Data-Driven Exploration of Selectivity and Off-Target Activities of Designated Chemical Probes
Abstract
1. Introduction
2. Results and Discussion
2.1. Qualifying Chemical Probes
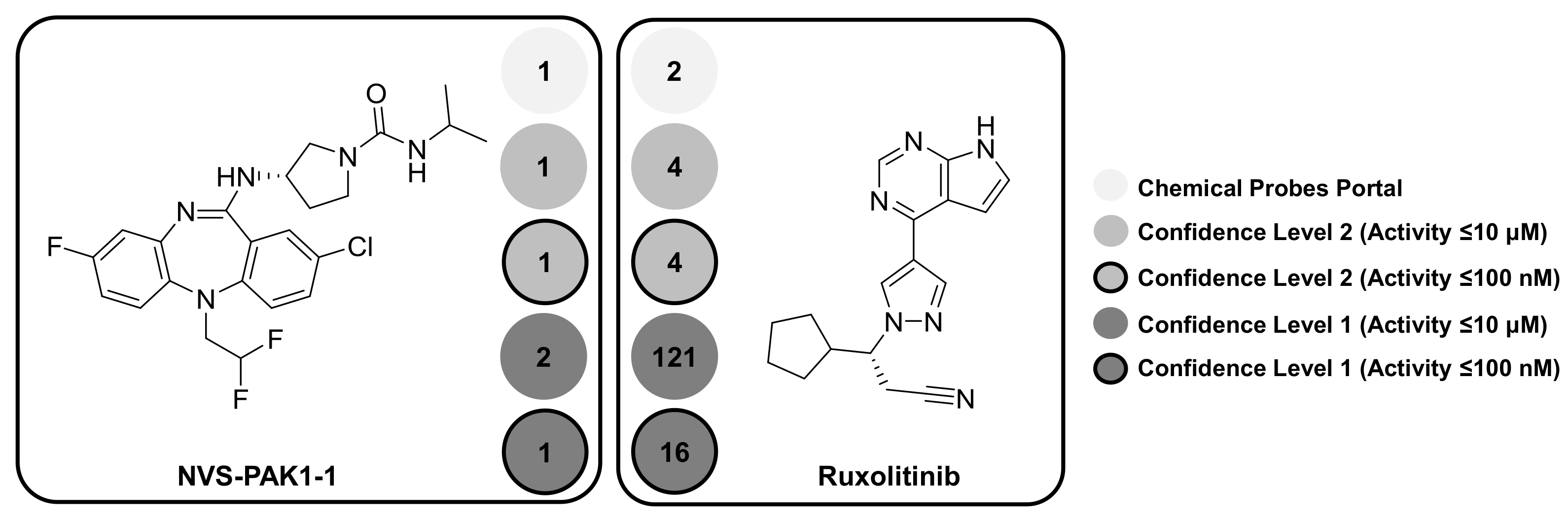
2.2. Selectivity Trends of Chemical Probes
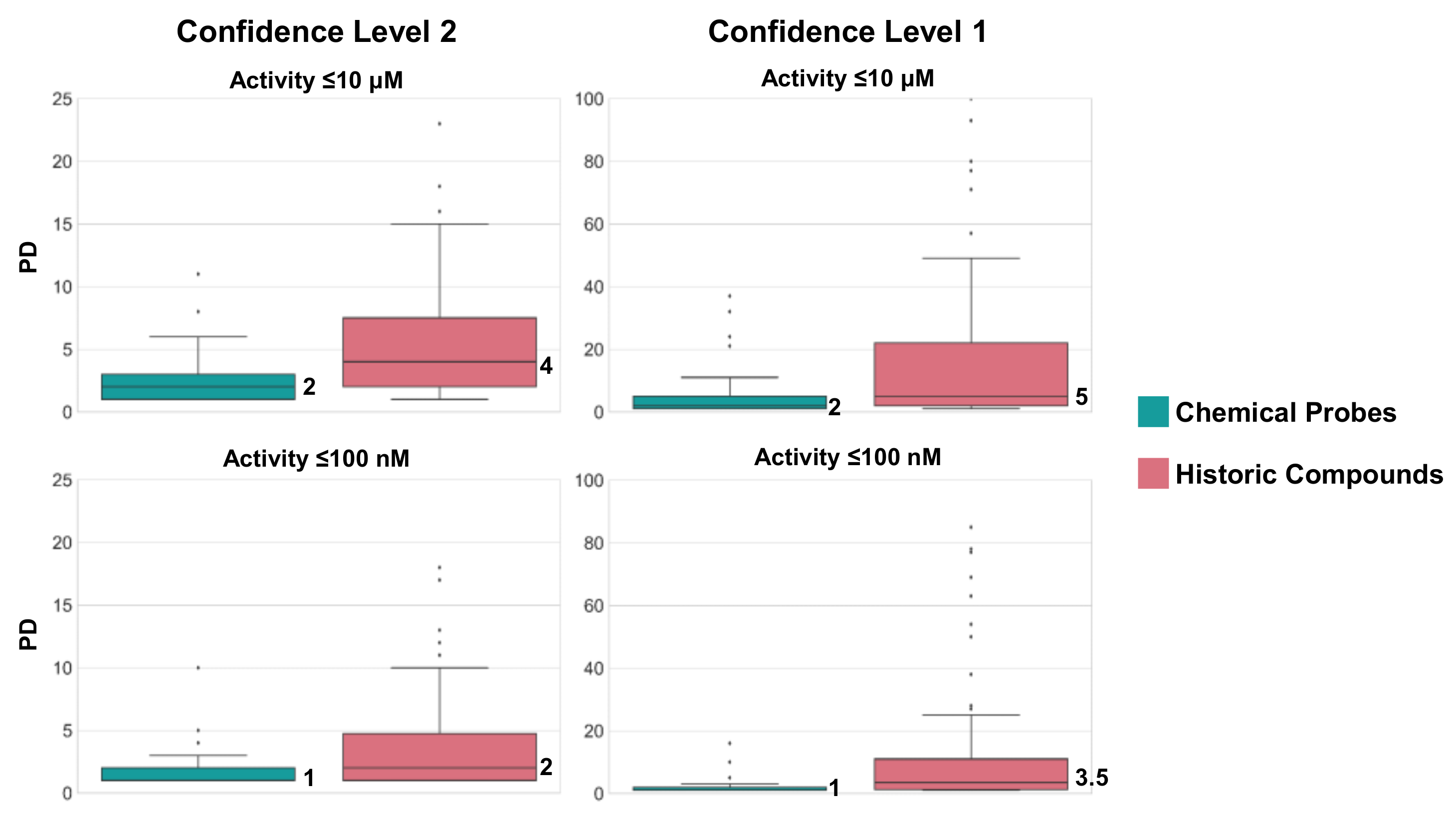
2.3. Chemical Probes and Historic Compounds
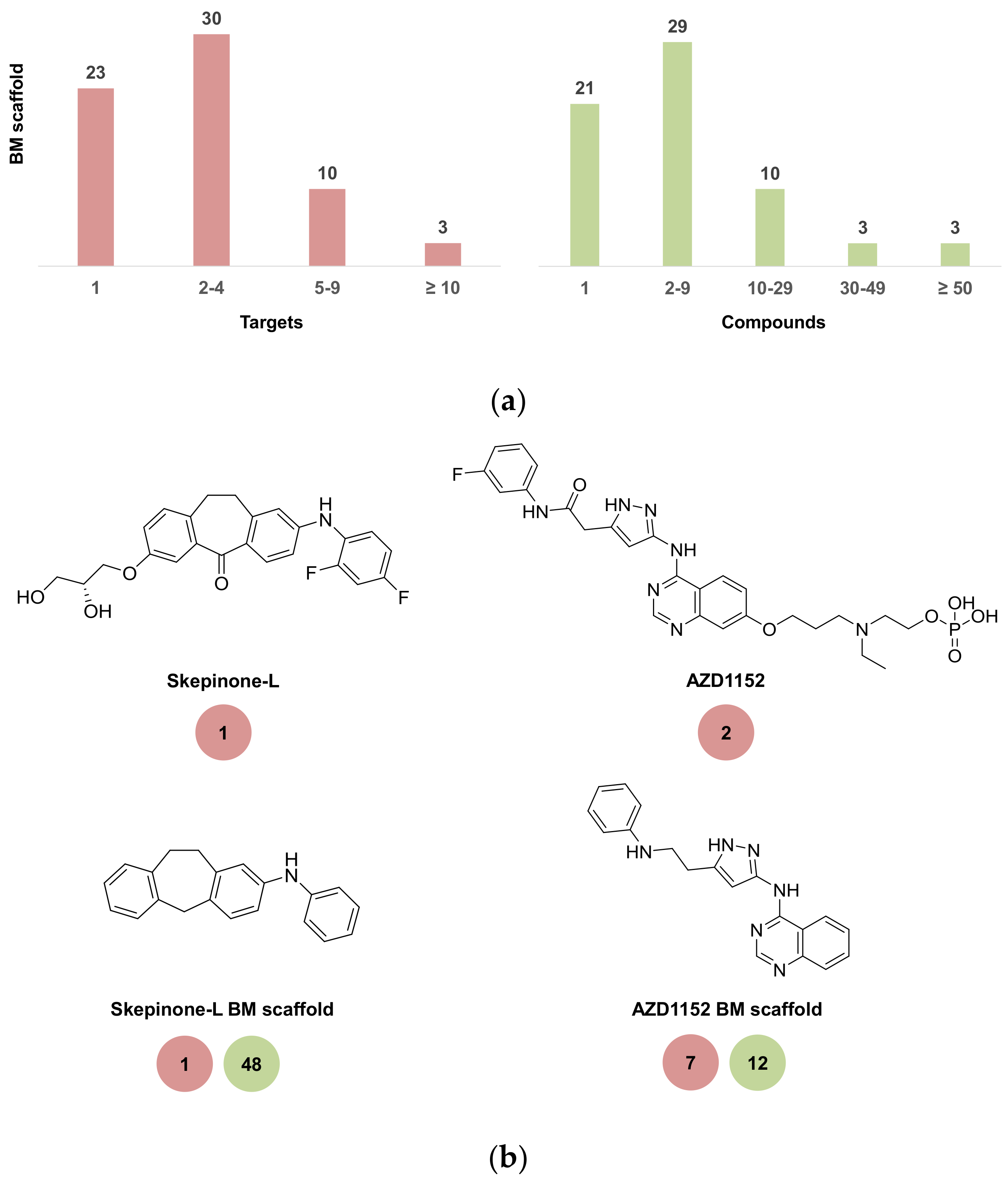
2.4. Scaffold Analysis of Chemical Probes
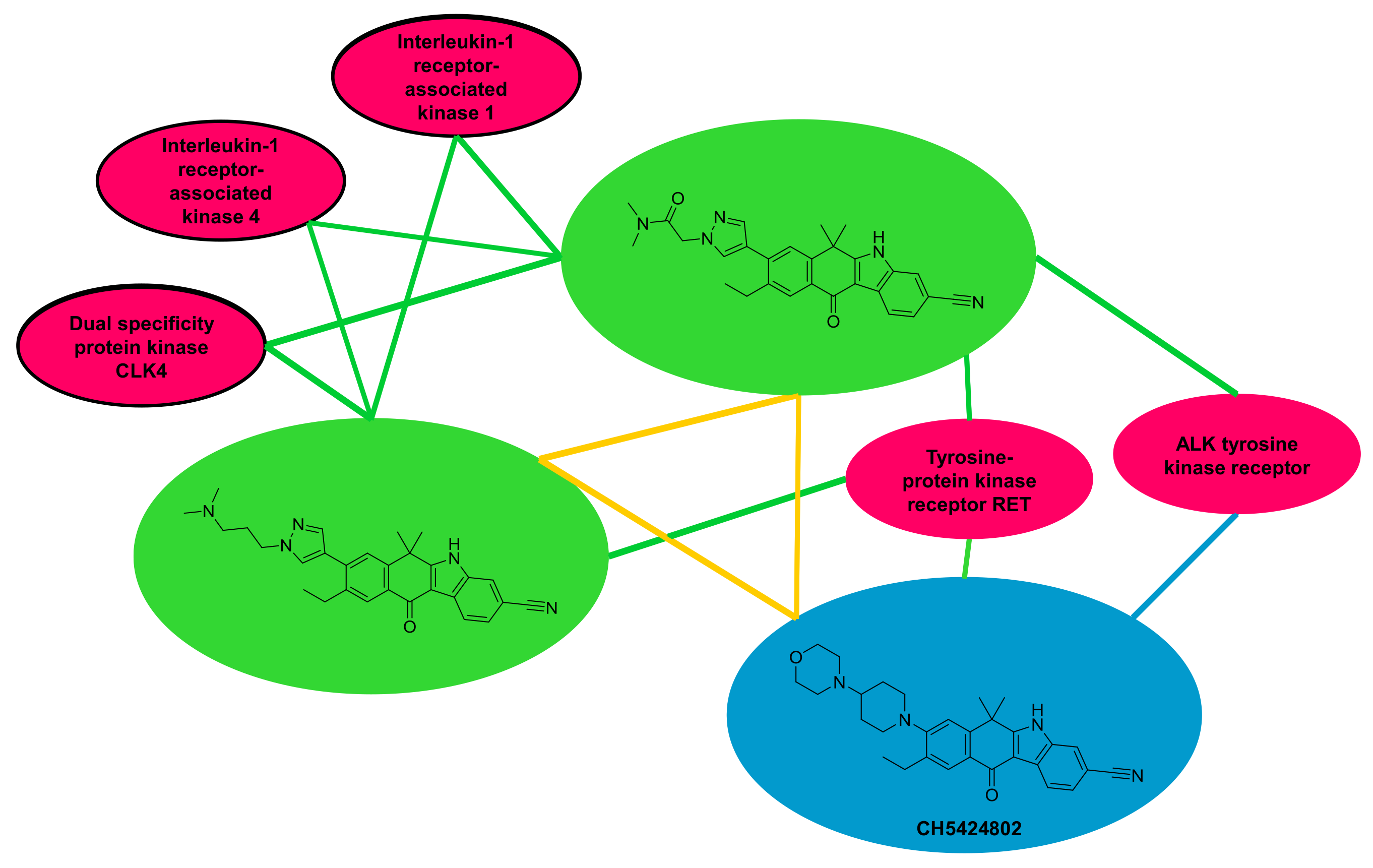
2.5. Off-Target Activity Assessment in Networks
2.6. Summary
3. Materials and Methods
3.1. Chemical Probes
3.2. Activity Data, Confidence Levels, and Historic Compounds
3.3. Bioactive Compounds, Scaffold Analysis, and Off-Target Predictions
Author Contributions
Conflicts of Interest
References
- International Human Genome Sequencing Consortium. Initial Sequencing and Analysis of the Human Genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- International Human Genome Sequencing Consortium. Finishing the Euchromatic Sequence of the Human Genome. Nature 2004, 431, 931–945. [Google Scholar] [CrossRef] [PubMed]
- Schenone, M.; Dančík, V.; Wagner, B.K.; Clemons, P.A. Target Identification and Mechanism of Action in Chemical Biology and Drug Discovery. Nat. Chem. Biol. 2013, 9, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Cornish, P.V.; Ha, T. A Survey of Single-Molecule Techniques in Chemical Biology. ACS Chem. Biol. 2007, 2, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Knowles, J.; Gromo, G. Target Selection in Drug Discovery. Nat. Rev. Drug Discov. 2003, 2, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.M.; Isserlin, R.; Bader, G.D.; Frye, S.V.; Willson, T.M.; Yu, F.H. Too Many Roads Not Taken. Nature 2011, 470, 163–165. [Google Scholar] [CrossRef] [PubMed]
- Oprea, T.I.; Bologa, C.G.; Brunak, S.; Campbell, A.; Gan, G.N.; Gaulton, A.; Gomez, S.M.; Guha, R.; Hersey, A.; Holmes, J.; et al. Unexplored Therapeutic Opportunities in the Human Genome. Nat. Rev. Drug Discov. 2018, 17, 317–332. [Google Scholar] [CrossRef] [PubMed]
- Bunnage, M.E.; Chekler, E.L.P.; Jones, L.H. Target Validation Using Chemical Probes. Nat. Chem. Biol. 2013, 9, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.H.; Bunnage, M.E. Applications of Chemogenomic Library Screening in Drug Discovery. Nat. Rev. Drug Discov. 2017, 16, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Simon, G.M.; Niphakis, M.J.; Cravatt, B.F. Determining Target Engagement in Living Systems. Nat. Chem. Biol. 2013, 9, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Frye, S.V. The Art of the Chemical Probe. Nat. Chem. Biol. 2010, 6, 159–161. [Google Scholar] [CrossRef] [PubMed]
- Workman, P.; Collins, I. Probing the Probes: Fitness Factors for Small Molecule Tools. Chem. Biol. 2010, 17, 561–577. [Google Scholar] [CrossRef] [PubMed]
- Arrowsmith, C.H.; Audia, J.E.; Austin, C.; Baell, J.; Bennett, J.; Blagg, J.; Bountra, C.; Brennan, P.E.; Brown, P.J.; Bunnage, M.E.; et al. The Promise and Peril of Chemical Probes. Nat. Chem. Biol. 2015, 11, 536–541. [Google Scholar] [CrossRef] [PubMed]
- Chemical Probes Portal. Available online: http://www.chemicalprobes.org/ (accessed on 14 July 2018).
- Structural Genomics Consortium. Available online: https://www.thesgc.org/ (accessed on 22 August 2018).
- Gaulton, A.; Bellis, L.J.; Bento, A.P.; Chambers, J.; Davies, M.; Hersey, A.; Light, Y.; McGlinchey, S.; Michalovich, D.; Al-Lazikani, B.; et al. ChEMBL: A Large-scale Bioactivity Database for Drug Discovery. Nucleic Acids Res. 2012, 40, D1100–D1107. [Google Scholar] [CrossRef] [PubMed]
- Bemis, G.W.; Murcko, M.A. The Properties of Known Drugs. 1. Molecular Frameworks. J. Med. Chem. 1996, 39, 2887–2893. [Google Scholar] [CrossRef] [PubMed]
- Kunimoto, R.; Bajorath, J. Design of a Tripartite Network for the Prediction of Drug Targets. J. Comput. Aided. Mol. Des. 2018, 32, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The Protein Kinase Complement of the Human Genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [PubMed]
- Miljković, F.; Bajorath, J. Exploring Selectivity of Multikinase Inhibitors across the Human Kinome. ACS Omega 2018, 3, 1147–1153. [Google Scholar] [CrossRef] [PubMed]
- Miljković, F.; Bajorath, J. Reconciling Selectivity Trends from a Comprehensive Kinase Inhibitor Profiling Campaign with Known Activity Data. ACS Omega 2018, 3, 3113–3119. [Google Scholar] [CrossRef] [PubMed]
- Miljković, F.; Bajorath, J. Evaluation of Kinase Inhibitor Selectivity Using Cell-Based Profiling Data. Mol. Inform. 2018, 37, 1800024. [Google Scholar] [CrossRef] [PubMed]
- Karpov, A.S.; Amiri, P.; Bellamacina, C.; Bellance, M.-H.; Breitenstein, W.; Daniel, D.; Denay, R.; Fabbro, D.; Fernandez, C.; Galuba, I.; et al. Optimization of a Dibenzodiazepine Hit to a Potent and Selective Allosteric PAK1 Inhibitor. ACS Med. Chem. Lett. 2015, 6, 776–781. [Google Scholar] [CrossRef] [PubMed]
- Quintás-Cardama, A.; Vaddi, K.; Liu, P.; Manshouri, T.; Li, J.; Scherle, P.A.; Caulder, E.; Wen, X.; Li, Y.; Waeltz, P.; et al. Preclinical Characterization of the Selective JAK1/2 Inhibitor INCB018424: Therapeutic Implications for the Treatment of Myeloproliferative Neoplasms. Blood 2010, 115, 3109–3117. [Google Scholar] [CrossRef] [PubMed]
- Koeberle, S.C.; Fischer, S.; Schollmeyer, D.; Schattel, V.; Grütter, C.; Rauh, D.; Laufer, S.A. Design, Synthesis, and Biological Evaluation of Novel Disubstituted Dibenzosuberones as Highly Potent and Selective Inhibitors of p38 Mitogen Activated Protein Kinase. J. Med. Chem. 2012, 55, 5868–5877. [Google Scholar] [CrossRef] [PubMed]
- Koeberle, S.C.; Romir, J.; Fischer, S.; Koeberle, A.; Schattel, V.; Albrecht, W.; Grütter, C.; Werz, O.; Rauh, D.; Stehle, T.; et al. Skepinone-L is a Selective p38 Mitogen-activated Protein Kinase Inhibitor. Nat. Chem. Biol. 2012, 8, 141–143. [Google Scholar] [CrossRef] [PubMed]
- Mortlock, A.A.; Foote, K.M.; Heron, N.M.; Jung, F.H.; Pasquet, G.; Lohmann, J.-J.M.; Warin, N.; Renaud, F.; De Savi, C.; Roberts, N.J.; et al. Discovery, Synthesis, and In Vivo Activity of a New Class of Pyrazoloquinazolines as Selective Inhibitors of Aurora B Kinase. J. Med. Chem. 2007, 50, 2213–2224. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ikezoe, T.; Nishioka, C.; Tasaka, T.; Taniguchi, A.; Kuwayama, Y.; Komatsu, N.; Bandobashi, K.; Togitani, K.; Koeffler, H.P.; et al. AZD1152, a Novel and Selective Aurora B Kinase Inhibitor, Induces Growth Arrest, Apoptosis, and Sensitization for Tubulin Depolymerizing Agent or Topoisomerase II Inhibitor in Human Acute Leukemia Cells In Vitro and In Vivo. Blood 2007, 110, 2034–2040. [Google Scholar] [CrossRef] [PubMed]
- Kenny, P.W.; Sadowski, J. Structure Modification in Chemical Databases. In Chemoinformatics in Drug Discovery; Wiley-Blackwell: Hoboken, NJ, USA, 2005; pp. 271–285. [Google Scholar]
- Stumpfe, D.; Dimova, D.; Bajorath, J. Computational Method for the Systematic Identification of Analog Series and Key Compounds Representing Series and Their Biological Activity Profiles. J. Med. Chem. 2016, 59, 7667–7676. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, H.; Tsukaguchi, T.; Hiroshima, S.; Kodama, T.; Kobayashi, T.; Fukami, T.A.; Oikawa, N.; Tsukuda, T.; Ishii, N.; Aoki, Y. CH5424802, a Selective ALK Inhibitor Capable of Blocking the Resistant Gatekeeper Mutant. Cancer Cell 2011, 19, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.-L.; Ideker, T. Cytoscape 2.8: New Features for Data Integration and Network Visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |


| Target Class | Chemical Probes | Target-Based Categories |
|---|---|---|
| Protein kinases | 33 | Chemical probes for kinase targets |
| Lipid kinases | 1 | |
| Epigenetics | 16 | Chemical probes for non-kinase targets |
| Other post-translation modification proteins | 13 | |
| Other proteins | 3 | |
| Structural proteins | 1 |
| Chemical Probe | Confidence Level 2 | Confidence Level 1 | ||
|---|---|---|---|---|
| ≤10 μM | ≤100 nM | ≤10 μM | ≤100 nM | |
| BMS-911543 | 6 (2) 1 | 3 | 6 (2) | 3 |
| BYL-719 | 4 (2) | 1 | 8 (3) | 1 |
| CCT244747 | 2 (1) | 1 | 2 (1) | 1 |
| CCT251545 | 4 (1) | 2 (1) | 4 (1) | 2 (1) |
| P505-15 | 6 (1) | 3 | 6 (1) | 3 |
| PF3644022 | 2 (1) | 1 | 2 (1) | 1 |
| Rapamycin | 5 (4) | 2 (1) | 7 (6) | 3 (2) |
| Ruxolitinib | 4 | 4 | 121 (1) | 16 |
| SGX-523 | 1 | 1 | 8 (1) | 1 |
| VX-745 | 3 (2) | 1 | 24 (2) | 2 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miljković, F.; Bajorath, J. Data-Driven Exploration of Selectivity and Off-Target Activities of Designated Chemical Probes. Molecules 2018, 23, 2434. https://doi.org/10.3390/molecules23102434
Miljković F, Bajorath J. Data-Driven Exploration of Selectivity and Off-Target Activities of Designated Chemical Probes. Molecules. 2018; 23(10):2434. https://doi.org/10.3390/molecules23102434
Chicago/Turabian StyleMiljković, Filip, and Jürgen Bajorath. 2018. "Data-Driven Exploration of Selectivity and Off-Target Activities of Designated Chemical Probes" Molecules 23, no. 10: 2434. https://doi.org/10.3390/molecules23102434
APA StyleMiljković, F., & Bajorath, J. (2018). Data-Driven Exploration of Selectivity and Off-Target Activities of Designated Chemical Probes. Molecules, 23(10), 2434. https://doi.org/10.3390/molecules23102434






