Application of Plant Viruses as a Biotemplate for Nanomaterial Fabrication
Abstract
1. Introduction
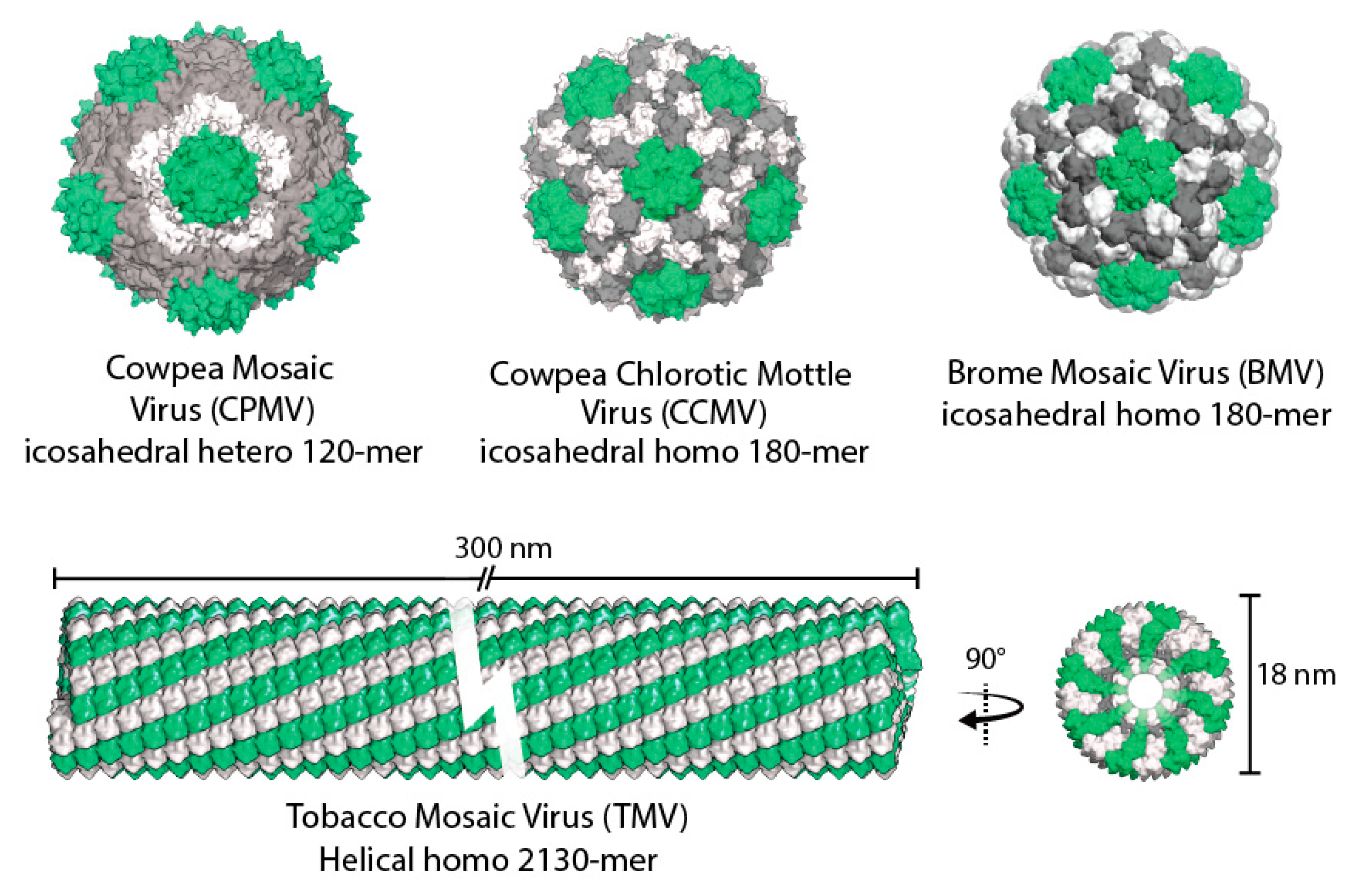
2. Plant Viruses as Biotemplates
2.1. Cowpea Chlorotic Mottle Virus
2.2. Cowpea Mosaic Virus
2.3. Brome Mosaic Virus
2.4. Tobacco Mosaic Virus
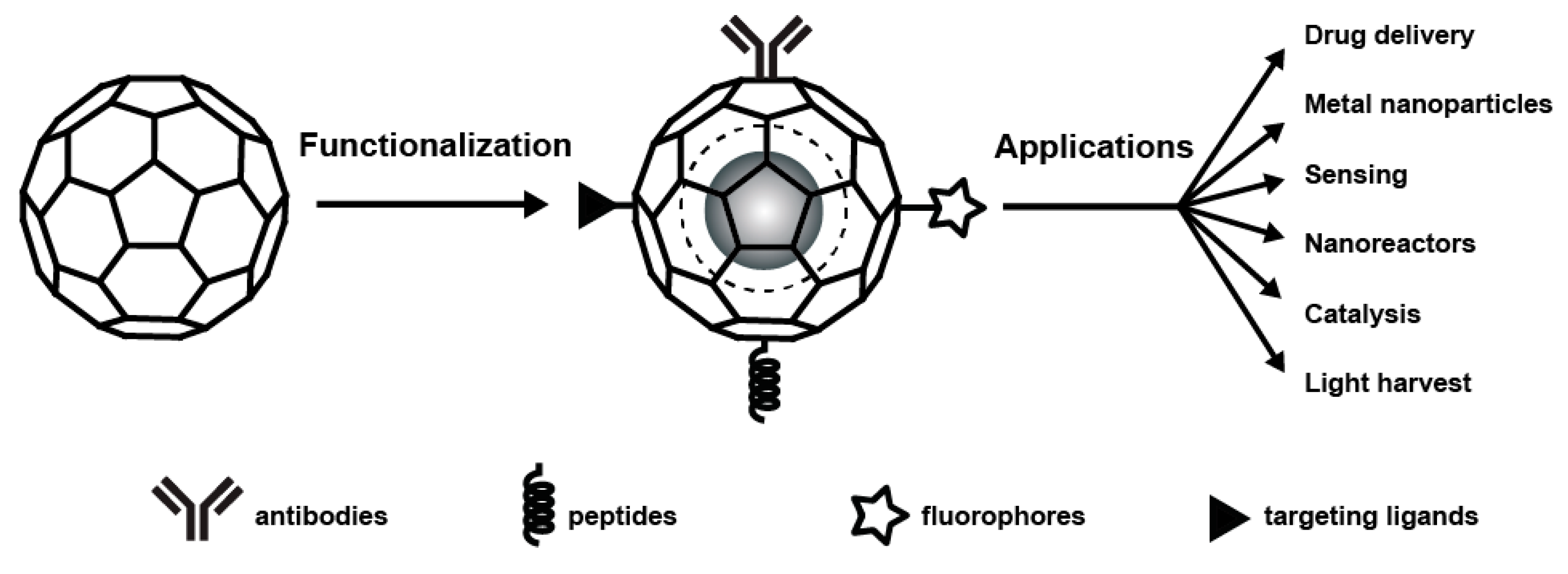
3. Chemical Modification and Genetic Engineering of Plant Viruses
4. PVN Assembly
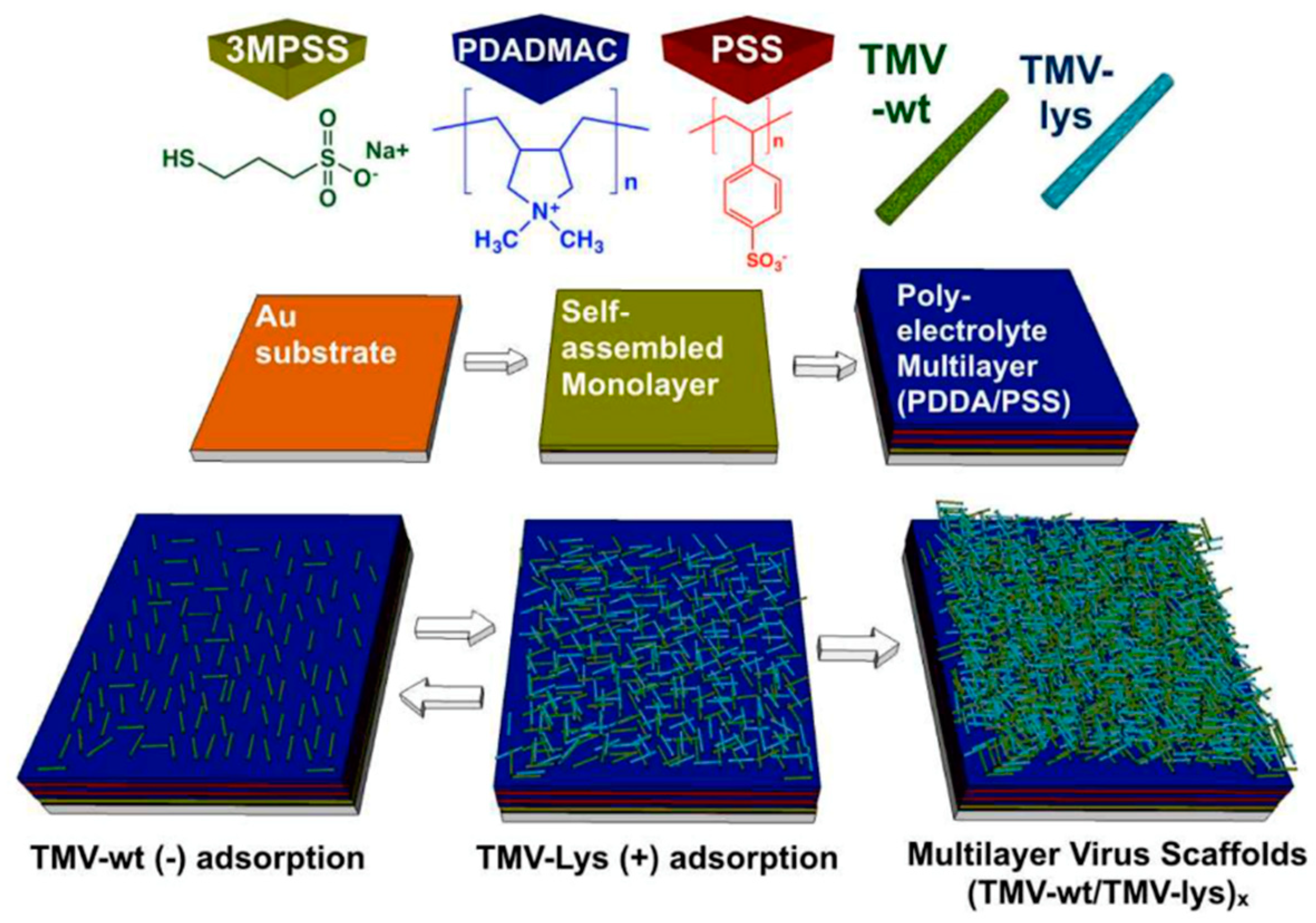
4.1. Two-Dimensional Surface Assemblies
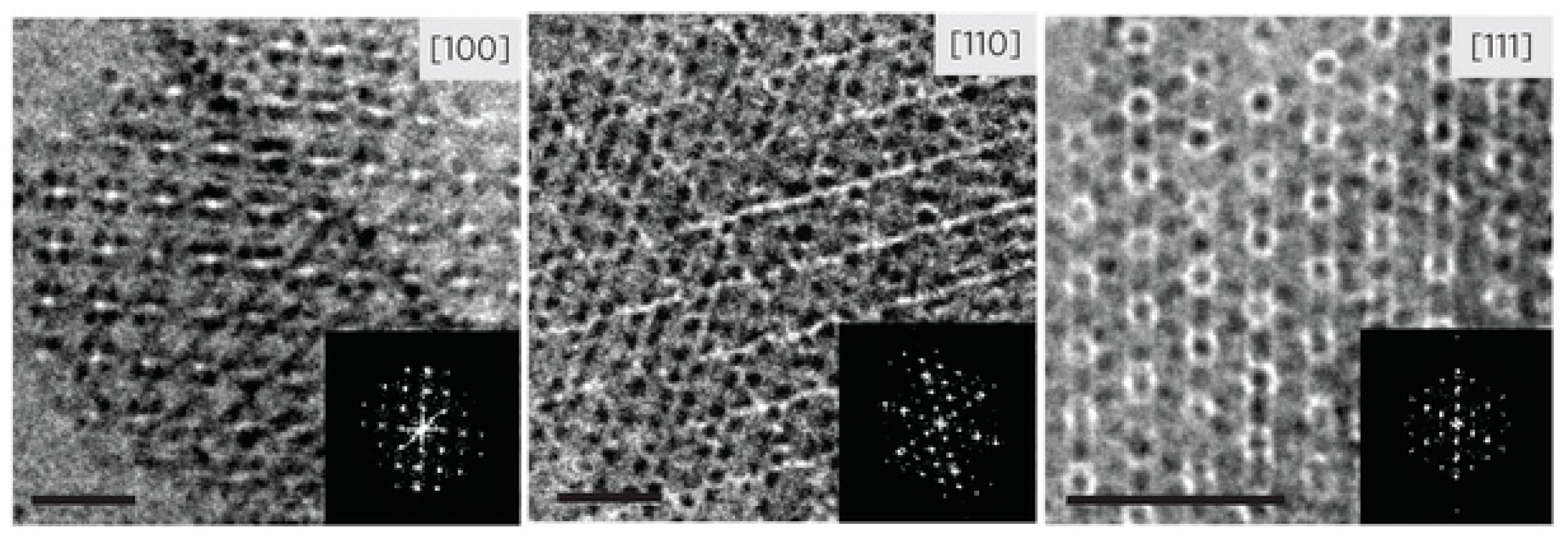
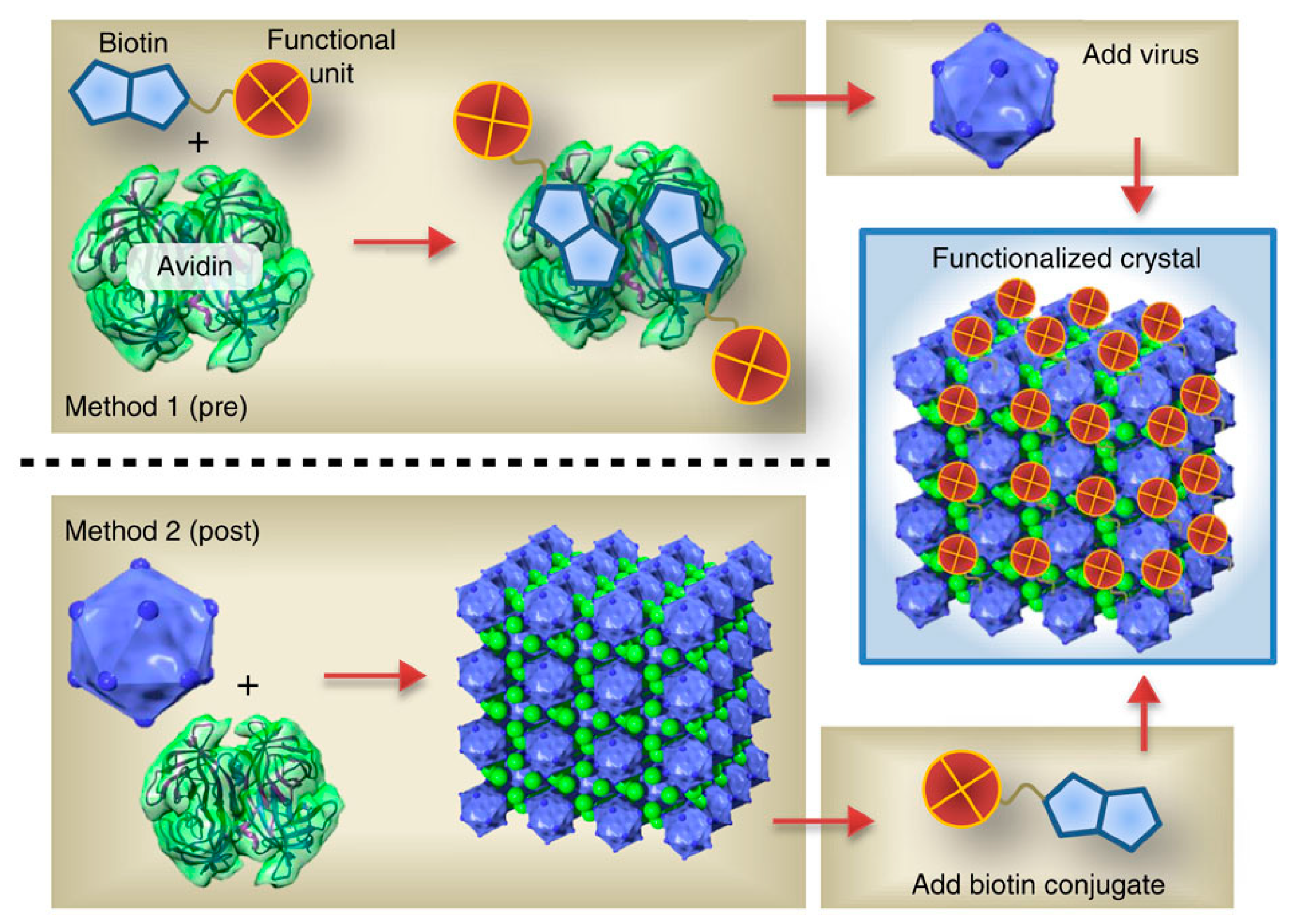
4.2. 3D Assemblies
5. Applications of Plant Viruses Based Nanomaterials
5.1. Biomedical Applications
5.1.1. Magnetic Resonance Imaging
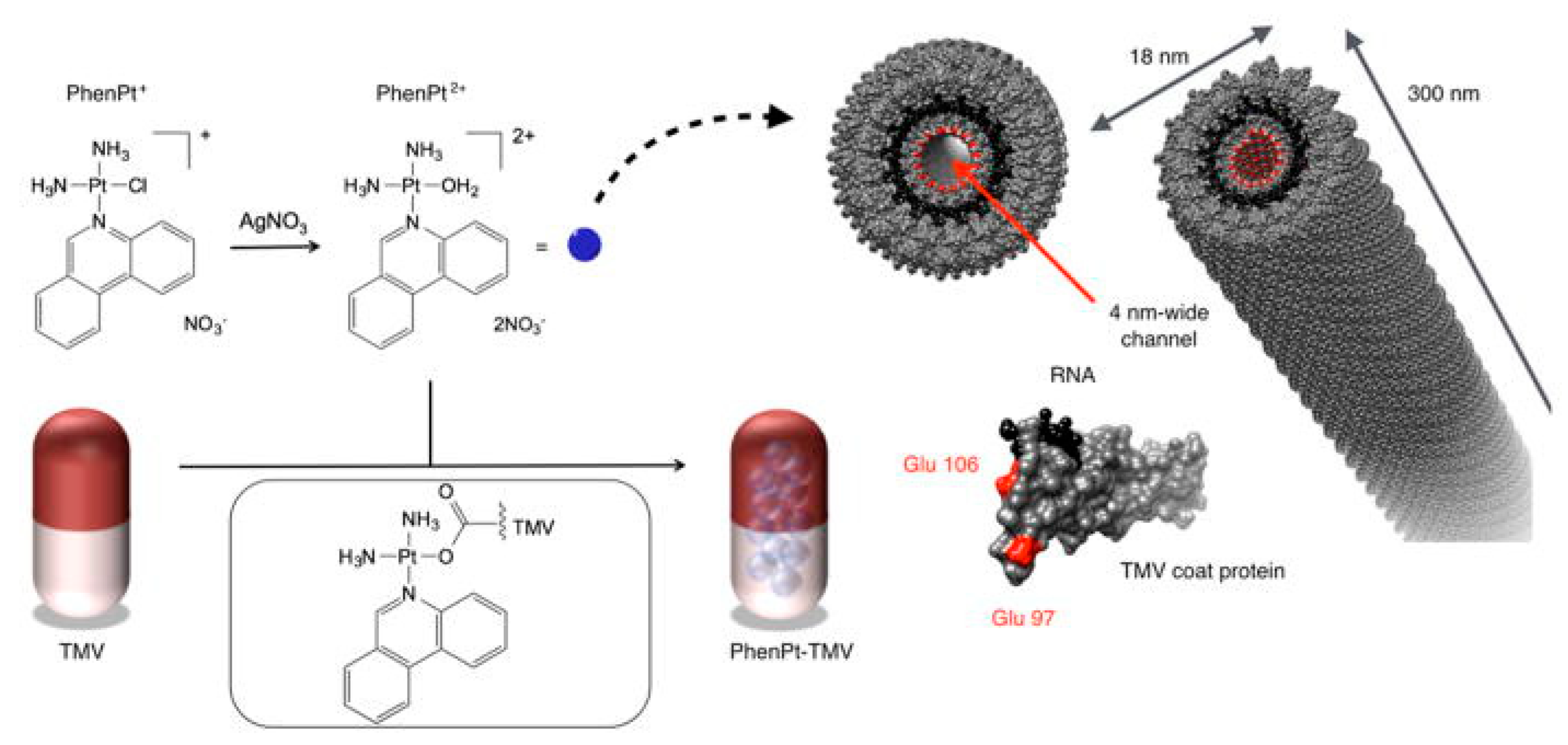
5.1.2. Therapeutic Delivery System
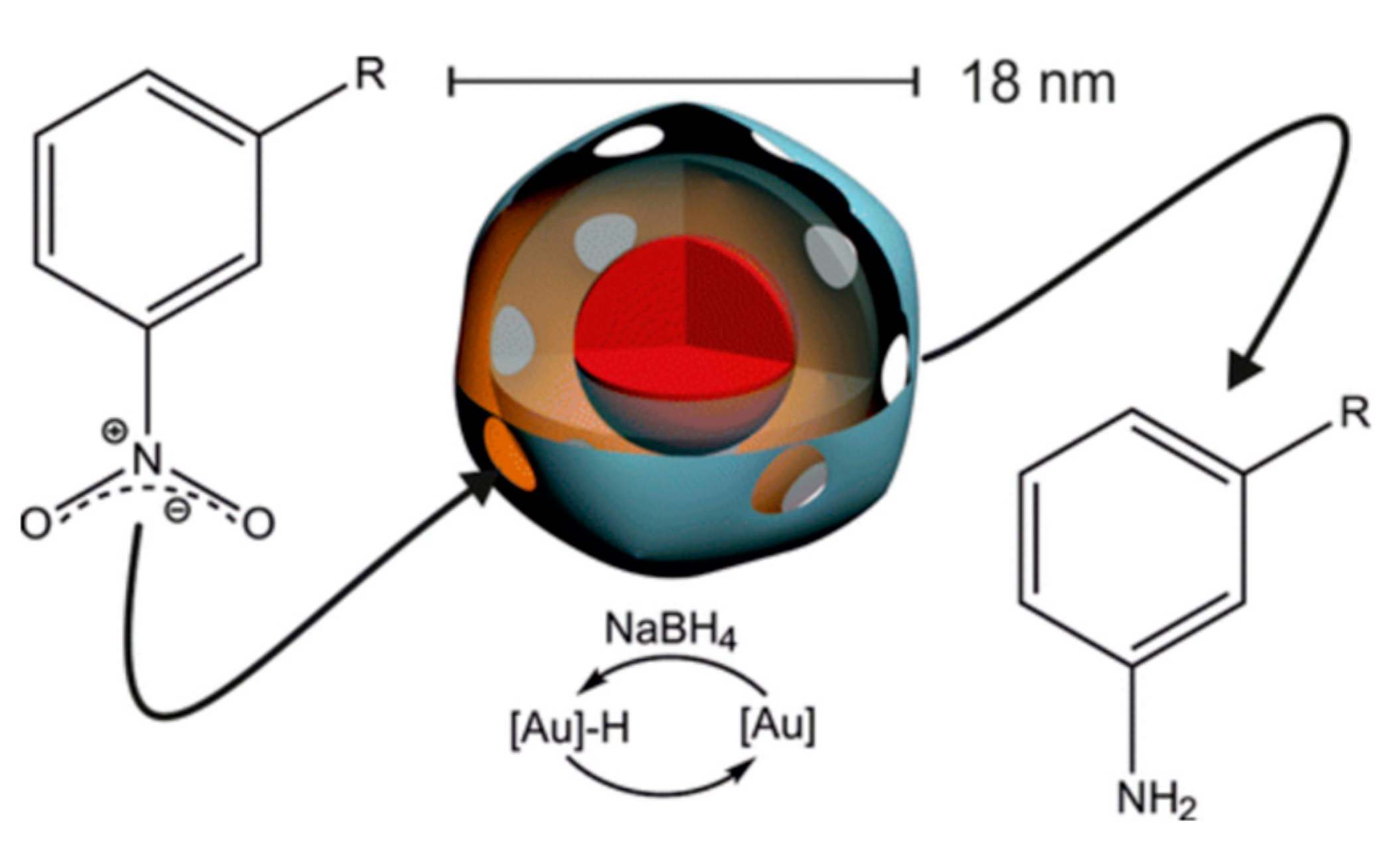
5.2. Enzymatic Nanoreactors and Catalysts
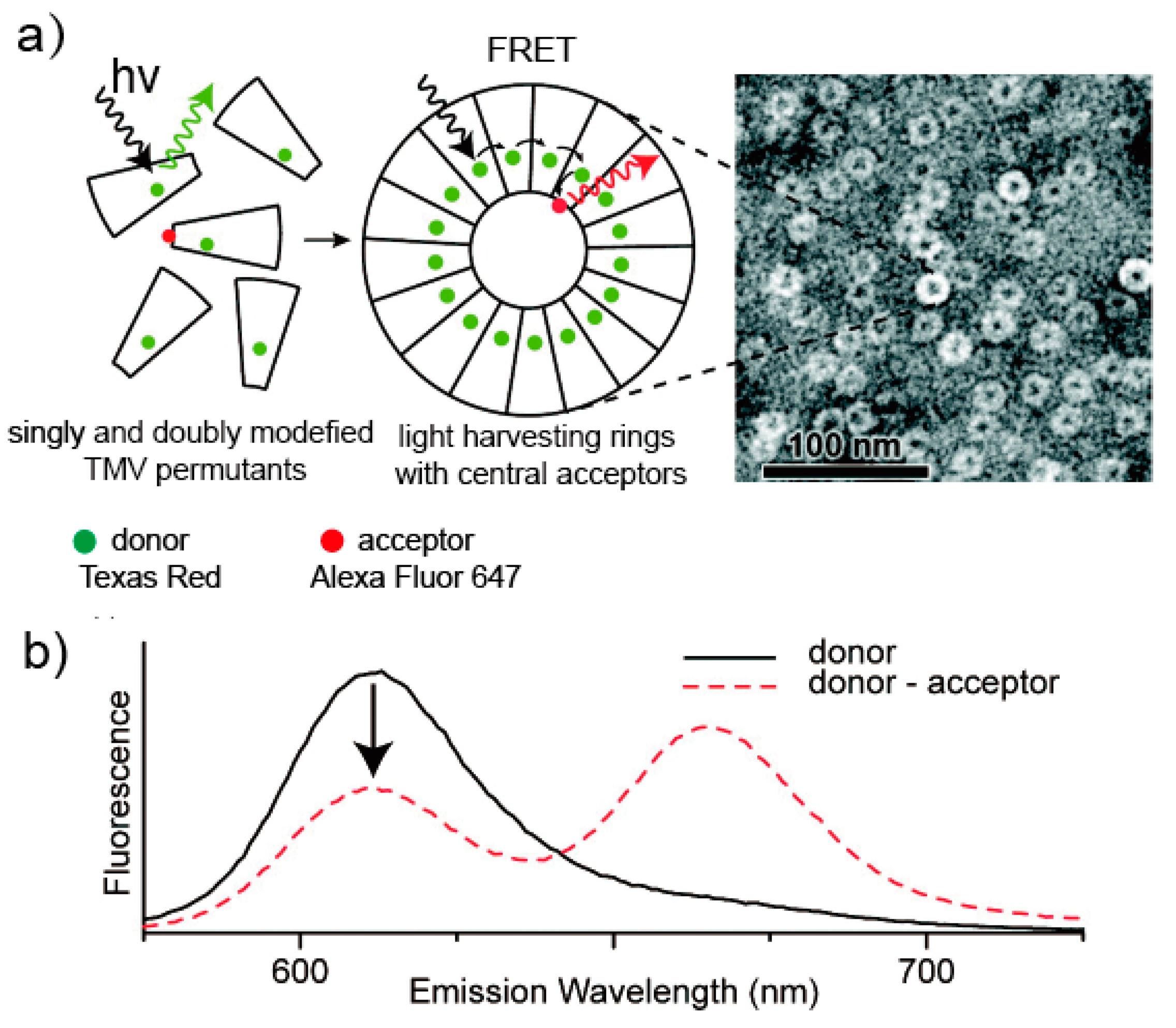
5.3. Light-Harvesting System Based on the TMV
5.4. Sensor Applications of Biomimetic Nanostructures
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Soto, C.M.; Ratna, B.R. Virus hybrids as nanomaterials for biotechnology. Curr. Opin. Biotechnol. 2010, 21, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, K.; Schneider, J.P. Self-assembling peptides and proteins for nanotechnological applications. Curr. Opin. Struct. Biol. 2004, 14, 480–486. [Google Scholar] [CrossRef] [PubMed]
- Seeman, N.C. Nanomaterials based on DNA. Annu. Rev. Biochem. 2010, 79, 65–87. [Google Scholar] [CrossRef] [PubMed]
- Czapar, A.E.; Steinmetz, N.F. Plant viruses and bacteriophages for drug delivery in medicine and biotechnology. Curr. Opin. Chem. Biol. 2017, 38, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Pulsipher, K.W.; Dmochowski, I.J. Ferritin: Versatile host, nanoreactor, and delivery agent. Isr. J. Chem. 2016, 56, 660–670. [Google Scholar] [CrossRef]
- Speir, J.A.; Munshi, S.; Wang, G.; Baker, T.S.; Johnson, J.E. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure 1995, 3, 63–78. [Google Scholar] [CrossRef]
- Johnson, J.E.; Speir, J.A. Quasi-equivalent viruses: A paradigm for protein assemblies. J. Mol. Biol. 1997, 269, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Douglas, T.; Young, M. Host–guest encapsulation of materials by assembled virus protein cages. Nature 1998, 393, 152–155. [Google Scholar] [CrossRef]
- Wang, Q.; Lin, T.; Tang, L.; Johnson, J.E.; Finn, M.G. Icosahedral virus particles as addressable nanoscale building blocks. Angew. Chem. Int. Ed. 2002, 41, 459–462. [Google Scholar] [CrossRef]
- Lin, T.; Chen, Z.; Usha, R.; Stauffacher, C.V.; Dai, J.B.; Schmidt, T.; Johnson, J.E. The refined crystal structure of cowpea mosaic virus at 2.8 Å resolution. Virology 1999, 265, 20–34. [Google Scholar] [CrossRef] [PubMed]
- Sainsbury, F.; Cañizares, M.C.; Lomonossoff, G.P. Cowpea mosaic virus: The plant virus-based biotechnology workhorse. Annu. Rev. Phytopathology 2010, 48, 437. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.J. Bionanoscience at the plant virus–inorganic chemistry interface. Inorg. Chim. Acta 2010, 363, 1070–1076. [Google Scholar] [CrossRef]
- Lane, L.C. The bromoviruses. Adv. Virus Res. 1974, 19, 151–220. [Google Scholar] [CrossRef] [PubMed]
- Ahlquist, P.; Luckow, V.; Kaesberg, P. Complete nucleotide sequence of brome mosaic virus RNA3. J. Mol. Biol. 1981, 153, 23–38. [Google Scholar] [CrossRef]
- Janda, M.; Ahlquist, P. Brome mosaic virus RNA replication protein 1a dramatically increases in vivo stability but not translation of viral genomic RNA3. Proc. Natl. Acad. Sci. USA 1998, 95, 2227–2232. [Google Scholar] [CrossRef] [PubMed]
- Ahlquist, P. Bromovirus RNA replication and transcription. Curr. Opin. Genet. Dev. 1992, 2, 71–76. [Google Scholar] [CrossRef]
- Sacher, R.; Ahlquist, P. Effects of deletions in the N-terminal basic arm of brome mosaic virus coat protein on RNA packaging and systemic infection. J. Virol. 1989, 63, 4545–4552. [Google Scholar] [PubMed]
- Díez, J.; Ishikawa, M.; Kaido, M.; Ahlquist, P. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc. Natl. Acad. Sci. USA 2000, 97, 3913–3918. [Google Scholar] [CrossRef] [PubMed]
- Cuillel, M.; Zulauf, M.; Jacrot, B. Self-assembly of brome mosaic virus protein into capsids: Initial and final states of aggregation. J. Mol. Biol. 1983, 164, 589–603. [Google Scholar] [CrossRef]
- Klug, A. The tobacco mosaic virus particle: Structure and assembly. Philos. Trans. Biol. Sci. 1999, 354, 531–535. [Google Scholar] [CrossRef] [PubMed]
- Pattanayek, R.; Stubbs, G. Structure of the U2 strain of tobacco mosaic virus refined at 3.5 Å resolution using X-ray fiber diffraction. J. Mol. Biol. 1992, 228, 516–528. [Google Scholar] [CrossRef]
- Stubbs, G. Tobacco mosaic virus particle structure and the initiation of disassembly. Philos. Trans. R. Soc. Lond. 1999, 354, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Alonso, J.M.; Gorzny, M.L.; Bittner, A.M. The physics of tobacco mosaic virus and virus-based devices in biotechnology. Trends Biotechnol. 2013, 31, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Knez, M.; Kadri, A.; Wege, C.; Gösele, U.; Jeske, H.; Nielsch, K. Atomic layer deposition on biological macromolecules: Metal oxide coating of tobacco mosaic virus and ferritin. Nano Lett. 2006, 6, 1172–1177. [Google Scholar] [CrossRef] [PubMed]
- Royston, E.; Lee, S.Y.; Culver, J.N.; Harris, M.T. Characterization of silica-coated tobacco mosaic virus. J. Colloid Interface Sci. 2006, 298, 706–712. [Google Scholar] [CrossRef] [PubMed]
- Górzny, M.Ł.; Walton, A.S.; WnęK, M.; Stockley, P.G.; Evans, S.D. Four-probe electrical characterization of Pt-coated TMV-based nanostructures. Nanotechnology 2008, 19, 165704. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Wang, C.; Zhang, W.; Luo, Z.; Tian, D.; Zhai, N.; Zhang, H.; Li, Z.; Jiang, X.; Tang, G. Au nanocrystals grown on a better-defined one-dimensional tobacco mosaic virus coated protein template genetically modified by a hexahistidine tag. Nanotechnology 2012, 23, 335602. [Google Scholar] [CrossRef] [PubMed]
- Wnęk, M.; Górzny, M.L.; Ward, M.B.; Wälti, C.; Davies, A.G.; Brydson, R.; Evans, S.D.; Stockley, P.G. Fabrication and characterization of gold nano-wires templated on virus-like arrays of tobacco mosaic virus coat proteins. Nanotechnology 2013, 24, 025605. [Google Scholar] [CrossRef] [PubMed]
- Meunier, S.; Strable, E.; Finn, M.G. Crosslinking of and coupling to viral capsid proteins by tyrosine oxidation. Chem. Biol. 2004, 11, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Pokorski, J.K.; Steinmetz, N.F. The art of engineering viral nanoparticles. Mol. Pharm. 2011, 8, 29–43. [Google Scholar] [CrossRef] [PubMed]
- Aljabali, A.A.; Barclay, J.E.; Butt, J.N.; Lomonossoff, G.P.; Evans, D.J. Redox-active ferrocene-modified cowpea mosaic virus nanoparticles. Dalton Trans. 2010, 39, 7569–7574. [Google Scholar] [CrossRef] [PubMed]
- Iha, R.K.; Wooley, K.L.; Nyström, A.M.; Burke, D.J.; Kade, M.J.; Hawker, C.J. Applications of orthogonal “click” chemistries in the synthesis of functional soft materials. Russ. Chem. Rev. 2009, 109, 5620–5686. [Google Scholar] [CrossRef] [PubMed]
- Molino, N.M.; Wang, S.-W. Caged protein nanoparticles for drug delivery. Curr. Opin. Biotechnol. 2014, 28, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Wen, A.M.; Shukla, S.; Saxena, P.; Aljabali, A.A.; Yildiz, I.; Dey, S.; Mealy, J.E.; Yang, A.C.; Evans, D.J.; Lomonossoff, G.P. Interior engineering of a viral nanoparticle and its tumor homing properties. Biomacromolecules 2012, 13, 3990–4001. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Li, F.; Dai, G.; Meng, C.; Wang, Q. Disulfide bond: Dramatically enhanced assembly capability and structural stability of tobacco mosaic virus nanorods. Biomacromolecules 2013, 14, 2593–2600. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Zhang, J.; Wang, Q. Site-selective nucleation and controlled growth of gold nanostructures in tobacco mosaic virus nanotubulars. Small 2015, 11, 2505–2509. [Google Scholar] [CrossRef] [PubMed]
- Strable, E.; Prasuhn, D.E., Jr.; Udit, A.K.; Brown, S.; Link, A.J.; Ngo, J.T.; Lander, G.; Quispe, J.; Potter, C.S.; Carragher, B. Unnatural amino acid incorporation into virus-like particles. Bioconjugate Chem. 2008, 19, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Carrico, Z.M.; Romanini, D.W.; Mehl, R.A.; Francis, M.B. Oxidative coupling of peptides to a virus capsid containing unnatural amino acids. Chem. Commun. 2008, 1205–1207. [Google Scholar] [CrossRef] [PubMed]
- Soto, C.M.; Blum, A.S.; Wilson, C.D.; Lazorcik, J.; Kim, M.; Gnade, B.; Ratna, B.R. Separation and recovery of intact gold-virus complex by agarose electrophoresis and electroelution: Application to the purification of cowpea mosaic virus and colloidal gold complex. Electrophoresis 2004, 25, 2901–2906. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lin, T.; Johnson, J.E.; Finn, M.G. Natural supramolecular building blocks. Cysteine-added mutants of cowpea mosaic virus. Chem. Biol. 2002, 9, 813–819. [Google Scholar] [CrossRef]
- Blum, A.S.; Soto, C.M.; Wilson, C.D.; Brower, T.L.; Pollack, S.K.; Schull, T.L.; Chatterji, A.; Lin, T.; Johnson, J.E.; Amsinck, C. An engineered virus as a scaffold for three-dimensional self-assembly on the nanoscale. Small 2005, 1, 702–706. [Google Scholar] [CrossRef] [PubMed]
- Sainsbury, F.; Lomonossoff, G.P. Extremely high-level and rapid transient protein production in plants without the use of viral replication. Plant Physiol. 2008, 148, 1212–1218. [Google Scholar] [CrossRef] [PubMed]
- Barick, K.C.; Bahadur, D. Self-assembly of colloidal nanoscale particles: Fabrication, properties and applications. J. Nanosci. Nanotechnol. 2010, 10, 668–689. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.J. The bionanoscience of plant viruses: Templates and synthons for new materials. J. Mater. Chem. 2008, 18, 3746–3754. [Google Scholar] [CrossRef]
- Kaur, G.; Valarmathi, M.T.; Potts, J.D.; Jabbari, E.; Sabo-Attwood, T.; Wang, Q. Regulation of osteogenic differentiation of rat bone marrow stromal cells on 2D nanorod substrates. Biomaterials 2010, 31, 1732–1741. [Google Scholar] [CrossRef]
- Zan, X.; Sitasuwan, P.; Powell, J.; Dreher, T.W.; Wang, Q. Polyvalent display of RGD motifs on turnip yellow mosaic virus for enhanced stem cell adhesion and spreading. Acta Biomater. 2012, 8, 2978–2985. [Google Scholar] [CrossRef]
- Sitasuwan, P.; Lee, L.A.; Bo, P.; Davis, E.N.; Lin, Y.; Wang, Q. A plant virus substrate induces early upregulation of BMP2 for rapid bone formation. Integr. Biol. 2012, 4, 651–660. [Google Scholar] [CrossRef]
- Sitasuwan, P.; Lee, L.A.; Li, K.; Nguyen, H.G.; Wang, Q. RGD-conjugated rod-like viral nanoparticles on 2D scaffold improve bone differentiation of mesenchymal stem cells. Front. Chem. 2014, 2, 31. [Google Scholar] [CrossRef]
- Tiu, B.D.B.; Tiu, S.B.; Wen, A.M.; Lam, P.; Steinmetz, N.F.; Advincula, R.C. Free-standing, nanopatterned janus membranes of conducting polymer–virus nanoparticle arrays. Langmuir 2016, 32, 6185–6193. [Google Scholar] [CrossRef]
- Yang, C.; Meldon, J.H.; Lee, B.; Yi, H. Investigation on the catalytic reduction kinetics of hexavalent chromium by viral-templated palladium nanocatalysts. Catal. Today 2014, 233, 108–116. [Google Scholar] [CrossRef]
- Uchida, M.; McCoy, K.; Fukuto, M.; Yang, L.; Yoshimura, H.; Miettinen, H.M.; LaFrance, B.; Patterson, D.P.; Schwarz, B.; Karty, J.A. Modular self-assembly of protein cage lattices for multistep catalysis. ACS Nano 2017, 12, 942–953. [Google Scholar] [CrossRef] [PubMed]
- Doni, G.; Kostiainen, M.A.; Danani, A.; Pavan, G.M. Generation-dependent molecular recognition controls self-assembly in supramolecular dendron−virus complexes. Nano Lett. 2010, 11, 723–728. [Google Scholar] [CrossRef] [PubMed]
- Steinmetz, N.F.; Calder, G.; Lomonossoff, G.P.; Evans, D.J. Plant viral capsids as nanobuilding blocks: Construction of arrays on solid supports. Langmuir 2006, 22, 10032–10037. [Google Scholar] [CrossRef] [PubMed]
- Suci, P.A.; Klem, M.T.; Arce, F.T.; Douglas, T.; Young, M. Assembly of multilayer films incorporating a viral protein cage architecture. Langmuir 2006, 22, 8891–8896. [Google Scholar] [CrossRef] [PubMed]
- Kostiainen, M.A.; Kasyutich, O.; Cornelissen, J.J.; Nolte, R.J. Self-assembly and optically triggered disassembly of hierarchical dendron–virus complexes. Nat. Chem. 2010, 2, 394. [Google Scholar] [CrossRef] [PubMed]
- Kostiainen, M.A.; Hiekkataipale, P.; Laiho, A.; Lemieux, V.; Seitsonen, J.; Ruokolainen, J.; Ceci, P. Electrostatic assembly of binary nanoparticle superlattices using protein cages. Nat. Nanotechnol. 2013, 8, 52. [Google Scholar] [CrossRef] [PubMed]
- Liljeström, V.; Mikkilä, J.; Kostiainen, M.A. Self-assembly and modular functionalization of three-dimensional crystals from oppositely charged proteins. Nat. Commun. 2014, 5, 4445. [Google Scholar] [CrossRef] [PubMed]
- Royston, E.; Ghosh, A.; Kofinas, P.; Harris, M.T.; Culver, J.N. Self-assembly of virus-structured high surface area nanomaterials and their application as battery electrodes. Langmuir 2008, 24, 906–912. [Google Scholar] [CrossRef] [PubMed]
- Namba, K.; Pattanayek, R.; Stubbs, G. Visualization of protein-nucleic acid interactions in a virus: Refined structure of intact tobacco mosaic virus at 2.9 å resolution by X-ray fiber diffraction. J. Mol. Biol. 1989, 208, 307–325. [Google Scholar] [CrossRef]
- Gerasopoulos, K.; Mccarthy, M.; Banerjee, P.; Fan, X.; Culver, J.N.; Ghodssi, R. Biofabrication methods for the patterned assembly and synthesis of viral nanotemplates. Nanotechnology 2010, 21, 055304. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Guo, J.; Brown, A.D.; Royston, E.; Wang, C.; Kofinas, P.; Culver, J.N. Virus-assembled flexible electrode-electrolyte interfaces for enhanced polymer-based battery applications. J. Nanomater. 2012, 2012, 5. [Google Scholar] [CrossRef]
- Tiu, B.D.B.; Kernan, D.L.; Tiu, S.B.; Wen, A.M.; Zheng, Y.; Pokorski, J.K.; Advincula, R.C.; Steinmetz, N.F. Electrostatic layer-by-layer construction of fibrous TMV biofilms. Nanoscale 2017, 9, 1580–1590. [Google Scholar] [CrossRef] [PubMed]
- Knez, M.; Sumser, M.P.; Bittner, A.M.; Wege, C.; Jeske, H.; Hoffmann, D.M.P.; Kuhnke, K.; Kern, K. Binding the tobacco mosaic virus to inorganic surfaces. Langmuir 2004, 20, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Lim, J.S.; Harris, M.T. Synthesis and application of virus-based hybrid nanomaterials. Biotechnol. Bioeng. 2012, 109, 16–30. [Google Scholar] [CrossRef] [PubMed]
- Qazi, S.; Liepold, L.O.; Abedin, M.J.; Johnson, B.; Prevelige, P.; Frank, J.A.; Douglas, T. P22 viral capsids as nanocomposite high-relaxivity MRI contrast agents. Mol. Pharm. 2012, 10, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Waters, E.A.; Wickline, S.A. Contrast agents for MRI. Basic Res. Cardiol. 2008, 103, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Prasuhn, D.E., Jr.; Yeh, R.M.; Obenaus, A.; Manchester, M.; Finn, M. Viral MRI contrast agents: Coordination of Gd by native virions and attachment of Gd complexes by azide–alkyne cycloaddition. Chem. Commun. 2007, 1269–1271. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Prasuhn, D.; Yeh, R.M.; Destito, G.; Rae, C.S.; Osborn, K.; Finn, M.; Manchester, M. Bio-distribution, toxicity and pathology of cowpea mosaic virus nanoparticles in vivo. J. Control. Release 2007, 120, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Millan, J.G.; Brasch, M.; Anaya-Plaza, E.; de la Escosura, A.; Velders, A.H.; Reinhoudt, D.N.; Torres, T.; Koay, M.S.; Cornelissen, J.J. Self-assembly triggered by self-assembly: Optically active, paramagnetic micelles encapsulated in protein cage nanoparticles. J. Inorg. Biochem. 2014, 136, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Bruckman, M.A.; Hern, S.; Jiang, K.; Flask, C.A.; Yu, X.; Steinmetz, N.F. Tobacco mosaic virus rods and spheres as supramolecular high-relaxivity MRI contrast agents. J. Mater. Chem. B 2013, 1, 1482–1490. [Google Scholar] [CrossRef] [PubMed]
- Bruckman, M.A.; Randolph, L.N.; Gulati, N.M.; Stewart, P.L.; Steinmetz, N.F. Silica-coated Gd(dota)-loaded protein nanoparticles enable magnetic resonance imaging of macrophages. J. Mater. Chem. B 2015, 3, 7503–7510. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Eber, F.J.; Nagarajan, A.S.; DiFranco, N.A.; Schmidt, N.; Wen, A.M.; Eiben, S.; Twyman, R.M.; Wege, C.; Steinmetz, N.F. The impact of aspect ratio on the biodistribution and tumor homing of rigid soft-matter nanorods. Adv. Healthc. Mater. 2015, 4, 874–882. [Google Scholar] [CrossRef] [PubMed]
- Bruckman, M.A.; Jiang, K.; Simpson, E.J.; Randolph, L.N.; Luyt, L.G.; Yu, X.; Steinmetz, N.F. Dual-modal magnetic resonance and fluorescence imaging of atherosclerotic plaques in vivo using VCAM-1 targeted tobacco mosaic virus. Nano Lett. 2014, 14, 1551–1558. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Wong, S.M.; Lim, L.Y. Application of plant viruses as nano drug delivery systems. Pharm. Res. 2010, 27, 2509–2513. [Google Scholar] [CrossRef] [PubMed]
- Paavonen, J.; Jenkins, D.; Bosch, F.X.; Naud, P.; Salmerón, J.; Wheeler, C.M.; Chow, S.N.; Apter, D.L.; Kitchener, H.C.; Castellsague, X. Efficacy of a prophylactic adjuvanted bivalent L1 virus-like-particle vaccine against infection with human papillomavirus types 16 and 18 in young women: An interim analysis of a phase III double-blind, randomised controlled trial. Lancet 2007, 369, 2161–2170. [Google Scholar] [CrossRef]
- Zhou, Y.; Maharaj, P.D.; Mallajosyula, J.K.; McCormick, A.A.; Kearney, C.M. In planta production of flock house virus transencapsidated RNA and its potential use as a vaccine. Mol. Biotechnol. 2015, 57, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Lizotte, P.; Wen, A.; Sheen, M.; Fields, J.; Rojanasopondist, P.; Steinmetz, N.; Fiering, S. In situ vaccination with cowpea mosaic virus nanoparticles suppresses metastatic cancer. Nat. Nanotechnol. 2016, 11, 295. [Google Scholar] [CrossRef] [PubMed]
- Lebel, M.-È.; Chartrand, K.; Tarrab, E.; Savard, P.; Leclerc, D.; Lamarre, A. Potentiating cancer immunotherapy using papaya mosaic virus-derived nanoparticles. Nano Lett. 2016, 16, 1826–1832. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.S.; Wang, M.W.; Fung, C.Y.; Chen, P.L.; Chang, C.F.; Huang, W.S.; Wang, J.Y.; Lin, P.Y.; Chang, D. Efficient gene transfer using the human JC virus-like particle that inhibits human colon adenocarcinoma growth in a nude mouse model. Gene Ther. 2010, 17, 1033. [Google Scholar] [CrossRef] [PubMed]
- Azizgolshani, O.; Garmann, R.F.; Cadena-Nava, R.; Knobler, C.M.; Gelbart, W.M. Reconstituted plant viral capsids can release genes to mammalian cells. Virology 2013, 441, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarczyk, S.J.; Sitaraman, K.; Young, H.A.; Hughes, S.H.; Chatterjee, D.K. Protein delivery using engineered virus-like particles. Proc. Natl. Acad. Sci. USA 2011, 108, 16998–17003. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.L.; Hubbard, L.C.; Hern, S.; Yildiz, I.; Gratzl, M.; Steinmetz, N.F. Shape matters: The diffusion rates of TMV rods and CPMV icosahedrons in a spheroid model of extracellular matrix are distinct. Biomater. Sci. 2013, 1, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Aljabali, A.A.; Shukla, S.; Lomonossoff, G.P.; Steinmetz, N.F.; Evans, D.J. CPMV-DOX delivers. Mol. Pharm. 2012, 10, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Wen, H.; Wen, Q.; Chen, X.; Wang, Y.; Xuan, W.; Liang, J.; Wan, S. Cucumber mosaic virus as drug delivery vehicle for doxorubicin. Biomaterials 2013, 34, 4632–4642. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, I.; Tsvetkova, I.; Wen, A.M.; Shukla, S.; Masarapu, M.H.; Dragnea, B.; Steinmetz, N.F. Engineering of brome mosaic virus for biomedical applications. RSC Adv. 2012, 2, 3670–3677. [Google Scholar] [CrossRef] [PubMed]
- Gandra, N.; Abbineni, G.; Qu, X.; Huai, Y.; Wang, L.; Mao, C. Bacteriophage bionanowire as a carrier for both cancer-targeting peptides and photosensitizers and its use in selective cancer cell killing by photodynamic therapy. Small 2013, 9, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Galaway, F.A.; Stockley, P.G. MS2 viruslike particles: A robust, semisynthetic targeted drug delivery platform. Mol. Pharm. 2012, 10, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Finbloom, J.A.; Han, K.; Aanei, I.L.; Hartman, E.C.; Finley, D.T.; Dedeo, M.T.; Fishman, M.; Downing, K.H.; Francis, M.B. Stable disk assemblies of a tobacco mosaic virus mutant as nanoscale scaffolds for applications in drug delivery. Bioconjugate Chem. 2016, 27, 2480–2485. [Google Scholar] [CrossRef] [PubMed]
- Hovlid, M.L.; Steinmetz, N.F.; Laufer, B.; Lau, J.L.; Kuzelka, J.; Wang, Q.; Hyypiä, T.; Nemerow, G.R.; Kessler, H.; Manchester, M. Guiding plant virus particles to integrin-displaying cells. Nanoscale 2012, 4, 3698–3705. [Google Scholar] [CrossRef] [PubMed]
- Chariou, P.L.; Lee, K.L.; Wen, A.M.; Gulati, N.M.; Stewart, P.L.; Steinmetz, N.F. Detection and imaging of aggressive cancer cells using an epidermal growth factor receptor (EGFR)-targeted filamentous plant virus-based nanoparticle. Bioconjugate Chem. 2015, 26, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Destito, G.; Yeh, R.; Rae, C.S.; Finn, M.G.; Manchester, M. Folic acid-mediated targeting of cowpea mosaic virus particles to tumor cells. Chem. Biol. 2007, 14, 1152–1162. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Low, P.S. Folate-mediated delivery of macromolecular anticancer therapeutic agents. Adv. Drug Deliv. Rev. 2002, 54, 675–693. [Google Scholar] [CrossRef]
- Raja, K.S.; Wang, Q.; Gonzalez, M.J.; Manchester, M.; Johnson, J.E.; Finn, M. Hybrid virus− polymer materials. 1. Synthesis and properties of PEG-decorated cowpea mosaic virus. Biomacromolecules 2003, 4, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Steinmetz, N.F.; Ablack, A.L.; Hickey, J.L.; Ablack, J.; Manocha, B.; Mymryk, J.S.; Luyt, L.G.; Lewis, J.D. Intravital imaging of human prostate cancer using viral nanoparticles targeted to gastrin-releasing peptide receptors. Small 2011, 7, 1664–1672. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Ablack, A.L.; Wen, A.M.; Lee, K.L.; Lewis, J.D.; Steinmetz, N.F. Increased tumor homing and tissue penetration of the filamentous plant viral nanoparticle potato virus X. Mol. Pharm. 2012, 10, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Steinmetz, N.F.; Manchester, M. Pegylated viral nanoparticles for biomedicine: The impact of PEG chain length on VNP cell interactions in vitro and ex vivo. Biomacromolecules 2009, 10, 784–792. [Google Scholar] [CrossRef] [PubMed]
- Tacar, O.; Sriamornsak, P.; Dass, C.R. Doxorubicin: An update on anticancer molecular action, toxicity and novel drug delivery systems. J. Pharm. Pharmacol. 2013, 65, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Bruckman, M.A.; Czapar, A.E.; VanMeter, A.; Randolph, L.N.; Steinmetz, N.F. Tobacco mosaic virus-based protein nanoparticles and nanorods for chemotherapy delivery targeting breast cancer. J. Control. Release 2016, 231, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Gao, S.; Wu, M.; Liu, X.; Qiao, J.; Zhou, Q.; Jiang, S.; Niu, Z. Tobacco mosaic virus-based 1D nanorod-drug carrier via the integrin-mediated endocytosis pathway. ACS Appl. Mater. Interfaces 2016, 8, 10800–10807. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Wong, S.M.; Lim, L.-Y. Folic acid-conjugated protein cages of a plant virus: A novel delivery platform for doxorubicin. Bioconjugate Chem. 2007, 18, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Park, G.Y.; Wilson, J.J.; Song, Y.; Lippard, S.J. Phenanthriplatin, a monofunctional DNA-binding platinum anticancer drug candidate with unusual potency and cellular activity profile. Proc. Natl. Acad. Sci. USA 2012, 109, 11987–11992. [Google Scholar] [CrossRef] [PubMed]
- Czapar, A.E.; Zheng, Y.-R.; Riddell, I.A.; Shukla, S.; Awuah, S.G.; Lippard, S.J.; Steinmetz, N.F. Tobacco mosaic virus delivery of phenanthriplatin for cancer therapy. ACS Nano 2016, 10, 4119–4126. [Google Scholar] [CrossRef] [PubMed]
- Brasch, M.; de la Escosura, A.; Ma, Y.; Uetrecht, C.; Heck, A.J.; Torres, T.; Cornelissen, J.J. Encapsulation of phthalocyanine supramolecular stacks into virus-like particles. J. Am. Chem. Soc. 2011, 133, 6878–6881. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.L.; Carpenter, B.L.; Wen, A.M.; Ghiladi, R.A.; Steinmetz, N.F. High aspect ratio nanotubes formed by tobacco mosaic virus for delivery of photodynamic agents targeting melanoma. ACS Biomater. Sci. Eng. 2016, 2, 838–844. [Google Scholar] [CrossRef] [PubMed]
- Wen, A.M.; Lee, K.L.; Cao, P.; Pangilinan, K.; Carpenter, B.L.; Lam, P.; Veliz, F.A.; Ghiladi, R.A.; Advincula, R.C.; Steinmetz, N.F. Utilizing viral nanoparticle/dendron hybrid conjugates in photodynamic therapy for dual delivery to macrophages and cancer cells. Bioconjugate Chem. 2016, 27, 1227–1235. [Google Scholar] [CrossRef] [PubMed]
- Suci, P.A.; Varpness, Z.; Gillitzer, E.; Douglas, T.; Young, M. Targeting and photodynamic killing of a microbial pathogen using protein cage architectures functionalized with a photosensitizer. Langmuir 2007, 23, 12280–12286. [Google Scholar] [CrossRef] [PubMed]
- Minten, I.J.; Claessen, V.I.; Blank, K.; Rowan, A.E.; Nolte, R.J.; Cornelissen, J.J. Catalytic capsids: The art of confinement. Chem. Sci. 2011, 2, 358–362. [Google Scholar] [CrossRef]
- Ueno, T.; Suzuki, M.; Goto, T.; Matsumoto, T.; Nagayama, K.; Watanabe, Y. Size-selective olefin hydrogenation by a PD nanocluster provided in an apo-ferritin cage. Angew. Chem. Int. Ed. 2004, 116, 2581–2584. [Google Scholar] [CrossRef]
- Comellas-Aragonès, M.; Engelkamp, H.; Claessen, V.I.; Sommerdijk, N.A.; Rowan, A.E.; Christianen, P.C.; Maan, J.C.; Verduin, B.J.; Cornelissen, J.J.; Nolte, R.J. A virus-based single-enzyme nanoreactor. Nat. Nanotechnol. 2007, 2, 635. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Traulsen, C.H.-H.; Cornelissen, J.J. Nitroarene reduction by a virus protein cage based nanoreactor. ACS Catal. 2016, 6, 3084–3091. [Google Scholar] [CrossRef]
- Schenk, A.S.; Eiben, S.; Goll, M.; Reith, L.; Kulak, A.N.; Meldrum, F.C.; Jeske, H.; Wege, C.; Ludwigs, S. Virus-directed formation of electrocatalytically active nanoparticle-based Co3O4 tubes. Nanoscale 2017, 9, 6334–6345. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.A.; Presley, A.D.; Francis, M.B. Self-assembling light-harvesting systems from synthetically modified tobacco mosaic virus coat proteins. J. Am. Chem. Soc. 2007, 129, 3104–3109. [Google Scholar] [CrossRef] [PubMed]
- Noriega, R.; Finley, D.T.; Haberstroh, J.; Geissler, P.L.; Francis, M.B.; Ginsberg, N.S. Manipulating excited-state dynamics of individual light-harvesting chromophores through restricted motions in a hydrated nanoscale protein cavity. J. Phys. Chem. B 2015, 119, 6963–6973. [Google Scholar] [CrossRef] [PubMed]
- Delor, M.; Dai, J.; Roberts, T.D.; Rogers, J.R.; Hamed, S.M.; Neaton, J.B.; Geissler, P.L.; Francis, M.B.; Ginsberg, N.S. Exploiting chromophore–protein interactions through linker engineering to tune photoinduced dynamics in a biomimetic light-harvesting platform. J. Am. Chem. Soc. 2018. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.-Z.; Miller, R.A.; Fleming, G.R.; Francis, M.B. Energy transfer dynamics in light-harvesting assemblies templated by the tobacco mosaic virus coat protein. J. Phys. Chem. B 2008, 112, 6887–6892. [Google Scholar] [CrossRef] [PubMed]
- Dedeo, M.T.; Duderstadt, K.E.; Berger, J.M.; Francis, M.B. Nanoscale protein assemblies from a circular permutant of the tobacco mosaic virus. Nano Lett. 2009, 10, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Schlick, T.L.; Ding, Z.; Kovacs, E.W.; Francis, M.B. Dual-surface modification of the tobacco mosaic virus. J. Am. Chem. Soc. 2005, 127, 3718–3723. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Fujitsuka, M.; Majima, T. Porphyrin light-harvesting arrays constructed in the recombinant tobacco mosaic virus scaffold. Chem.-A Eur. J. 2007, 13, 8660–8666. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Lin, L.; Sun, D.; Chen, H.; Yang, D.; Li, Q. Bio-inspired synthesis of metal nanomaterials and applications. Chem. Soc. Rev. 2015, 44, 6330–6374. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Yi, H. An integrated approach for enhanced protein conjugation and capture with viral nanotemplates and hydrogel microparticle platforms via rapid bioorthogonal reactions. Langmuir 2014, 30, 7762–7770. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Liu, A.; Cao, B. Virus-based chemical and biological sensing. Angew. Chem. Int. Ed. 2009, 48, 6790–6810. [Google Scholar] [CrossRef] [PubMed]
- Koch, C.; Wabbel, K.; Eber, F.J.; Krolla-Sidenstein, P.; Azucena, C.; Gliemann, H.; Eiben, S.; Geiger, F.; Wege, C. Modified TMV particles as beneficial scaffolds to present sensor enzymes. Front. Plant Sci. 2015, 6, 1137. [Google Scholar] [CrossRef] [PubMed]
- Elghanian, R.; Storhoff, J.J.; Mucic, R.C.; Letsinger, R.L.; Mirkin, C.A. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science 1997, 277, 1078–1081. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Han, M.S.; Mirkin, C.A. Colorimetric detection of mercuric ion (Hg2+) in aqueous media using DNA-functionalized gold nanoparticles. Angew. Chem. Int. Ed. 2007, 119, 4171–4174. [Google Scholar] [CrossRef]
- Lee, J.-S.; Ulmann, P.A.; Han, M.S.; Mirkin, C.A. A DNA−gold nanoparticle-based colorimetric competition assay for the detection of cysteine. Nano Lett. 2008, 8, 529–533. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lu, Y. A colorimetric lead biosensor using dnazyme-directed assembly of gold nanoparticles. J. Am. Chem. Soc. 2003, 125, 6642–6643. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lu, Y. Rational design of “turn-on” allosteric dnazyme catalytic beacons for aqueous mercury ions with ultrahigh sensitivity and selectivity. Angew. Chem. Int. Ed. 2007, 119, 7731–7734. [Google Scholar] [CrossRef]
- Liu, J.; Lu, Y. Adenosine-dependent assembly of aptazyme-functionalized gold nanoparticles and its application as a colorimetric biosensor. Anal. Chem. 2004, 76, 1627–1632. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lu, Y. Fast colorimetric sensing of adenosine and cocaine based on a general sensor design involving aptamers and nanoparticles. Angew. Chem. Int. Ed. 2006, 118, 96–100. [Google Scholar] [CrossRef]
- Jiang, X.; Sun, C.; Guo, Y.; Nie, G.; Xu, L. Peroxidase-like activity of apoferritin paired gold clusters for glucose detection. Biosens. Bioelectron. 2015, 64, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Chen, B.; Zhang, H.; Sun, Y.; Wang, J.; Zhang, J.; Fu, Y. BSA-stabilized pt nanozyme for peroxidase mimetics and its application on colorimetric detection of mercury (II) ions. Biosens. Bioelectron. 2015, 66, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Zang, F.; Gerasopoulos, K.; Fan, X.Z.; Brown, A.D.; Culver, J.N.; Ghodssi, R. An electrochemical sensor for selective tnt sensing based on tobacco mosaic virus-like particle binding agents. Chem. Commun. 2014, 50, 12977–12980. [Google Scholar] [CrossRef] [PubMed]
- Zang, F.; Gerasopoulos, K.; Brown, A.D.; Culver, J.N.; Ghodssi, R. Capillary microfluidics-assembled virus-like particle bionanoreceptor interfaces for label-free biosensing. ACS Appl. Mater. Interfaces 2017, 9, 8471–8479. [Google Scholar] [CrossRef] [PubMed]
- Bruckman, M.A.; Liu, J.; Koley, G.; Li, Y.; Benicewicz, B.; Niu, Z.; Wang, Q. Tobacco mosaic virus based thin film sensor for detection of volatile organic compounds. J. Mater. Chem. 2010, 20, 5715–5719. [Google Scholar] [CrossRef]
- Sapsford, K.E.; Soto, C.M.; Blum, A.S.; Chatterji, A.; Lin, T.; Johnson, J.E.; Ligler, F.S.; Ratna, B.R. A cowpea mosaic virus nanoscaffold for multiplexed antibody conjugation: Application as an immunoassay tracer. Biosens. Bioelectron. 2006, 21, 1668–1673. [Google Scholar] [CrossRef] [PubMed]
- Seetharam, R.N.; Szuchmacher Blum, A.; Soto, C.M.; Whitley, J.L.; Sapsford, K.E.; Chatterji, A.; Lin, T.; Johnson, J.E.; Guerra, C.; Satir, P.; et al. Long term storage of virus templated fluorescent materials for sensing applications. Nanotechnology 2008, 19, 105504. [Google Scholar] [CrossRef] [PubMed]
- Soto, C.M.; Blum, A.S.; Vora, G.J.; Lebedev, N.; Meador, C.E.; Won, A.P.; Chatterji, A.; Johnson, J.E.; Ratna, B.R. Fluorescent signal amplification of carbocyanine dyes using engineered viral nanoparticles. J. Am. Chem. Soc. 2006, 128, 5184–5189. [Google Scholar] [CrossRef] [PubMed]
- Vora, G.J.; Meador, C.E.; Anderson, G.P.; Taitt, C.R. Comparison of detection and signal amplification methods for DNA microarrays. Mol. Cell. Probes 2008, 22, 294–300. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Dong, Y.; Zhou, J.; Li, X.; Wang, F. Application of Plant Viruses as a Biotemplate for Nanomaterial Fabrication. Molecules 2018, 23, 2311. https://doi.org/10.3390/molecules23092311
Zhang Y, Dong Y, Zhou J, Li X, Wang F. Application of Plant Viruses as a Biotemplate for Nanomaterial Fabrication. Molecules. 2018; 23(9):2311. https://doi.org/10.3390/molecules23092311
Chicago/Turabian StyleZhang, Yu, Yixin Dong, Jinhua Zhou, Xun Li, and Fei Wang. 2018. "Application of Plant Viruses as a Biotemplate for Nanomaterial Fabrication" Molecules 23, no. 9: 2311. https://doi.org/10.3390/molecules23092311
APA StyleZhang, Y., Dong, Y., Zhou, J., Li, X., & Wang, F. (2018). Application of Plant Viruses as a Biotemplate for Nanomaterial Fabrication. Molecules, 23(9), 2311. https://doi.org/10.3390/molecules23092311




