KMUP-1, a GPCR Modulator, Attenuates Triglyceride Accumulation Involved MAPKs/Akt/PPARγ and PKA/PKG/HSL Signaling in 3T3-L1 Preadipocytes
Abstract
1. Introduction
2. Results
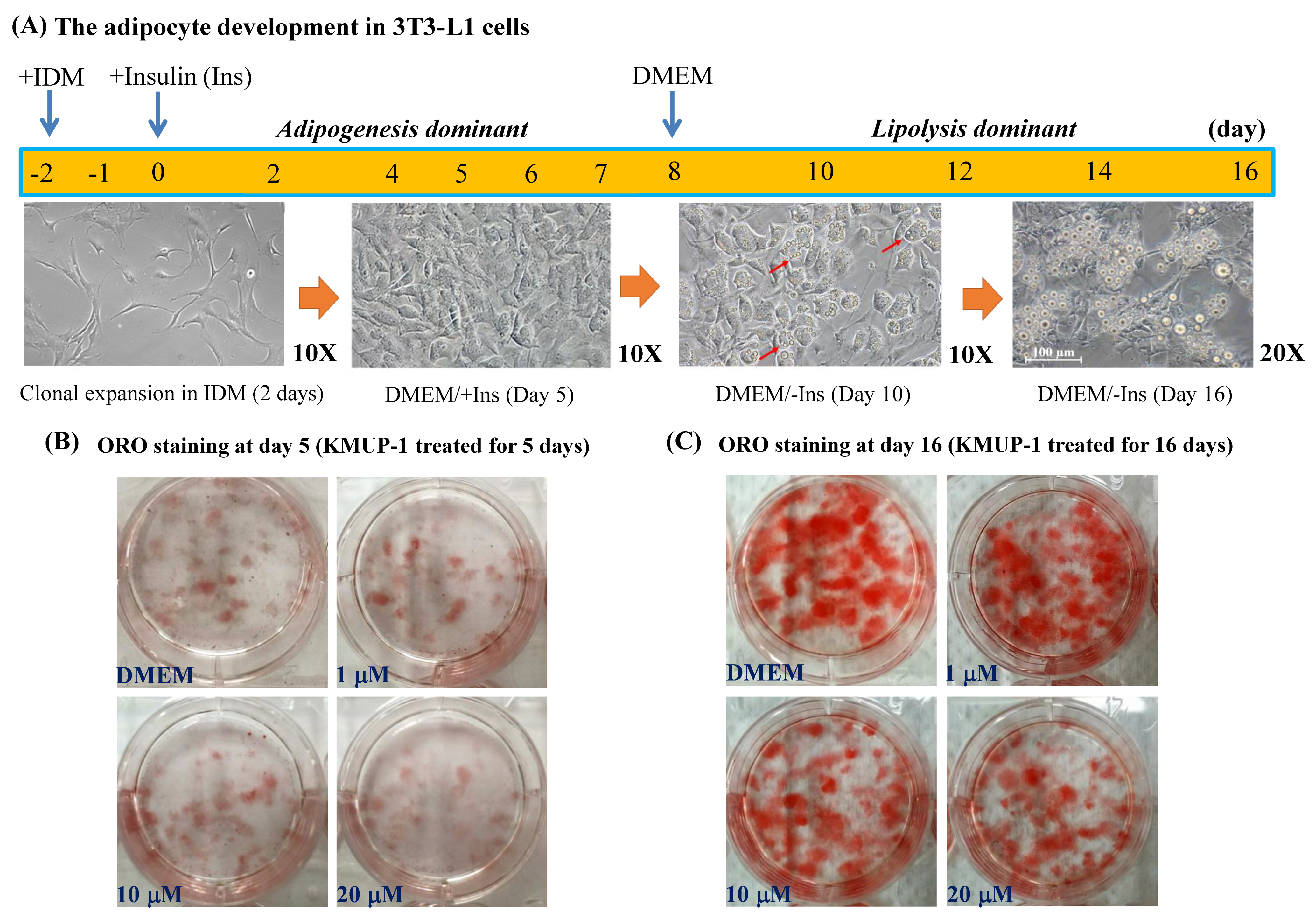
2.1. KMUP-1 and Ractopamine Affected the Adipocyte Development
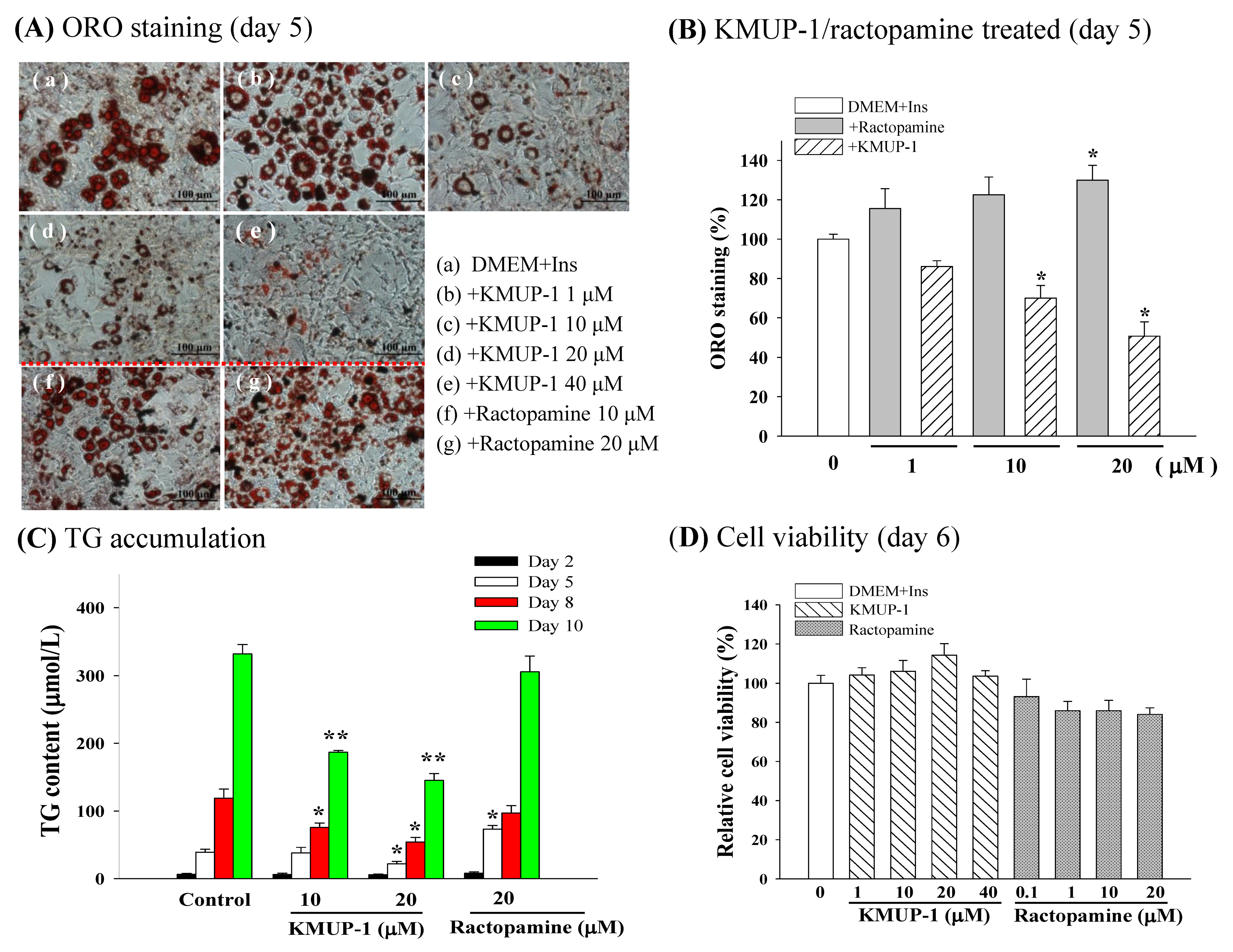
2.2. ORO Staining, TG Content, and Cell Viability
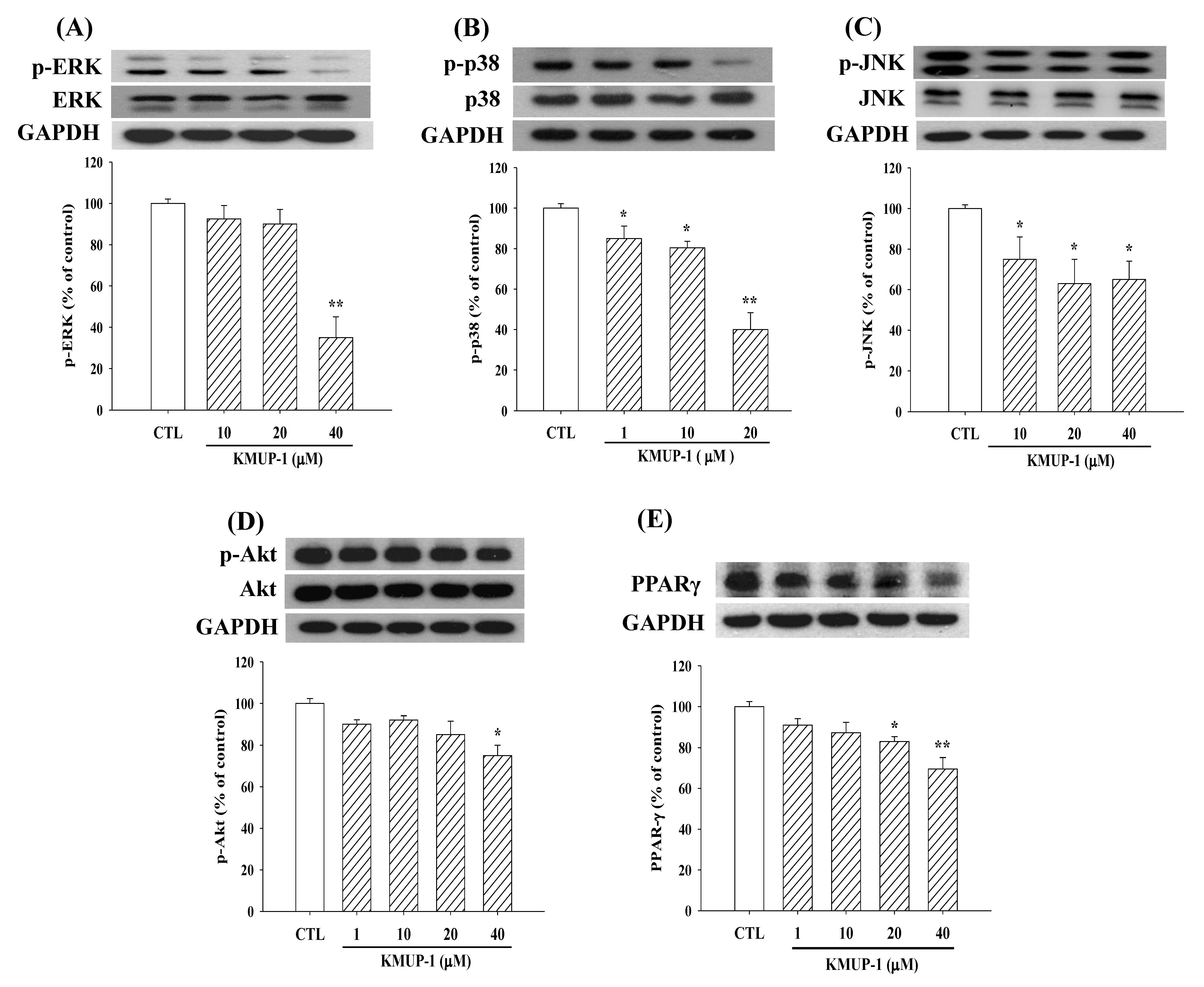
2.3. KMUP-1 Affected MAPKs/p-Akt/PPARγ Expression
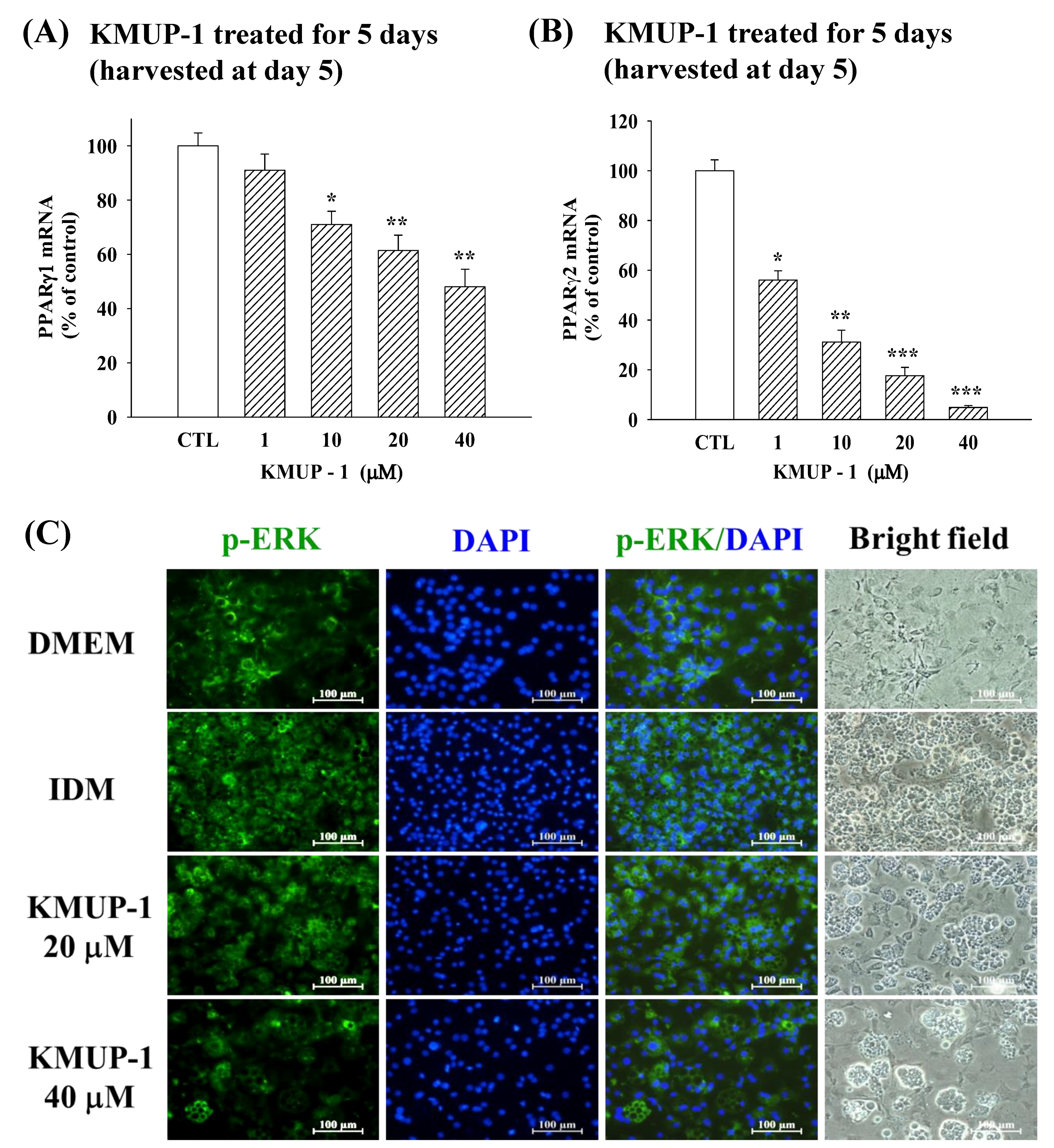
2.4. KMUP-1 Influenced PPARγ1/PPARγ2 mRNA and p-ERK Immunoreactivity
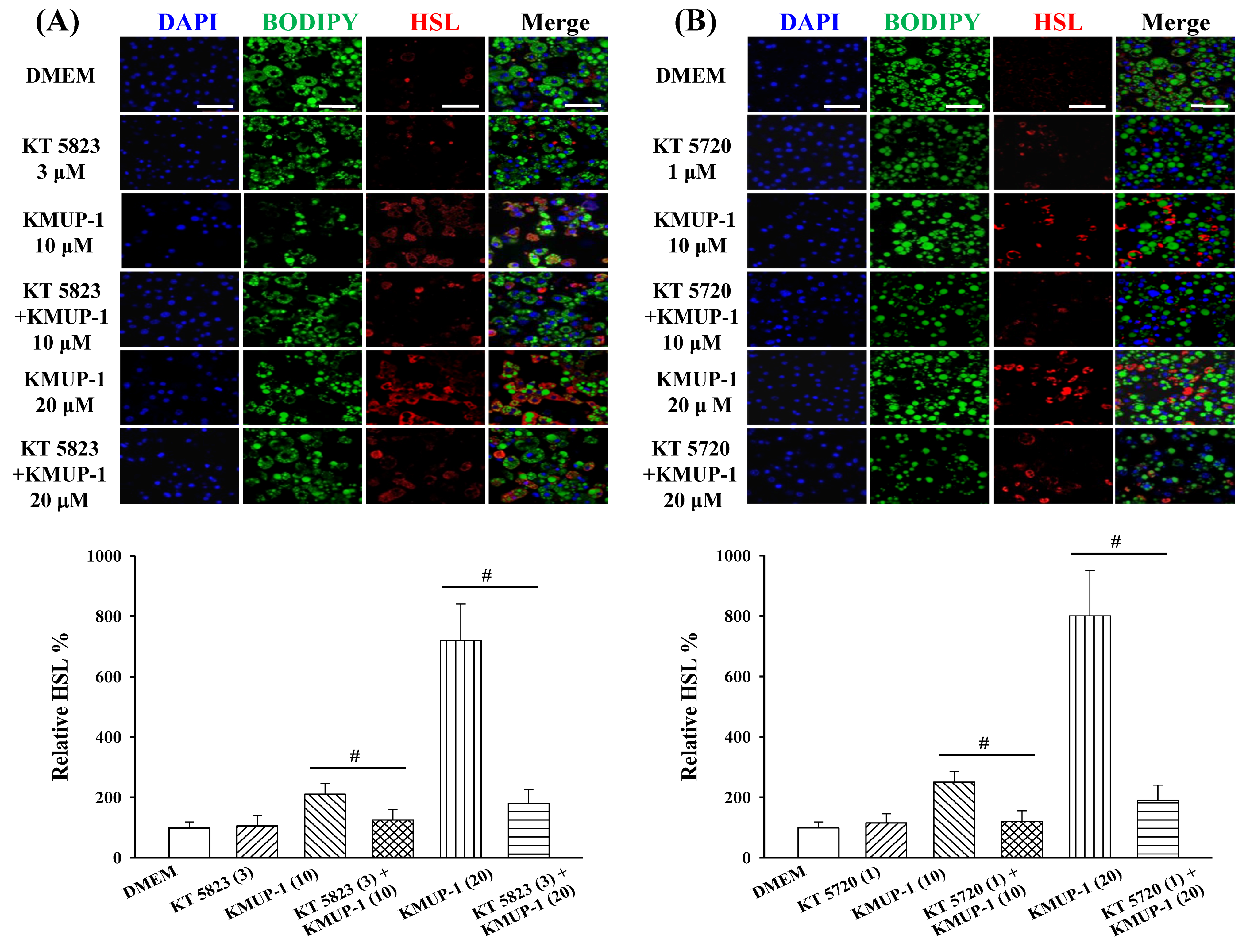
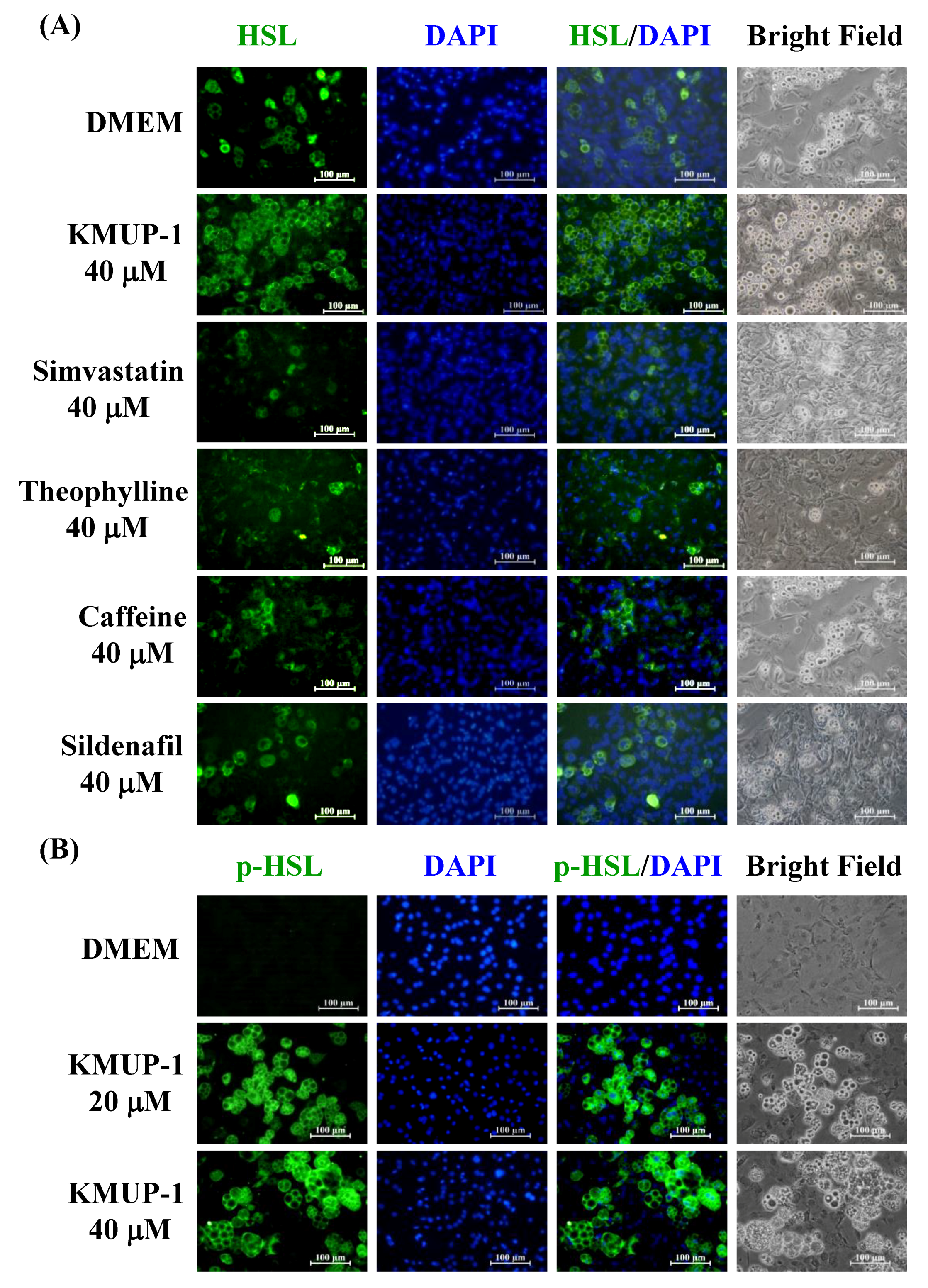
2.5. KMUP-1 Influenced HSL/p-HSL Immunoreactivity
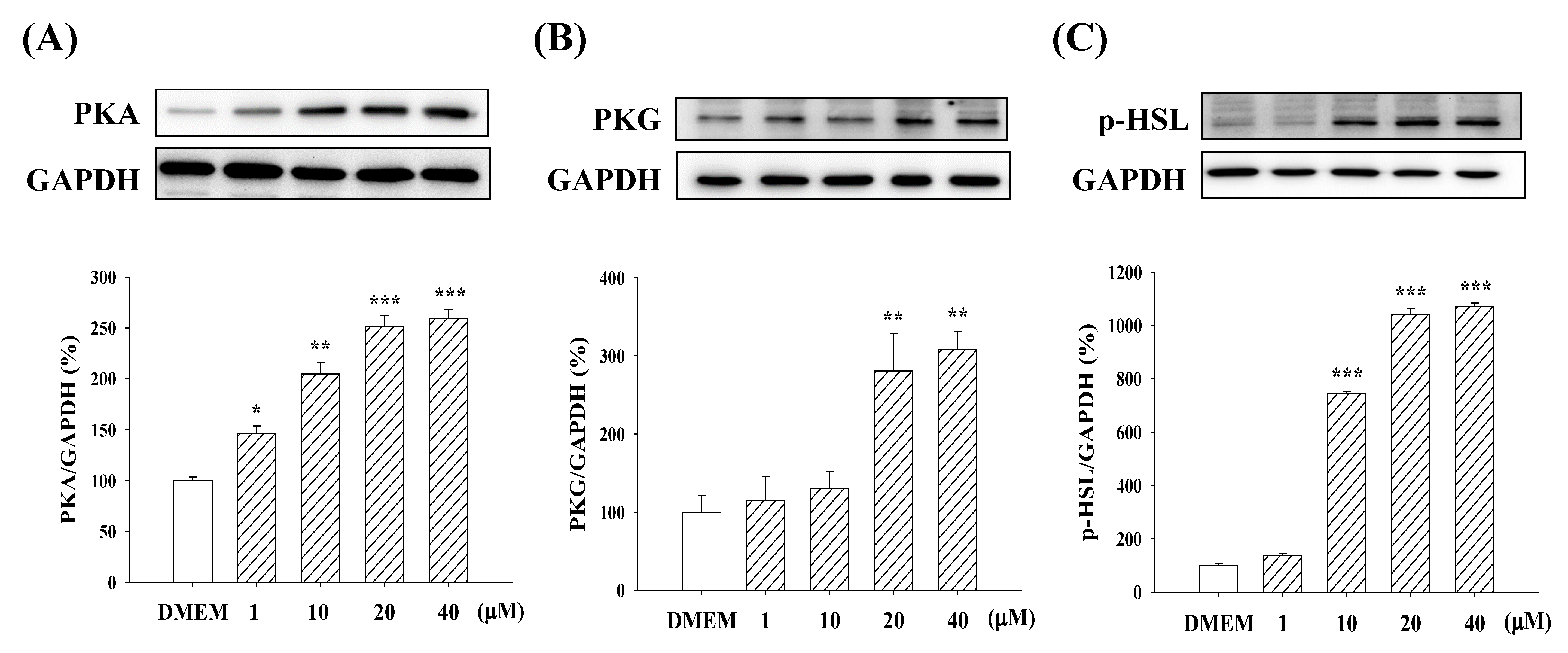
2.6. Expression of PKA/PKG and p-HSL
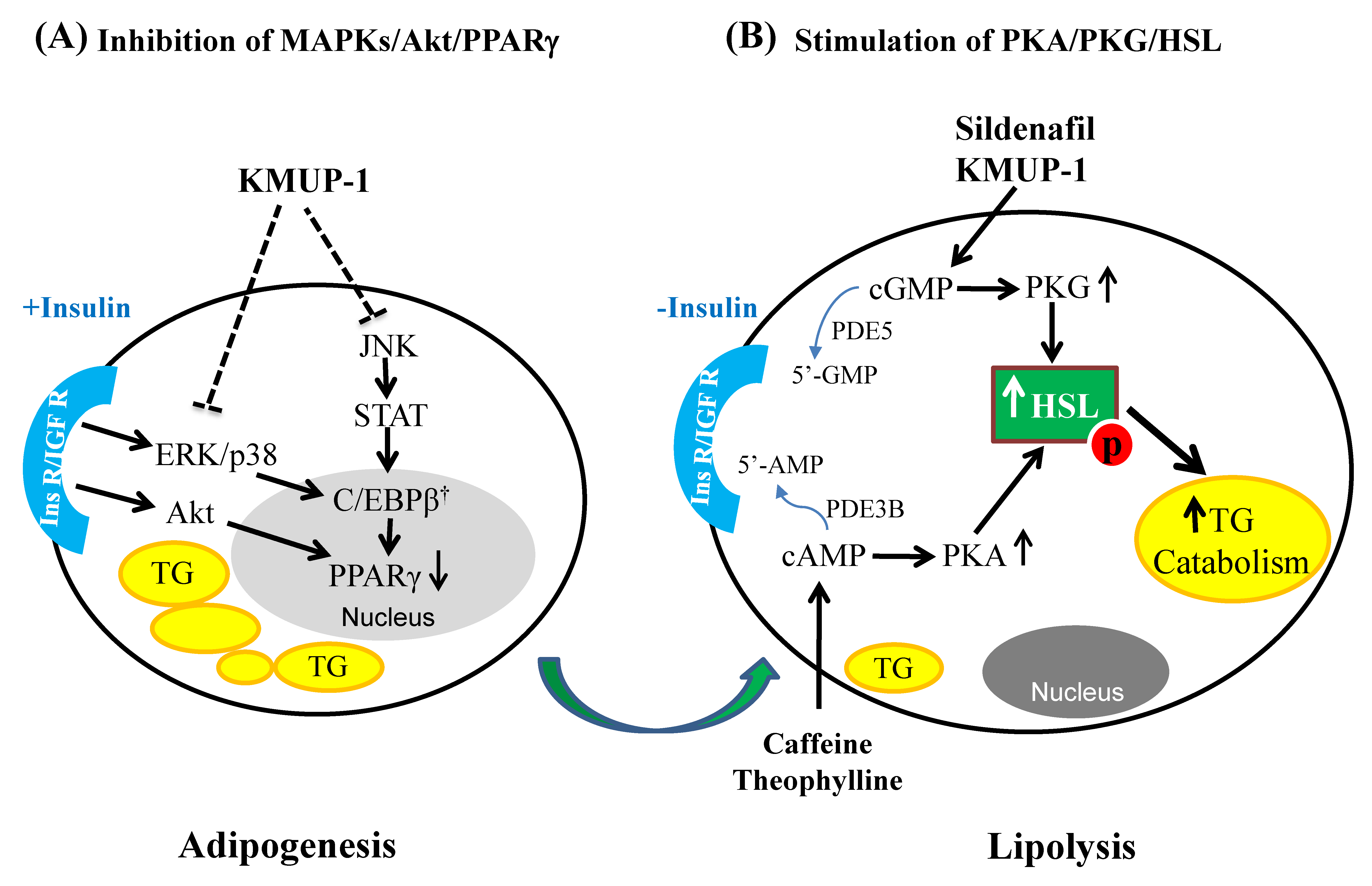
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Oil Red O Staining, TG Content and Cell Viability
4.3. Western Blotting Analysis
4.4. Real-Time Quantitative Polymerase Chain Reaction (qPCR)
4.5. Immunofluorescent Staining
4.6. Reagents
4.7. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kuo, K.K.; Wu, B.N.; Liu, C.P.; Yang, T.Y.; Kao, L.P.; Wu, J.R.; Lai, W.T.; Chen, I.J. Xanthine-based KMUP-1 improves HDL via PPARgamma/SR-B1, LDL via LDLRs, and HSL via PKA/PKG for hepatic fat loss. J. Lipid Res. 2015, 56, 2070–2084. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.N.; Kuo, K.K.; Chen, Y.H.; Chang, C.T.; Huang, H.T.; Chai, C.Y.; Dai, Z.K.; Chen, I.J. Theophylline-Based KMUP-1 Improves Steatohepatitis via MMP-9/IL-10 and Lipolysis via HSL/p-HSL in Obese Mice. Int. J. Mol. Sci. 2016, 17, 1345. [Google Scholar] [CrossRef] [PubMed]
- Yeh, J.L.; Hsu, J.H.; Wu, P.J.; Liou, S.F.; Liu, C.P.; Chen, I.J.; Wu, B.N.; Dai, Z.K.; Wu, J.R. KMUP-1 attenuates isoprenaline-induced cardiac hypertrophy in rats through NO/cGMP/PKG and ERK1/2/calcineurin A pathways. Br. J. Pharmacol. 2010, 159, 1151–1160. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.H.; Dai, Z.K.; Wu, B.N.; Yeh, J.L.; Chai, C.Y.; Chu, K.S.; Liu, C.P.; Chen, I.J. The xanthine derivative KMUP-1 inhibits models of pulmonary artery hypertension via increased NO and cGMP-dependent inhibition of RhoA/Rho kinase. Br. J. Pharmacol. 2010, 160, 971–986. [Google Scholar] [CrossRef] [PubMed]
- Rizzatti, V.; Boschi, F.; Pedrotti, M.; Zoico, E.; Sbarbati, A.; Zamboni, M. Lipid droplets characterization in adipocyte differentiated 3T3-L1 cells: Size and optical density distribution. Eur. J. Histochem. 2013, 57, e24. [Google Scholar] [CrossRef] [PubMed]
- Sohle, J.; Knott, A.; Holtzmann, U.; Siegner, R.; Gronniger, E.; Schepky, A.; Gallinat, S.; Wenck, H.; Stab, F.; Winnefeld, M. White Tea extract induces lipolytic activity and inhibits adipogenesis in human subcutaneous (pre)-adipocytes. Nutr. Metab. (Lond) 2009, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Rayalam, S.; Della-Fera, M.A.; Baile, C.A. Phytochemicals and regulation of the adipocyte life cycle. J. Nutr. Biochem. 2008, 19, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Abel, E.D. A new twist in the function of the cardiac lipid droplet. Nat. Med. 2011, 17, 1045–1046. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Nakatani, K.; Morioka, K.; Urakawa, H.; Maruyama, N.; Kitagawa, N.; Katsuki, A.; Araki-Sasaki, R.; Hori, Y.; Gabazza, E.C.; et al. Nitric oxide stimulates glucose transport through insulin-independent GLUT4 translocation in 3T3-L1 adipocytes. Eur. J. Endocrinol. 2003, 149, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Lafontan, M.; Moro, C.; Berlan, M.; Crampes, F.; Sengenes, C.; Galitzky, J. Control of lipolysis by natriuretic peptides and cyclic GMP. Trends Endocrinol. Metab. 2008, 19, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ji, J.; Yan, G.; Wu, J.; Sun, X.; Shen, J.; Jiang, H.; Wang, H. Sildenafil promotes adipogenesis through a PKG pathway. Biochem. Biophys. Res Commun. 2010, 396, 1054–1059. [Google Scholar] [CrossRef] [PubMed]
- Ayala, J.E.; Bracy, D.P.; Julien, B.M.; Rottman, J.N.; Fueger, P.T.; Wasserman, D.H. Chronic treatment with sildenafil improves energy balance and insulin action in high fat-fed conscious mice. Diabetes 2007, 56, 1025–1033. [Google Scholar] [CrossRef] [PubMed]
- Rios, N.B.; Esparragon, F.R.; Rodriguez Perez, J.C. Telmisartan-induced eNOS gene expression is partially independent of its PPAR-gamma agonist property. Clin. Invest. Med. 2012, 35, 55–64. [Google Scholar] [CrossRef]
- Armani, A.; Marzolla, V.; Rosano, G.M.; Fabbri, A.; Caprio, M. Phosphodiesterase type 5 (PDE5) in the adipocyte: A novel player in fat metabolism? Trends Endocrinol. Metab. 2011, 22, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Chaves, V.E.; Frasson, D.; Kawashita, N.H. Several agents and pathways regulate lipolysis in adipocytes. Biochimie 2011, 93, 1631–1640. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, M.; Osuga, J.; Yahagi, N.; Okazaki, H.; Tamura, Y.; Igarashi, M.; Takase, S.; Harada, K.; Okazaki, S.; Iizuka, Y.; et al. Hormone-sensitive lipase is involved in hepatic cholesteryl ester hydrolysis. J. Lipid Res. 2008, 49, 1829–1838. [Google Scholar] [CrossRef] [PubMed]
- Jeninga, E.H.; Bugge, A.; Nielsen, R.; Kersten, S.; Hamers, N.; Dani, C.; Wabitsch, M.; Berger, R.; Stunnenberg, H.G.; Mandrup, S.; et al. Peroxisome proliferator-activated receptor gamma regulates expression of the anti-lipolytic G-protein-coupled receptor 81 (GPR81/Gpr81). J. Biol. Chem. 2009, 284, 26385–26393. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, J.Z.; Hu, J.X.; Wu, H.; Li, Y.L.; Chen, H.L.; Bai, H.; Hai, C.X. ROS-activated p38 MAPK/ERK-Akt cascade plays a central role in palmitic acid-stimulated hepatocyte proliferation. Free Radic. Biol. Med. 2011, 51, 539–551. [Google Scholar] [CrossRef] [PubMed]
- Kogure, A.; Sakane, N.; Takakura, Y.; Umekawa, T.; Yoshioka, K.; Nishino, H.; Yamamoto, T.; Kawada, T.; Yoshikawa, T.; Yoshida, T. Effects of caffeine on the uncoupling protein family in obese yellow KK mice. Clin. Exp. Pharmacol. Physiol. 2002, 29, 391–394. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.R.; Yoon, B.K.; Park, H.; Seok, J.W.; Choi, H.; Yu, J.H.; Choi, Y.; Song, S.J.; Kim, A.; Kim, J.W. Caffeine inhibits adipogenesis through modulation of mitotic clonal expansion and the AKT/GSK3 pathway in 3T3-L1 adipocytes. BMB Rep. 2016, 49, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Yang, L.; Xu, F.; Lin, L.; Zheng, G. Combination therapy with catechins and caffeine inhibits fat accumulation in 3T3-L1 cells. Exp. Ther. Med. 2017, 13, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Hauner, H.; Skurk, T.; Wabitsch, M. Cultures of human adipose precursor cells. Methods Mol. Biol. 2001, 155, 239–247. [Google Scholar] [PubMed]
- Bost, F.; Aouadi, M.; Caron, L.; Binetruy, B. The role of MAPKs in adipocyte differentiation and obesity. Biochimie 2005, 87, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Engelman, J.A.; Lisanti, M.P.; Scherer, P.E. Specific inhibitors of p38 mitogen-activated protein kinase block 3T3-L1 adipogenesis. J. Biol. Chem. 1998, 273, 32111–32120. [Google Scholar] [CrossRef] [PubMed]
- Jaeschke, A.; Czech, M.P.; Davis, R.J. An essential role of the JIP1 scaffold protein for JNK activation in adipose tissue. Genes Dev. 2004, 18, 1976–1980. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Liao, K. Protein kinase B/AKT 1 plays a pivotal role in insulin-like growth factor-1 receptor signaling induced 3T3-L1 adipocyte differentiation. J. Biol. Chem. 2004, 279, 35914–35922. [Google Scholar] [CrossRef] [PubMed]
- DiPilato, L.M.; Ahmad, F.; Harms, M.; Seale, P.; Manganiello, V.; Birnbaum, M.J. The Role of PDE3B phosphorylation in the inhibition of lipolysis by insulin. Mol. Cell Biol. 2015, 35, 2752–2760. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, B.; Gogg, S.; Hedjazifar, S.; Jenndahl, L.; Hammarstedt, A.; Smith, U. Inflammation and impaired adipogenesis in hypertrophic obesity in man. Am. J. Physiol. Endocrinol. Metab. 2009, 297, E999–E1003. [Google Scholar] [CrossRef] [PubMed]
- Fernyhough, M.E.; Okine, E.; Hausman, G.; Vierck, J.L.; Dodson, M.V. PPARgamma and GLUT-4 expression as developmental regulators/markers for preadipocyte differentiation into an adipocyte. Domest. Anim. Endocrinol. 2007, 33, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Keshet, R.; Bryansker Kraitshtein, Z.; Shanzer, M.; Adler, J.; Reuven, N.; Shaul, Y. c-Abl tyrosine kinase promotes adipocyte differentiation by targeting PPAR-gamma 2. Proc. Natl. Acad. Sci. USA 2014, 111, 16365–16370. [Google Scholar] [CrossRef] [PubMed]
- De Almeida, V.V.; Nuñez, A.J.C.; Miyada, V.S. Ractopamine as a metabolic modifier feed additive for finishing pigs: A review. Braz. Arch. Biol. Techn. 2012, 55, 445–456. [Google Scholar] [CrossRef]
- Mita, T.; Furuhashi, M.; Hiramitsu, S.; Ishii, J.; Hoshina, K.; Ishimura, S.; Fuseya, T.; Watanabe, Y.; Tanaka, M.; Ohno, K.; et al. FABP4 is secreted from adipocytes by adenyl cyclase-PKA- and guanylyl cyclase-PKG-dependent lipolytic mechanisms. Obesity 2015, 23, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Ray, H.; Pinteur, C.; Frering, V.; Beylot, M.; Large, V. Depot-specific differences in perilipin and hormone-sensitive lipase expression in lean and obese. Lipids Health Dis. 2009, 8, 58. [Google Scholar] [CrossRef] [PubMed]
- Bray, G.A.; Greenway, F.L. Current and potential drugs for treatment of obesity. Endocr. Rev. 1999, 20, 805–875. [Google Scholar] [CrossRef] [PubMed]
- Mitschke, M.M.; Hoffmann, L.S.; Gnad, T.; Scholz, D.; Kruithoff, K.; Mayer, P.; Haas, B.; Sassmann, A.; Pfeifer, A.; Kilic, A. Increased cGMP promotes healthy expansion and browning of white adipose tissue. FASEB J. 2013, 27, 1621–1630. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.-P.; Chau, P.-C.; Chang, C.-T.; An, L.-M.; Yeh, J.-L.; Chen, I.-J.; Wu, B.-N. KMUP-1, a GPCR Modulator, Attenuates Triglyceride Accumulation Involved MAPKs/Akt/PPARγ and PKA/PKG/HSL Signaling in 3T3-L1 Preadipocytes. Molecules 2018, 23, 2433. https://doi.org/10.3390/molecules23102433
Liu C-P, Chau P-C, Chang C-T, An L-M, Yeh J-L, Chen I-J, Wu B-N. KMUP-1, a GPCR Modulator, Attenuates Triglyceride Accumulation Involved MAPKs/Akt/PPARγ and PKA/PKG/HSL Signaling in 3T3-L1 Preadipocytes. Molecules. 2018; 23(10):2433. https://doi.org/10.3390/molecules23102433
Chicago/Turabian StyleLiu, Chung-Pin, Pei-Chun Chau, Chain-Ting Chang, Li-Mei An, Jwu-Lai Yeh, Ing-Jun Chen, and Bin-Nan Wu. 2018. "KMUP-1, a GPCR Modulator, Attenuates Triglyceride Accumulation Involved MAPKs/Akt/PPARγ and PKA/PKG/HSL Signaling in 3T3-L1 Preadipocytes" Molecules 23, no. 10: 2433. https://doi.org/10.3390/molecules23102433
APA StyleLiu, C.-P., Chau, P.-C., Chang, C.-T., An, L.-M., Yeh, J.-L., Chen, I.-J., & Wu, B.-N. (2018). KMUP-1, a GPCR Modulator, Attenuates Triglyceride Accumulation Involved MAPKs/Akt/PPARγ and PKA/PKG/HSL Signaling in 3T3-L1 Preadipocytes. Molecules, 23(10), 2433. https://doi.org/10.3390/molecules23102433





