✓ | ✓ | ✓ |
| SeNPsMPV1-5_120_e | ✓ | ✓ | ✓ | ✓ | ✓ |
| SeNPsMPV1-10_120_e | ✓ | ✓ | ✓ | ✓ | ✓ |
| SeNSsMPV1-G_e | ✓ | ✓ | ✓ | - | ✓ |
| SeNSsMPV1-P_e | ✓ | ✓ | ✓ | - | ✓ |
| Organic Material Samples | dH (nm) | PdI |
|---|---|---|
| OM_SeNPsMPV1-0.5_120_e | 140 ± 23 | 0.135 |
| OM_SeNPsMPV1-2_120_e | 131 ± 13 | 0.173 |
| OM_SeNPsMPV1-5_120_e | 155 ± 20 | 0.167 |
| OM_SeNPsMPV1-10_120_e | 167 ± 35 | 0.181 |
| OM_SeNSsMPV1-G_e | 143 ± 17 | 0.110 |
| OM_SeNSsMPV1-P_e | 152 ± 23 | 0.105 |
| Biogenic Extracts | ζ (mV) | Organic Material Samples | ζ (mV) |
|---|---|---|---|
| SeNPsMPV1-0.5_120_e | −18 ± 1 | OM_SeNPsMPV1-0.5_120_e | −18 ± 3 |
| SeNPsMPV1-2_120_e | −21 ± 2 | OM_SeNPsMPV1-2_120_e | −13 ± 4 |
| SeNPsMPV1-5_120_e | −22 ± 1 | OM_SeNPsMPV1-5_120_e | −19 ± 2 |
| SeNPsMPV1-10_120_e | −16 ± 3 | OM_SeNPsMPV1-10_120_e | −12 ± 4 |
| SeNSsMPV1-G_e | −2 ± 2 | OM_SeNSsMPV1-G_e | −6 ± 5 |
| SeNSsMPV1-P_e | 3 ± 1 | OM_SeNSsMPV1-P_e | 4 ± 2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piacenza, E.; Presentato, A.; Bardelli, M.; Lampis, S.; Vallini, G.; Turner, R.J. Influence of Bacterial Physiology on Processing of Selenite, Biogenesis of Nanomaterials and Their Thermodynamic Stability. Molecules 2019, 24, 2532. https://doi.org/10.3390/molecules24142532
Piacenza E, Presentato A, Bardelli M, Lampis S, Vallini G, Turner RJ. Influence of Bacterial Physiology on Processing of Selenite, Biogenesis of Nanomaterials and Their Thermodynamic Stability. Molecules. 2019; 24(14):2532. https://doi.org/10.3390/molecules24142532
Chicago/Turabian StylePiacenza, Elena, Alessandro Presentato, Marta Bardelli, Silvia Lampis, Giovanni Vallini, and Raymond J. Turner. 2019. "Influence of Bacterial Physiology on Processing of Selenite, Biogenesis of Nanomaterials and Their Thermodynamic Stability" Molecules 24, no. 14: 2532. https://doi.org/10.3390/molecules24142532
APA StylePiacenza, E., Presentato, A., Bardelli, M., Lampis, S., Vallini, G., & Turner, R. J. (2019). Influence of Bacterial Physiology on Processing of Selenite, Biogenesis of Nanomaterials and Their Thermodynamic Stability. Molecules, 24(14), 2532. https://doi.org/10.3390/molecules24142532









