Influence of the Bioactive Diet Components on the Gene Expression Regulation
Abstract
:1. Introduction
2. Mechanisms of Diet Components and Gene Expression Interaction
2.1. Chromatin Structure (Including DNA Methylation, Histone Modification, Telomere Length)
2.2. Non-Coding RNA (microRNA and lnc-RNA)
3. Activation of Transcription Factors by Nutrients
4. The Influence of Bioactive Diet Components on Diseases
5. Application of Bioactive Diet Components in Dietician’s Work
Limitations to the Studies on the Influence of the Bioactive Diet Components on the Gene Expression Regulation
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AAR | amino acid response |
| AARE | amino acid response element |
| AID/APOBEC proteins | activation-induced cytidine deaminase/apolipoprotein B mRNA-editing enzyme complex |
| ACC | acetyl-CoA carboxylase |
| AP-1 | activator protein 1 |
| ATF4 | activating transcription factor 4 |
| BANCR | BRAF-Activated Non-Protein Coding RNA |
| bHLH | Zip-basic-helix–loop–helix–leucine zipper |
| CAC | colitis-associated cancer |
| C/EBP | enhancer-binding protein |
| ChoRE | carbohydrate response element motifs |
| ChREBP | carbohydrate-responsive element binding protein |
| COX-2 | cyclooxygenase-2 |
| DHA | docosahexaenoic acid |
| DNA-BER | DNA-Base Excision Repair |
| DNMT | DNA methyltransferase |
| EGCG | Epigallocatechin-3-gallate |
| eIF2 | eukaryotic initiation factor 2 |
| EPA | eicosapentaenoic acid |
| ER | endoplasmic reticulum |
| FAs | fatty acids |
| FAS | fatty acid synthase |
| FASKOL | fatty acid synthase knockout in liver |
| FOXP3 | forkhead box P3 |
| FPP | farnesyl-diphosphate |
| FXR | farnesoid X receptor |
| GCN2 | general control nonderepressible 2 |
| GI barrier | gastrointestinal barrier |
| HAT | histone acetylase |
| HDAC | histone deacetylase |
| HDM | histone demethylases |
| HMG-CoA | 3-hydroxy-3-methylglutaryl-CoA |
| HMT | methyltransferases |
| HNF | 4-hepatic nuclear factor |
| IBD | inflammatory bowel disease |
| LDL | low-density lipoproteins |
| LDLRAP1 | low-density lipoprotein receptor adapter protein 1 |
| lncRNAs | long non-coding RNAs |
| LXR | liver X receptor |
| MBD proteins | methyl-CpG binding proteins |
| mRNA | messenger RNA |
| miRNA | microRNA |
| MTHFR | methylenetetrahydrofolate reductase |
| MTRR | methionine synthase reductase |
| NF-κB | nuclear factor kappa-light chain-enhancer of activated B cells |
| NSRE | nutrient-sensing response elements |
| PKC | protein Kinase C |
| PPAR | peroxisome proliferator-activated receptor |
| PUFA | polyunsaturated fatty acids |
| RXR | retinoid X receptor |
| SAM | S-adenosyl-methionine |
| SAH | S-adenosylhomocysteine |
| SIDT1 | defective-1 transmembrane family member 1 |
| SIRT | sirtuin |
| SNP | single-nucleotide polymorphism |
| SREBP | sterol regulatory element-binding proteins |
| STAT3 | signal transducer and activator of transcription 3 |
| Tet | ten-eleven translocation enzymes |
| TL | telomere length |
| TRAIL | TNF-related apoptosis-inducing ligand |
| Tregs | regulatory T cells |
| TFs | transcription factors |
| TR | thyroid hormone receptor |
| UHRF proteins | ubiquitin-like, containing PHD and RING finger domain protein |
| 5hmC | 5-hydroxymethylcytosine |
| 5mC | 5-methyl cytosine |
References
- Subbiah, M.T. Nutrigenetics and nutraceuticals: The next wave riding on personalized medicine. Transl. Res. 2007, 149, 55–61. [Google Scholar] [CrossRef]
- Panagiotou, G.; Nielsen, J. Nutritional systems biology: Definitions and approaches. Annu. Rev. Nutr. 2009, 29, 329–339. [Google Scholar] [CrossRef]
- Phillips, C.M. Nutrigenetics and metabolic disease: Current status and implications for personalised nutrition. Nutrients 2013, 5, 32–57. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, E.; Davis, C.; Milner, J. Nutrigenomics, proteomics, metabolomics, and the practice of dietetics. J. Am. Diet Assoc. 2006, 106, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Stover, P.J. Human nutrition and genetic variation. Food Nutr. Bull. 2007, 28 (Suppl. 1), S101–S115. [Google Scholar] [CrossRef]
- Tishkoff, S.A.; Verrelli, B.C. Role of evolutionary history on haplotype block structure in the human genome: Implications for disease mapping. Curr. Opin. Genet. Dev. 2003, 13, 569–575. [Google Scholar] [CrossRef] [PubMed]
- El-Sohemy, A. Nutrigenetics. Forum Nutr. 2007, 60, 25–30. [Google Scholar]
- Kaput, J.; Rodriguez, R.L. Nutritional genomics: The next frontier in the postgenomic era. Physiol. Genomics 2004, 16, 166–177. [Google Scholar] [CrossRef]
- Mooser, V.; Ordovas, J.M. ‘Omic’ approaches and lipid metabolism: Are these new technologies holding their promises? Curr. Opin. Lipidol. 2003, 14, 115–119. [Google Scholar] [CrossRef]
- Ferguson, L.R. Nutrigenomics: Integrating genomic approaches into nutrition research. Mol. Diagn. Ther. 2006, 10, 101–108. [Google Scholar] [CrossRef]
- Afman, L.; Muller, M. Nutrigenomics: From molecular nutrition to prevention of disease. J. Am. Diet Assoc. 2006, 106, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, L.R. Nutrigenomics approaches to functional foods. J. Am. Diet Assoc. 2009, 109, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Jaenisch, R.; Bird, A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003, 33, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Hardy, T.M.; Tollefsbol, T.O. Epigenetic diet: Impact on the epigenome and cancer. Epigenomics 2011, 3, 503–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirk, H.; Cefalu, W.T.; Ribnicky, D.; Liu, Z.; Eilertsen, K.J. Botanicals as epigenetic modulators for mechanisms contributing to development of metabolic syndrome. Metabolism 2008, 57 (Suppl. 1), S16–S23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fila, M.; Chojnacki, C.; Chojnacki, J.; Blasiak, J. Is an “Epigenetic Diet” for Migraines Justified? The Case of Folate and DNA Methylation. Nutrients 2019, 11, 2763. [Google Scholar] [CrossRef] [Green Version]
- Ling, C.; Rönn, T. Epigenetics in Human Obesity and Type 2 Diabetes. Cell Metab. 2019, 29, 1028–1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N. Epigenetic modulation of DNA methylation by nutrition and its mechanisms in animals. Anim. Nutr. 2015, 1, 144–151. [Google Scholar] [CrossRef]
- Choi, S.-W. Nutrients and DNA methylation. In Nutrients and Epigenetics; CRC Press: Boca Raton, FL, USA, 2009. [Google Scholar]
- Ly, A.; Hoyt, L.; Crowell, J.; Kim, Y.-I. Folate and DNA methylation. Antioxid. Redox Signal. 2012, 17, 302–326. [Google Scholar] [CrossRef]
- McNulty, H.; Pentieva, K. Folate bioavailability. Proc. Nutr. Soc. 2004, 63, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Nasir, A.; Bullo, M.M.H.; Ahmed, Z.; Imtiaz, A.; Yaqoob, E.; Jadoon, M.; Ahmed, H.; Afreen, A.; Yaqoob, S. Nutrigenomics: Epigenetics and cancer prevention: A comprehensive review. Crit. Rev. Food Sci. Nutr. 2020, 60, 1375–1387. [Google Scholar] [CrossRef] [PubMed]
- Minor, E.A.; Court, B.L.; Young, J.I.; Wang, G. Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J. Biol. Chem. 2013, 288, 13669–13674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moradi Sarabi, M.; Naghibalhossaini, F. The impact of polyunsaturated fatty acids on DNA methylation and expression of DNMTs in human colorectal cancer cells. Biomed. Pharmacother. 2018, 101, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Qionglin, H.; Mingming, M.; Yu, Z.; Qingjin, Y.; Junjie, Z.; Xiaoxia, Y.; Lijian, Z.; Chun, C. The Anticancer Role of Omega-3 Polyunsaturated Fatty Acids was Closely Associated with the Increase in Genomic DNA Hydroxymethylation. Anti-Cancer Agents Med. Chem. 2019, 19, 330–336. [Google Scholar]
- Farias, N.; Ho, N.; Butler, S.; Delaney, L.; Morrison, J.; Shahrzad, S.; Coomber, B.L. The effects of folic acid on global DNA methylation and colonosphere formation in colon cancer cell lines. J. Nutr. Biochem. 2015, 26, 818–826. [Google Scholar] [CrossRef] [Green Version]
- Lubecka-Gajewska, K.; Kaufman-Szymczyk, A.; Stefanska, B.; Fabianowska-Majewska, K. Folic acid enforces DNA methylation-mediated transcriptional silencing of PTEN, APC and RARbeta2 tumour suppressor genes in breast cancer. Biochem. Biophys. Res. Commun. 2012, 430, 2623–2628. [Google Scholar]
- Ham, M.-S.; Lee, J.-K.; Kim, K.-C. S-adenosyl methionine specifically protects the anticancer effect of 5-FU via DNMTs expression in human A549 lung cancer cells. Mol. Clin. Oncol. 2013, 1, 373–378. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Zhao, L.-Z.; Hong, L.; Shan, C.; Shi, W.; Cai, W. Alteration in methylation pattern of GATA-4 promoter region in vitamin A-deficient offspring’s heart. J. Nutr. Biochem. 2013, 24, 1373–1380. [Google Scholar] [CrossRef]
- Qiu, W.; Lin, J.; Zhu, Y.; Zhang, J.; Zeng, L.; Su, M.; Tian, Y. Kaempferol Modulates DNA Methylation and Downregulates DNMT3B in Bladder Cancer. Cell. Physiol. Biochem. 2017, 41, 1325–1335. [Google Scholar] [CrossRef]
- Weng, Y.-P.; Hung, P.-F.; Ku, W.-Y.; Chang, C.-Y.; Wu, B.-H.; Wu, M.-H.; Yao, J.-Y.; Yang, J.-R.; Lee, C.-H. The inhibitory activity of gallic acid against DNA methylation: Application of gallic acid on epigenetic therapy of human cancers. Oncotarget 2017, 9, 361–374. [Google Scholar] [CrossRef] [Green Version]
- Sheng, J.; Shi, W.; Guo, H.; Long, W.; Wang, Y.; Qi, J.; Liu, J.; Xu, Y. The Inhibitory Effect of (-)-Epigallocatechin-3-Gallate on Breast Cancer Progression via Reducing SCUBE2 Methylation and DNMT Activity. Molecules 2019, 24, 2899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.H.; Zhang, B.L.; Zhang, X.M.; Tong, J.D.; Gu, Y.H.; Guo, L.L.; Jin, H.M. EGCG Attenuates Renal Damage via Reversing Klotho Hypermethylation in Diabetic db/db Mice and HK-2 Cells. Oxid. Med. Cell. Longev. 2020, 2020, 6092715. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Kim, Y.; Kim, Y. Effects of β-carotene on Expression of Selected MicroRNAs, Histone Acetylation, and DNA Methylation in Colon Cancer Stem Cells. J. Cancer Prev. 2019, 24, 224–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewinska, A.; Adamczyk-Grochala, J.; Deregowska, A.; Wnuk, M. Sulforaphane-Induced Cell Cycle Arrest and Senescence are accompanied by DNA Hypomethylation and Changes in microRNA Profile in Breast Cancer Cells. Theranostics 2017, 7, 3461–3477. [Google Scholar] [CrossRef]
- Su, X.; Wang, S.; Zhang, H.; Yang, G.; Bai, Y.; Liu, P.; Meng, L.; Jiang, X.; Xin, Y. Sulforaphane Prevents Angiotensin II-Induced Cardiomyopathy by Activation of Nrf2 Through Epigenetic Modification. SSRN Electron. J. 2020, 15, 405–417. [Google Scholar] [CrossRef]
- Ceccarelli, V.; Ronchetti, S.; Marchetti, M.C.; Calvitti, M.; Riccardi, C.; Grignani, F.; Vecchini, A. Molecular mechanisms underlying eicosapentaenoic acid inhibition of HDAC1 and DNMT expression and activity in carcinoma cells. Biochim. Biophys. Acta BBA Gene Regul. Mech. 2020, 1863, 194481. [Google Scholar] [CrossRef]
- Chatterjee, B.; Ghosh, K.; Kanade, S.R. Resveratrol modulates epigenetic regulators of promoter histone methylation and acetylation that restores BRCA1, p53, p21CIP1 in human breast cancer cell lines. BioFactors 2019, 45, 818–829. [Google Scholar] [CrossRef]
- Izquierdo-Torres, E.; Hernández-Oliveras, A.; Meneses-Morales, I.; Rodríguez, G.; Fuentes-García, G.; Zarain-Herzberg, Á. Resveratrol up-regulates ATP2A3 gene expression in breast cancer cell lines through epigenetic mechanisms. Int. J. Biochem. Cell Biol. 2019, 113, 37–47. [Google Scholar] [CrossRef]
- Dai, L.; Chen, L.; Wang, W.; Lin, P. Resveratrol inhibits ACHN cells via regulation of histone acetylation. Pharm. Biol. 2020, 58, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Alrafas, H.R.; Busbee, P.B.; Chitrala, K.N.; Nagarkatti, M.; Nagarkatti, P. Alterations in the Gut Microbiome and Suppression of Histone Deacetylases by Resveratrol Are Associated with Attenuation of Colonic Inflammation and Protection Against Colorectal Cancer. J. Clin. Med. 2020, 9, 1796. [Google Scholar] [CrossRef]
- Pandey, M.; Kaur, P.; Shukla, S.; Abbas, A.; Fu, P.; Gupta, S. Plant flavone apigenin inhibits HDAC and remodels chromatin to induce growth arrest and apoptosis in human prostate cancer cells: In vitro and in vivo study. Mol. Carcinogenes. 2012, 51, 952–962. [Google Scholar] [CrossRef] [Green Version]
- Yan, W.; Wu, T.H.Y.; Leung, S.S.Y.; To, K.K.W. Flavonoids potentiated anticancer activity of cisplatin in non-small cell lung cancer cells in vitro by inhibiting histone deacetylases. Life Sci. 2020, 258, 118211. [Google Scholar] [CrossRef]
- Attoub, S.; Hassan, A.H.; Vanhoecke, B.; Iratni, R.; Takahashi, T.; Gaben, A.M.; Bracke, M.; Awad, S.; John, A.; Kamalboor, H.A.; et al. Inhibition of cell survival, invasion, tumor growth and histone deacetylase activity by the dietary flavonoid luteolin in human epithelioid cancer cells. Eur. J. Pharmacol. 2011, 651, 18–25. [Google Scholar] [CrossRef]
- Wang, S.-W.; Chen, Y.-R.; Chow, J.-M.; Chien, M.-H.; Yang, S.-F.; Wen, Y.-C.; Lee, W.-J.; Tseng, T.-H. Stimulation of Fas/FasL-mediated apoptosis by luteolin through enhancement of histone H3 acetylation and c-Jun activation in HL-60 leukemia cells. Mol. Carcinogenes. 2018, 57, 866–877. [Google Scholar] [CrossRef]
- Pal-Bhadra, M.; Ramaiah, M.J.; Reddy, T.L.; Krishnan, A.; Pushpavalli, S.N.; Babu, K.S.; Tiwari, A.K.; Rao, J.M.; Yadav, J.S.; Bhadra, U. Plant HDAC inhibitor chrysin arrest cell growth and induce p21WAF1 by altering chromatin of STAT response element in A375 cells. BMC Cancer 2012, 12, 180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anantharaju, P.G.; Reddy, D.B.; Padukudru, M.A.; Chitturi, C.M.K.; Vimalambike, M.G.; Madhunapantula, S.V. Induction of colon and cervical cancer cell death by cinnamic acid derivatives is mediated through the inhibition of Histone Deacetylases (HDAC). PLoS ONE 2017, 12, e0186208. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.-G.; Ko, E.-B.; Choi, K.-C. Gallic acid, a phenolic acid, hinders the progression of prostate cancer by inhibition of histone deacetylase 1 and 2 expression. J. Nutr. Biochem. 2020, 84, 108444. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.; Quan, J.; Liu, L.; Xu, Z.; Zhu, J.; Huang, X.; Tian, J. Epigallocatechin gallate reverses cTnI-low expression-induced age-related heart diastolic dysfunction through histone acetylation modification. J. Cell. Mol. Med. 2017, 21, 2481–2490. [Google Scholar] [CrossRef] [Green Version]
- Deb, G.; Shankar, E.; Thakur, V.S.; Ponsky, L.E.; Bodner, D.R.; Fu, P.; Gupta, S. Green tea–induced epigenetic reactivation of tissue inhibitor of matrix metalloproteinase-3 suppresses prostate cancer progression through histone-modifying enzymes. Mol. Carcinogenes. 2019, 58, 1194–1207. [Google Scholar] [CrossRef]
- Moradzadeh, M.; Roustazadeh, A.; Tabarraei, A.; Erfanian, S.; Sahebkar, A. Epigallocatechin-3-gallate enhances differentiation of acute promyelocytic leukemia cells via inhibition of PML-RARα and HDAC1. Phytother. Res. 2018, 32, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, R.; Zhan, Z.; Li, X.; Zhou, F.; Xing, A.; Jiang, C.; Chen, Y.; An, L. Beneficial Effects of Sulforaphane Treatment in Alzheimer’s Disease May Be Mediated through Reduced HDAC1/3 and Increased P75NTR Expression. Front. Aging Neurosci. 2017, 9, 121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitsiogianni, M.; Trafalis, D.T.; Franco, R.; Zoumpourlis, V.; Pappa, A.; Panayiotidis, M.I. Sulforaphane and iberin are potent epigenetic modulators of histone acetylation and methylation in malignant melanoma. Eur. J. Nutr. 2021, 60, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.L.; Kala, R.; Tollefsbol, T.O. Mechanisms for the Inhibition of Colon Cancer Cells by Sulforaphane through Epigenetic Modulation of MicroRNA-21 and Human Telomerase Reverse Transcriptase (hTERT) Down-regulation. Curr. Cancer Drug Targets 2018, 18, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Hossain, S. Effect of Histone Deacetylase Inhibitors on Vitamin D Dependent Gene Expression in Human Colorectal and Breast Cancers. FASEB J. 2017, 31 (Suppl. 1), 644–647. [Google Scholar]
- Udroiu, I.; Marinaccio, J.; Sgura, A. Epigallocatechin-3-gallate induces telomere shortening and clastogenic damage in glioblastoma cells. Environ. Mol. Mutagenes. 2019, 60, 683–692. [Google Scholar] [CrossRef]
- Fang, M.; Chen, D.; Yang, C.S. Dietary polyphenols may affect DNA methylation. J. Nutr. 2007, 137 (Suppl. 1), 223S–228S. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.J.; Shim, J.Y.; Zhu, B.T. Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Mol. Pharmacol. 2005, 68, 1018–1030. [Google Scholar] [CrossRef] [Green Version]
- Nandakumar, V.; Vaid, M.; Katiyar, S.K. (−)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p16INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis 2011, 32, 537–544. [Google Scholar] [CrossRef] [Green Version]
- Meeran, S.M.; Patel, S.N.; Tollefsbol, T.O. Sulforaphane causes epigenetic repression of hTERT expression in human breast cancer cell lines. PLoS ONE 2010, 5, e11457. [Google Scholar] [CrossRef]
- Mukherjee, N.; Kumar, A.; Ghosh, R. DNA Methylation and Flavonoids in Genitourinary Cancers. Curr. Pharmacol. Rep. 2015, 1, 112–120. [Google Scholar] [CrossRef] [Green Version]
- Fini, L.; Selgrad, M.; Fogliano, V.; Graziani, G.; Romano, M.; Hotchkiss, E.; Daoud, Y.A.; De Vol, E.B.; Boland, R.; Ricciardiello, L. Annurca apple polyphenols have potent demethylating activity and can reactivate silenced tumor suppressor genes in colorectal cancer cells. J. Nutr. 2007, 137, 2622–2628. [Google Scholar] [CrossRef] [Green Version]
- King-Batoon, A.; Leszczynska, J.M.; Klein, C.B. Modulation of gene methylation by genistein or lycopene in breast cancer cells. Environ. Mol. Mutagenesis 2008, 49, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Crescenti, A.; Solà, R.; Valls, R.M.; Caimari, A.; Del Bas, J.M.; Anguera, A.; Anglés, N.; Arola, L. Cocoa Consumption Alters the Global DNA Methylation of Peripheral Leukocytes in Humans with Cardiovascular Disease Risk Factors: A Randomized Controlled Trial. PLoS ONE 2013, 8, e65744. [Google Scholar] [CrossRef] [PubMed]
- Dolinoy, D.C.; Weidman, J.R.; Waterland, R.A.; Jirtle, R.L. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ. Health Perspect. 2006, 114, 567–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dan, J.; Chen, T. Genetic Studies on Mammalian DNA Methyltransferases. Adv. Exp. Med. Biol. 2016, 945, 123–150. [Google Scholar] [PubMed]
- Ding, Y.B.; He, J.L.; Liu, X.Q.; Chen, X.M.; Long, C.L.; Wang, Y.X. Expression of DNA methyltransferases in the mouse uterus during early pregnancy and susceptibility to dietary folate deficiency. Reproduction 2012, 144, 91–100. [Google Scholar] [CrossRef]
- Rees, W.D.; Hay, S.M.; Brown, D.S.; Antipatis, C.; Palmer, R.M. Maternal protein deficiency causes hypermethylation of DNA in the livers of rat fetuses. J. Nutr. 2000, 130, 1821–1826. [Google Scholar] [CrossRef] [Green Version]
- Gluckman, P.D.; Hanson, M.A.; Cooper, C.; Thornburg, K.L. Effect of in utero and early-life conditions on adult health and disease. N. Engl. J. Med. 2008, 359, 61–73. [Google Scholar] [CrossRef] [Green Version]
- Heijmans, B.T.; Tobi, E.W.; Stein, A.D.; Putter, H.; Blauw, G.J.; Susser, E.S.; Slagboom, P.E.; Lumey, L.H. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl. Acad. Sci. USA 2008, 105, 17046–17049. [Google Scholar] [CrossRef] [Green Version]
- Painter, R.C.; Osmond, C.; Gluckman, P.; Hanson, M.; Phillips, D.I.W.; Roseboom, T.J. Transgenerational effects of prenatal exposure to the Dutch famine on neonatal adiposity and health in later life. BJOG Int. J. Obstet. Gynaecol. 2008, 115, 1243–1249. [Google Scholar] [CrossRef]
- Tobi, E.W.; Lumey, L.H.; Talens, R.P.; Kremer, D.; Putter, H.; Stein, A.D.; Slagboom, P.E.; Heijmans, B.T. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum. Mol. Genet. 2009, 18, 4046–4053. [Google Scholar] [CrossRef] [PubMed]
- Dudley, K.J.; Sloboda, D.M.; Connor, K.L.; Beltrand, J.; Vickers, M.H. Offspring of mothers fed a high fat diet display hepatic cell cycle inhibition and associated changes in gene expression and DNA methylation. PLoS ONE 2011, 6, e21662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marco, A.; Kisliouk, T.; Tabachnik, T.; Meiri, N.; Weller, A. Overweight and CpG methylation of the Pomc promoter in offspring of high-fat-diet-fed dams are not “reprogrammed” by regular chow diet in rats. FASEB J. 2014, 28, 4148–4157. [Google Scholar] [CrossRef] [Green Version]
- Vucetic, Z.; Kimmel, J.; Totoki, K.; Hollenbeck, E.; Reyes, T.M. Maternal high-fat diet alters methylation and gene expression of dopamine and opioid-related genes. Endocrinology 2010, 151, 4756–4764. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Daniel, M.; Tollefsbol, T.O. Epigenetic regulation of caloric restriction in aging. BMC Med. 2011, 9, 98. [Google Scholar] [CrossRef] [Green Version]
- McKay, J.A.; Mathers, J.C. Diet. induced epigenetic changes and their implications for health. Acta Physiol. 2011, 202, 103–118. [Google Scholar] [CrossRef]
- Milagro, F.I.; Campion, J.; Cordero, P.; Goyenechea, E.; Gomez-Uriz, A.M.; Abete, I.; Zulet, M.A.; Martinez, J.A. A dual epigenomic approach for the search of obesity biomarkers: DNA methylation in relation to diet-induced weight loss. FASEB J 2011, 25, 1378–1389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouchard, L.; Rabasa-Lhoret, R.; Faraj, M.; Lavoie, M.-E.; Mill, J.; Pérusse, L.; Vohl, M.-C. Differential epigenomic and transcriptomic responses in subcutaneous adipose tissue between low and high responders to caloric restriction. Am. J. Clin. Nutr. 2010, 91, 309–320. [Google Scholar] [CrossRef] [Green Version]
- Hjort, L.; Jørgensen, S.W.; Gillberg, L.; Hall, E.; Brøns, C.; Frystyk, J.; Vaag, A.A.; Ling, C. 36 h fasting of young men influences adipose tissue DNA methylation of LEP and ADIPOQ in a birth weight-dependent manner. Clin. Epigen. 2017, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, D.R.; Bultman, S.J. Metaboloepigenetics: Interrelationships between energy metabolism and epigenetic control of gene expression. J. Cell. Physiol. 2012, 227, 3169–3177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenfeld, J.A.; Wang, Z.; Schones, D.E.; Zhao, K.; DeSalle, R.; Zhang, M.Q. Determination of enriched histone modifications in non-genic portions of the human genome. BMC Genomics 2009, 10, 143. [Google Scholar] [CrossRef] [Green Version]
- Ciesielski, O.; Biesiekierska, M.; Balcerczyk, A. Epigallocatechin-3-gallate (EGCG) Alters Histone Acetylation and Methylation and Impacts Chromatin Architecture Profile in Human Endothelial Cells. Molecules 2020, 25, 2326. [Google Scholar] [CrossRef]
- Dashwood, R.H.; Myzak, M.C.; Ho, E. Dietary HDAC inhibitors: Time to rethink weak ligands in cancer chemoprevention? Carcinogenesis 2006, 27, 344–349. [Google Scholar] [CrossRef]
- Hu, J.; Shen, T.; Xie, J.; Wang, S.; He, Y.; Zhu, F. Curcumin modulates covalent histone modification and TIMP1 gene activation to protect against vascular injury in a hypertension rat model. Exp. Ther. Med. 2017, 14, 5896–5902. [Google Scholar] [CrossRef]
- Kang, J.; Lin, C.; Chen, J.; Liu, Q. Copper induces histone hypoacetylation through directly inhibiting histone acetyltransferase activity. Chem. Biol. Interact. 2004, 148, 115–123. [Google Scholar] [CrossRef]
- Li, Y.; Liu, L.; Andrews, L.G.; Tollefsbol, T.O. Genistein depletes telomerase activity through cross-talk between genetic and epigenetic mechanisms. Int. J. Cancer 2009, 125, 286–296. [Google Scholar] [CrossRef] [Green Version]
- North, B.J.; Verdin, E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 2004, 5, 224. [Google Scholar] [CrossRef] [Green Version]
- Guarente, L.; Picard, F. Calorie restriction—The SIR2 connection. Cell 2005, 120, 473–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehedint, M.G.; Niculescu, M.D.; Craciunescu, C.N.; Zeisel, S.H. Choline deficiency alters global histone methylation and epigenetic marking at the Re1 site of the calbindin 1 gene. FASEB J. 2010, 24, 184–195. [Google Scholar] [CrossRef] [Green Version]
- Babizhayev, M.A.; Savel’yeva, E.L.; Moskvina, S.N.; Yegorov, Y.E. Telomere length is a biomarker of cumulative oxidative stress, biologic age, and an independent predictor of survival and therapeutic treatment requirement associated with smoking behavior. Am. J. Ther. 2011, 18, e209–e226. [Google Scholar] [CrossRef]
- Daniel, M.; Peek, G.W.; Tollefsbol, T.O. Regulation of the human catalytic subunit of telomerase (hTERT). Gene 2012, 498, 135–146. [Google Scholar] [CrossRef] [Green Version]
- Mirabello, L.; Huang, W.Y.; Wong, J.Y.; Chatterjee, N.; Reding, D.; Crawford, E.D.; De Vivo, I.; Hayes, R.B.; Savage, S.A. The association between leukocyte telomere length and cigarette smoking, dietary and physical variables, and risk of prostate cancer. Aging Cell 2009, 8, 405–413. [Google Scholar] [CrossRef] [Green Version]
- Ornish, D.; Lin, J.; Daubenmier, J.; Weidner, G.; Epel, E.; Kemp, C.; Magbanua, M.J.; Marlin, R.; Yglecias, L.; Carroll, P.R.; et al. Increased telomerase activity and comprehensive lifestyle changes: A pilot study. Lancet Oncol. 2008, 9, 1048–1057. [Google Scholar] [CrossRef]
- Nettleton, J.A.; Diez-Roux, A.; Jenny, N.S.; Fitzpatrick, A.L.; Jacobs, D.R., Jr. Dietary patterns, food groups, and telomere length in the Multi-Ethnic Study of Atherosclerosis (MESA). Am. J. Clin. Nutr. 2008, 88, 1405–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crous-Bou, M.; Molinuevo, J.-L.; Sala-Vila, A. Plant.-Rich Dietary Patterns, Plant. Foods and Nutrients, and Telomere Length. Adv. Nutr. 2019, 10 (Suppl. 4), S296–S303. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Ibarra, M.J.; Hernández, J.; Caire-Juvera, G. Diet, physical activity and telomere length in adults. Nutr. Hosp. 2019, 36, 1403–1417. [Google Scholar]
- Quintanilha, B.J.; Reis, B.Z.; Duarte, G.B.S.; Cozzolino, S.M.F.; Rogero, M.M. Nutrimiromics: Role of microRNAs and Nutrition in Modulating Inflammation and Chronic Diseases. Nutrients 2017, 9, 1168. [Google Scholar] [CrossRef] [PubMed]
- Plotnikova, O.; Baranova, A.; Skoblov, M. Comprehensive Analysis of Human microRNA–mRNA Interactome. Front. Genet. 2019, 10, 933. [Google Scholar] [CrossRef]
- Mar-Aguilar, F.; Arreola-Triana, A.; Mata-Cardona, D.; Gonzalez-Villasana, V.; Rodríguez-Padilla, C.; Reséndez-Pérez, D. Evidence of transfer of miRNAs from the diet to the blood still inconclusive. PeerJ 2020, 8, e9567. [Google Scholar] [CrossRef]
- Chen, X.; Zen, K.; Zhang, C.-Y. Reply to Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat. Biotechnol. 2013, 31, 967–969. [Google Scholar] [CrossRef] [PubMed]
- Snow, J.W.; Hale, A.E.; Isaacs, S.K.; Baggish, A.L.; Chan, S.Y. Ineffective delivery of diet-derived microRNAs to recipient animal organisms. RNA Biol. 2013, 10, 1107–1116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tosar, J.; Rovira, C.; Naya, H.; Cayota, A. Mining of public sequencing databases supports a non-dietary origin for putative foreign miRNAs: Underestimated effects of contamination in NGS. RNA 2014, 20, 754–757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2012, 22, 107–126. [Google Scholar] [CrossRef] [PubMed]
- Chin, A.R.; Fong, M.Y.; Somlo, G.; Wu, J.; Swiderski, P.; Wu, X.; Wang, S.E. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res. 2016, 26, 217–228. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Zhang, F.; Dong, L.; Wu, H.; Xu, J.; Li, H.; Wang, J.; Zhou, Z.; Liu, C.; Wang, Y.; et al. SIDT1-dependent absorption in the stomach mediates host uptake of dietary and orally administered microRNAs. Cell Res. 2021, 31, 247–258. [Google Scholar] [CrossRef]
- Elhassan, M.O.; Christie, J.; Duxbury, M.S. Homo sapiens systemic RNA interference-defective-1 transmembrane family member 1 (SIDT1) protein mediates contact-dependent small RNA transfer and microRNA-21-driven chemoresistance. J. Biol. Chem. 2012, 287, 5267–5277. [Google Scholar] [CrossRef] [Green Version]
- Simardeep, K.; Suresh, K. Crosstalk between food components and microRNAs: Role in metabolism, nutrition, health and diseases. Integr. Food Nutr. Metab. 2020, 7, 1–11. [Google Scholar] [CrossRef]
- Qiu, B.; Xu, X.; Yi, P.; Hao, Y. Curcumin reinforces MSC-derived exosomes in attenuating osteoarthritis via modulating the miR-124/NF-kB and miR-143/ROCK1/TLR9 signalling pathways. J. Cell. Mol. Med. 2020, 24, 10855–10865. [Google Scholar] [CrossRef]
- Li, Y.; Sun, W.; Han, N.; Zou, Y.; Yin, D. Curcumin inhibits proliferation, migration, invasion and promotes apoptosis of retinoblastoma cell lines through modulation of miR-99a and JAK/STAT pathway. BMC Cancer 2018, 18, 1230. [Google Scholar] [CrossRef] [Green Version]
- Otsuka, K.; Yamamoto, Y.; Ochiya, T. Regulatory role of resveratrol, a microRNA-controlling compound, in HNRNPA1 expression, which is associated with poor prognosis in breast cancer. Oncotarget 2018, 9, 24718–24730. [Google Scholar] [CrossRef] [Green Version]
- Fu, J.; Shrivastava, A.; Shrivastava, S.K.; Srivastava, R.K.; Shankar, S. Triacetyl resveratrol upregulates miRNA-200 and suppresses the Shh pathway in pancreatic cancer: A potential therapeutic agent. Int. J. Oncol. 2019, 54, 1306–1316. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Jiang, H.; Chen, Y.; Ren, F. Resveratrol chemosensitizes adriamycin-resistant breast cancer cells by modulating miR-122-5p. J. Cell. Biochem. 2019, 120, 16283–16292. [Google Scholar] [CrossRef]
- Karimi Dermani, F.; Saidijam, M.; Amini, R.; Mahdavinezhad, A.; Heydari, K.; Najafi, R. Resveratrol Inhibits Proliferation, Invasion, and Epithelial–Mesenchymal Transition by Increasing miR-200c Expression in HCT-116 Colorectal Cancer Cells. J. Cell. Biochem. 2017, 118, 1547–1555. [Google Scholar] [CrossRef]
- Tomé-Carneiro, J.; Larrosa, M.; Yáñez-Gascón, M.J.; Dávalos, A.; Gil-Zamorano, J.; Gonzálvez, M.; García-Almagro, F.J.; Ruiz Ros, J.A.; Tomás-Barberán, F.A.; Espín, J.C.; et al. One-year supplementation with a grape extract containing resveratrol modulates inflammatory-related microRNAs and cytokines expression in peripheral blood mononuclear cells of type 2 diabetes and hypertensive patients with coronary artery disease. Pharmacol. Res. 2013, 72, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Cui, L. Resveratrol suppresses melanoma by inhibiting NF-κB/miR-221 and inducing TFG expression. Arch. Dermatol. Res. 2017, 309, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, H.; Wang, H.; Shi, J. Quercetin attenuates high glucose-induced injury in human retinal pigment epithelial cell line ARPE-19 by up-regulation of miR-29b. J. Biochem. 2020, 167, 495–502. [Google Scholar] [CrossRef]
- Tao, S.-F.; He, H.-F.; Chen, Q. Quercetin inhibits proliferation and invasion acts by up-regulating miR-146a in human breast cancer cells. Mol. Cell. Biochem. 2015, 402, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chang, B.; Zheng, G.; Du, S.; Li, X. Quercetin stimulates osteogenic differentiation of bone marrow stromal cells through miRNA-206/connexin 43 pathway. Am. J. Transl. Res. 2020, 12, 2062–2070. [Google Scholar]
- Tofigh, R.; Tutunchi, S.; Akhavan, S.; Panahi, G. The effects of Quercetin on miRNA-21 expression in MCF-7 cells. Arch. Med Lab. Sci. 2017. [Google Scholar] [CrossRef]
- Wang, F.; Ke, Y.; Yang, L.; Wang, F.J. Quercetin protects human oral keratinocytes from lipopolysaccharide-induced injury by downregulating microRNA-22. Hum. Exp. Toxicol. 2020, 39, 096032712091829. [Google Scholar] [CrossRef]
- Wang, X.; Xue, X.; Wang, H.; Xu, F.; Xin, Z.; Wang, K.; Cui, M.; Qin, W. Quercetin inhibits human microvascular endothelial cells viability, migration and tube-formation in vitro through restraining microRNA-216a. J. Drug Target. 2020, 28, 609–616. [Google Scholar] [CrossRef]
- Alshammari, G.; Al-Qahtani, W.; Alfaris, N.; Alzahrani, N.; Alkhateeb, M.; Yahya, M. Quercetin prevents cadmium chloride-induced hepatic steatosis and fibrosis by downregulating the transcription of miR-21. BioFactors 2021, 47, 489–505. [Google Scholar] [CrossRef]
- Youness, R.; Assal, R.; Ezzat, S.; Gad, M.; Abdel Motaal, A. A methoxylated quercetin glycoside harnesses HCC tumor progression in a TP53/miR-15/miR-16 dependent manner. Nat. Prod. Res. 2018, 34, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Fang, Z.; Zha, Z.; Sun, Q.; Wang, H.; Sun, M.; Qiao, B. Quercetin inhibits cell viability, migration and invasion by regulating miR-16/HOXA10 axis in oral cancer. Eur. J. Pharmacol. 2019, 847, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-M.; Yang, P.-W.; Feng, X.-J.; Zhu, Y.-W.; Qiu, F.-J.; Hu, X.-D.; Zhang, S.-H. Apigenin Inhibits the Growth of Hepatocellular Carcinoma Cells by Affecting the Expression of microRNA Transcriptome. Front. Oncol. 2021, 11, 657665. [Google Scholar] [CrossRef] [PubMed]
- Aida, R.; Hagiwara, K.; Okano, K.; Nakata, K.; Obata, Y.; Yamashita, T.; Yoshida, K.; Hagiwara, H. miR-34a-5p might have an important role for inducing apoptosis by down-regulation of SNAI1 in apigenin-treated lung cancer cells. Mol. Biol. Rep. 2021, 48, 2291–2297. [Google Scholar] [CrossRef]
- Zhao, X.; Zhou, H.-B.; Liu, J.; Xie, J.; Hu, R. Apigenin suppresses proliferation, invasion, and epithelial–mesenchymal transition of cervical carcinoma cells by regulation of miR-152/BRD4 axis. Kaohsiung J. Med Sci. 2021, 37, 583–593. [Google Scholar] [CrossRef]
- Cheng, Y.; Han, X.; Mo, F.; Zeng, H.; Zhao, Y.; Wang, H.; Zheng, Y.; Ma, X. Apigenin inhibits the growth of colorectal cancer through down-regulation of E2F1/3 by miRNA-215-5p. Phytomedicine 2021, 89, 153603. [Google Scholar] [CrossRef]
- Jiang, Z.-Q.; Li, M.-H.; Qin, Y.-M.; Jiang, H.-Y.; Zhang, X.; Wu, M.-H. Luteolin Inhibits Tumorigenesis and Induces Apoptosis of Non-Small Cell Lung Cancer Cells via Regulation of MicroRNA-34a-5p. Int. J. Mol. Sci. 2018, 19, 447. [Google Scholar] [CrossRef] [Green Version]
- Gao, G.; Ge, R.; Li, Y.; Liu, S. Luteolin exhibits anti-breast cancer property through up-regulating miR-203. Artif. Cells Nanomed. Biotechnol. 2019, 47, 3265–3271. [Google Scholar] [CrossRef] [Green Version]
- Yang, P.-W.; Lu, Z.-Y.; Pan, Q.; Chen, T.-T.; Feng, X.-J.; Wang, S.-M.; Pan, Y.-C.; Zhu, M.-H.; Zhang, S.-H. MicroRNA-6809-5p mediates luteolin-induced anticancer effects against hepatoma by targeting flotillin 1. Phytomedicine 2019, 57, 18–29. [Google Scholar] [CrossRef]
- Zhou, Y.; Ding, B.-Z.; Lin, Y.-P.; Wang, H.-B. MiR-34a, as a suppressor, enhance the susceptibility of gastric cancer cell to luteolin by directly targeting HK1. Gene 2018, 644, 56–65. [Google Scholar] [CrossRef]
- Magura, J.; Moodley, R.; Mackraj, I. The effect of hesperidin and luteolin isolated from Eriocephalus africanus on apoptosis, cell cycle and miRNA expression in MCF-7. J. Biomol. Struct. Dyn. 2020. [Google Scholar] [CrossRef]
- Liu, X.; Meng, J. Luteolin alleviates LPS-induced bronchopneumonia injury in vitro and in vivo by down-regulating microRNA-132 expression. Biomed. Pharmacother. 2018, 106, 1641–1649. [Google Scholar] [CrossRef]
- Mg, S.; Son, S.; Seo, H.; Lee, J.; Kim, C.k.; Kuh, H.-J.; Park, J.K. Luteolin-regulated MicroRNA-301-3p targets caspase-8 and modulates TRAIL sensitivity in PANC-1 cells. Anticancer Res. 2020, 40, 723–731. [Google Scholar]
- Han, X.; Liu, C.-F.; Gao, N.; Zhao, J.; Xu, J. Kaempferol suppresses proliferation but increases apoptosis and autophagy by up-regulating microRNA-340 in human lung cancer cells. Biomed. Pharmacother. 2018, 108, 809–816. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Cui, M.; Li, C.; Li, H.; Dai, Y.; Cui, K.; Li, Z. Kaempferol Reverses Aerobic Glycolysis via miR-339-5p-Mediated PKM Alternative Splicing in Colon Cancer Cells. J. Agric. Food Chem. 2021, 69, 3060–3068. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Liu, X.; Li, H.; Yan, Y.; Hong, X.; Lin, Z. Kaempferol inhibits proliferation, migration, and invasion of liver cancer HepG2 cells by down-regulation of microRNA-21. Int. J. Immunopathol. Pharmacol. 2018, 32, 2058738418814341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, R.; Hao, P.; Yu, G.; Liu, C.; Yu, C.; Huang, Y.; Wang, Y. Kaempferol protects chondrogenic ATDC5 cells against inflammatory injury triggered by lipopolysaccharide through down-regulating miR-146a. Int. Immunopharmacol. 2019, 69, 373–381. [Google Scholar] [CrossRef]
- Cui, S.; Tang, J.; Wang, S.; Li, L. Kaempferol protects lipopolysaccharide-induced inflammatory injury in human aortic endothelial cells (HAECs) by regulation of miR-203. Biomed. Pharmacother. 2019, 115, 108888. [Google Scholar] [CrossRef]
- Javan, N.; Khadem Ansari, M.H.; Dadashpour, M.; Khojastehfard, M.; Bastami, M.; Rahmati-Yamchi, M.; Zarghami, N. Synergistic Antiproliferative Effects of Co-nanoencapsulated Curcumin and Chrysin on MDA-MB-231 Breast Cancer Cells Through Upregulating miR-132 and miR-502c. Nutr. Cancer 2019, 71, 1201–1213. [Google Scholar] [CrossRef]
- Mohammadian, F.; Pilehvar-Soltanahmadi, Y.; Zarghami, F.; Akbarzadeh, A.; Zarghami, N. Upregulation of miR-9 and Let-7a by nanoencapsulated chrysin in gastric cancer cells. Artif. Cells Nanomed. Biotechnol. 2016, 45, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohammadian, F.; Pilehvar-Soltanahmadi, Y.; Mofarrah, M.; Dastani-Habashi, M.; Zarghami, N. Down regulation of miR-18a, miR-21 and miR-221 genes in gastric cancer cell line by chrysin-loaded PLGA-PEG nanoparticles. Artif. Cells Nanomed. Biotechnol. 2016, 44, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-M.; Wang, B.-W.; Pan, C.-M.; Fang, W.-J.; Chua, S.-K.; Cheng, W.-P.; Shyu, K.-G. Chrysin boosts KLF2 expression through suppression of endothelial cell-derived exosomal microRNA-92a in the model of atheroprotection. Eur. J. Nutr. 2021. [Google Scholar] [CrossRef] [PubMed]
- Salem, A.M.; Ragheb, A.S.; Hegazy, M.G.A.; Matboli, M.; Eissa, S. Caffeic Acid Modulates miR-636 Expression in Diabetic Nephropathy Rats. Indian J. Clin. Biochem. 2019, 34, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Arffa, M.; Zapf, M.; Kothari, A.; Chang, V.; Gupta, G.; Ding, X.; Al-Gayyar, M.; Syn, W.; Elsherbiny, N.; Kuo, P.; et al. Epigallocatechin-3-Gallate Upregulates miR-221 to Inhibit Osteopontin-Dependent Hepatic Fibrosis. PLoS ONE 2016, 11, e0167435. [Google Scholar] [CrossRef] [PubMed]
- Mekky, R.Y.; El-Ekiaby, N.; El Sobky, S.A.; Elemam, N.M.; Youness, R.A.; El-Sayed, M.; Hamza, M.T.; Esmat, G.; Abdelaziz, A.I. Epigallocatechin gallate (EGCG) and miR-548m reduce HCV entry through repression of CD81 receptor in HCV cell models. Arch. Virol. 2019, 164, 1587–1595. [Google Scholar] [CrossRef]
- Yamada, S.; Tsukamoto, S.; Huang, Y.; Makio, A.; Kumazoe, M.; Yamashita, S.; Tachibana, H. Epigallocatechin-3-O-gallate up-regulates microRNA-let-7b expression by activating 67-kDa laminin receptor signaling in melanoma cells. Sci. Rep. 2016, 6, 19225. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Yu, S.; Yu, Y.; Xu, G. (−)-Epigallocatechin-3-gallate suppresses prostate cancer cell growth via activating miR-520a-3p. Rev. Bras. Farmacogn. 2020, 30, 528–536. [Google Scholar] [CrossRef]
- Zhang, C.; Liang, R.; Gan, X.; Yang, X.; Chen, L.; Jian, J. MicroRNA-384-5p/Beclin-1 As Potential Indicators For Epigallocatechin Gallate Against Cardiomyocytes Ischemia Reperfusion Injury By Inhibiting Autophagy Via PI3K/Akt Pathway. Drug Des. Dev. Ther. 2019, 13, 3607–3623. [Google Scholar] [CrossRef] [Green Version]
- Zan, L.; Chen, Q.; Zhang, L.; Li, X. Epigallocatechin gallate (EGCG) suppresses growth and tumorigenicity in breast cancer cells by downregulation of miR-25. Bioengineered 2019, 10, 374–382. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Cheng, D.; Yang, J.; Wu, R.; Li, W.; Kong, A.-N. Sulforaphane epigenetically demethylates the CpG sites of the miR-9-3 promoter and reactivates miR-9-3 expression in human lung cancer A549 cells. J. Nutr. Biochem. 2018, 56, 109–115. [Google Scholar] [CrossRef]
- Yin, L.; Xiao, X.; Georgikou, C.; Luo, Y.; Liu, L.; Gladkich, J.; Gross, W.; Herr, I. Sulforaphane Induces miR135b-5p and Its Target Gene, RASAL2, thereby Inhibiting the Progression of Pancreatic Cancer. Mol. Ther. Oncolytics 2019, 14, 74–81. [Google Scholar] [CrossRef] [Green Version]
- Feng, M.-H.; Li, J.-W.; Sun, H.-T.; He, S.-Q.; Pang, J. Sulforaphane inhibits the activation of hepatic stellate cell by miRNA-423-5p targeting suppressor of fused. Hum. Cell 2019, 32, 403–410. [Google Scholar] [CrossRef]
- Georgikou, C.; Yin, L.; Gladkich, J.; Xiao, X.; Sticht, C.; Torre, C.d.l.; Gretz, N.; Gross, W.; Schäfer, M.; Karakhanova, S.; et al. Inhibition of miR30a-3p by sulforaphane enhances gap junction intercellular communication in pancreatic cancer. Cancer Lett. 2020, 469, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Koolivand, M.; Ansari, M.; Piroozian, F.; Moein, S.; MalekZadeh, K. Alleviating the progression of acute myeloid leukemia (AML) by sulforaphane through controlling miR-155 levels. Mol. Biol. Rep. 2018, 45, 2491–2499. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhao, Z.; Li, M.; Liu, M.; Bahena, A.; Zhang, Y.; Zhang, Y.; Nambiar, C.; Liu, G. Sulforaphane promotes apoptosis, and inhibits proliferation and self-renewal of nasopharyngeal cancer cells by targeting STAT signal through miRNA-124-3p. Biomed. Pharmacother. 2018, 103, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-Y.; Kim, Y.-M.; Hong, S. Astaxanthin suppresses the metastasis of colon cancer by inhibiting the MYC-mediated downregulation of microRNA-29a-3p and microRNA-200a. Sci. Rep. 2019, 9, 9457. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Gong, X.; Rubin, L.P.; Choi, S.-W.; Kim, Y. β-Carotene 15,15′-oxygenase inhibits cancer cell stemness and metastasis by regulating differentiation-related miRNAs in human neuroblastoma. J. Nutr. Biochem. 2019, 69, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhao, W.; Hao, J.; Ruihua, R. MicroRNA-let-7f-1 is induced by lycopene and inhibits cell proliferation and triggers apoptosis in prostate cancer. Mol. Med. Rep. 2016, 13, 2708–2714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Xue, B.; Du, C.; Zhang, L.; Wang, Y.; Zhang, Y.; Li, J. Docosahexaenoic acid supresses breast cancer cell proliferation and migration by promoting the expression of miR-99a and targeting mTOR signaling. Arab. J. Chem. 2021, 14, 103298. [Google Scholar] [CrossRef]
- Bai, X.; Shao, J.; Zhou, S.; Zhao, Z.; Li, F.; Xiang, R.; Zhao, A.; Pan, J. Inhibition of lung cancer growth and metastasis by DHA and its metabolite, RvD1, through miR-138-5p/FOXC1 pathway. J. Exp. Clin. Cancer Res. 2019. [Google Scholar] [CrossRef]
- Dai, X.; Li, M.; Geng, F. Omega-3 Polyunsaturated Fatty Acids Eicosapentaenoic Acid and Docosahexaenoic Acid Enhance Dexamethasone Sensitivity in Multiple Myeloma Cells by the p53/miR-34a/Bcl-2 Axis. Biochemistry 2017, 82, 826–833. [Google Scholar] [CrossRef]
- Javadian, M.; Shekari, N.; Soltani - Zangbar, M.S.; Mohammadi, A.; Mansoori, B.; Maralbashi, S.; Shanehbandi, D.; Baradaran, B.; Darabi, M.; Kazemi, T. Docosahexaenoic acid suppresses migration of triple-negative breast cancer cell through targeting metastasis-related genes and microRNA under normoxic and hypoxic conditions. J. Cell. Biochem. 2020, 121, 2416–2427. [Google Scholar] [CrossRef]
- LeMay-Nedjelski, L.; Ennis, J.; Taibi, A.; Comelli, E.; Thompson, L. Omega-3 Polyunsaturated Fatty Acids Time-Dependently Reduce Cell Viability and Oncogenic MicroRNA-21 Expression in Estrogen Receptor-Positive Breast Cancer Cells (MCF-7). Int. J. Mol. Sci. 2018, 19, 244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, X.; Xu, C.; Xu, Q.; Lang, D. Docosahexaenoic acid inhibits vascular smooth muscle cell migration and proliferation by decreasing microRNA-155 expression levels. Mol. Med. Rep. 2020, 22, 3396–3404. [Google Scholar]
- Shekari, N.; Javadian, M.; Ghaffari, S.; Baradaran, B.; Darabi, M.; Kazemi, T. DHA Abolishes the Detrimental Effect of Docetaxel on Downregulation of the MICA via Decreasing the Expression Level of MicroRNA-20a in Gastric Cancer. J. Gastrointest. Cancer 2020, 51, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Zhen, J.; Lu, Z. Synergetic Neuroprotective Effect of Docosahexaenoic Acid and Aspirin in SH-Y5Y by Inhibiting miR-21 and Activating RXRα and PPARα. DNA Cell Biol. 2017, 36, 482–489. [Google Scholar] [CrossRef]
- Karkeni, E.; Bonnet, L.; Marcotorchino, J.; Tourniaire, F.; Astier, J.; Ye, J.; Landrier, J.-F. Vitamin D limits inflammation-linked microRNA expression in adipocytes in vitro and in vivo: A new mechanism for the regulation of inflammation by vitamin D. Epigenetics 2018, 13, 156–162. [Google Scholar] [CrossRef] [Green Version]
- Giangreco, A.A.; Vaishnav, A.; Wagner, D.; Finelli, A.; Fleshner, N.; Van der Kwast, T.; Vieth, R.; Nonn, L. Tumor Suppressor microRNAs, miR-100 and -125b, Are Regulated by 1,25-dihydroxyvitamin D in Primary Prostate Cells and in Patient Tissue. Cancer Prev. Res. 2013, 6, 483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, S.; Wall, D.; Curran, C.; Newell, J.; Kerin, M.; Dwyer, R. MicroRNA-10a is reduced in breast cancer and regulated in part through retinoic acid. BMC Cancer 2015, 15, 345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, Y.; Liu, Z.; Yang, G.; Gao, D.; Niu, X. MicroRNA expression profiles in rats with selenium deficiency and the possible role of the Wnt/β-catenin signaling pathway in cardiac dysfunction. Int. J. Mol. Med. 2015, 35, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.M.; Liang, D.; Jin, J.; Li, D.J.; Zhang, Y.C.; Gao, Z.Y.; He, Y.T. Research progress on the relationship between zinc deficiency, related microRNAs, and esophageal carcinoma. Thorac. Cancer 2017, 8, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, B.; Pfaffl, M.W.; Dumpler, J.; von Mutius, E.; Ege, M.J. microRNA in native and processed cow’s milk and its implication for the farm milk effect on asthma. J. Allergy Clin. Immunol. 2016, 137, 1893–1895.e13. [Google Scholar] [CrossRef] [Green Version]
- Tingö, L.; Ahlberg, E.; Johansson, L.; Pedersen, S.A.; Chawla, K.; Sætrom, P.; Cione, E.; Simpson, M.R. Non-Coding RNAs in Human Breast Milk: A Systematic Review. Front. Immunol. 2021, 12, 3522. [Google Scholar] [CrossRef]
- Kupsco, A.; Prada, D.; Valvi, D.; Hu, L.; Petersen, M.S.; Coull, B.; Grandjean, P.; Weihe, P.; Baccarelli, A.A. Human milk extracellular vesicle miRNA expression and associations with maternal characteristics in a population-based cohort from the Faroe Islands. Sci. Rep. 2021, 11, 5840. [Google Scholar] [CrossRef]
- Carrillo-Lozano, E.; Sebastián-Valles, F.; Knott-Torcal, C. Circulating microRNAs in Breast Milk and Their Potential Impact on the Infant. Nutrients 2020, 12, 3066. [Google Scholar] [CrossRef]
- Beaver, L.M.; Kuintzle, R.; Buchanan, A.; Wiley, M.W.; Glasser, S.T.; Wong, C.P.; Johnson, G.S.; Chang, J.H.; Lohr, C.V.; Williams, D.E.; et al. Long noncoding RNAs and sulforaphane: A target for chemoprevention and suppression of prostate cancer. J. Nutr. Biochem. 2017, 42, 72–83. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Lin, Y.; Xu, J.; Chu, H.; Hao, S.; Liu, X.; Song, X.; Jiang, L.; Zheng, H. Luteolin suppresses tumor progression through lncRNA BANCR and its downstream TSHR/CCND1 signaling in thyroid carcinoma. Int. J. Clin. Exp. Pathol. 2017, 10, 9591–9598. [Google Scholar] [PubMed]
- Hu, D.-L.; Wang, G.; Yu, J.; Zhang, L.-H.; Huang, Y.-F.; Wang, D.; Zhou, H.-H. Epigallocatechin-3-gallate modulates long non-coding RNA and mRNA expression profiles in lung cancer cells. Mol. Med. Rep. 2019, 19, 1509–1520. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Chen, X.; Jiang, J.; Wan, X.; Wang, Y.; Xu, P. Epigallocatechin gallate reverses gastric cancer by regulating the long noncoding RNA LINC00511/miR-29b/KDM2A axis. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2020, 1866, 165856. [Google Scholar] [CrossRef]
- Luo, Y.; Yan, B.; Liu, L.; Yin, L.; Ji, H.; An, X.; Gladkich, J.; Qi, Z.; Delatorre, C.; Herr, I. Sulforaphane Inhibits the Expression of Long Noncoding RNA H19 and its Target APOBEC3G and Thereby Pancreatic Cancer Progression. Cancers 2021, 13, 827. [Google Scholar] [CrossRef]
- Jin, T.; Guo, Y.; Huang, Z.; Zhang, Q.; Huang, Z.; Zhang, Y.; Huang, Z. Vitamin D inhibits the proliferation of Oral Squamous Cell Carcinoma by suppressing lncRNA LUCAT1 through the MAPK pathway. J. Cancer 2020, 11, 5971–5981. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, S.; Guo, B. Vitamin D Suppresses Ovarian Cancer Growth and Invasion by Targeting Long Non-Coding RNA CCAT2. Int. J. Mol. Sci. 2020, 21, 2334. [Google Scholar] [CrossRef] [Green Version]
- Zuo, S.; Wu, L.; Wang, Y.; Yuan, X. Long Non-coding RNA MEG3 Activated by Vitamin D Suppresses Glycolysis in Colorectal Cancer via Promoting c-Myc Degradation. Front. Oncol. 2020, 10, 274. [Google Scholar] [CrossRef] [Green Version]
- Haro, D.; Marrero, P.F.; Relat, J. Nutritional Regulation of Gene Expression: Carbohydrate-, Fat- and Amino Acid-Dependent Modulation of Transcriptional Activity. Int. J. Mol. Sci. 2019, 20, 1386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.-J.; Cha, J.-Y. Recent insights into the role of ChREBP in intestinal fructose absorption and metabolism. BMB Rep. 2018, 51, 429–436. [Google Scholar] [PubMed] [Green Version]
- Havula, E.; Hietakangas, V. Sugar sensing by ChREBP/Mondo-Mlx-new insight into downstream regulatory networks and integration of nutrient-derived signals. Curr. Opin. Cell Biol. 2018, 51, 89–96. [Google Scholar] [CrossRef] [Green Version]
- Poupeau, A.; Postic, C. Cross-regulation of hepatic glucose metabolism via ChREBP and nuclear receptors. Biochim. Biophys. Acta 2011, 1812, 995–1006. [Google Scholar] [CrossRef] [Green Version]
- Meng, J.; Feng, M.; Dong, W.; Zhu, Y.; Li, Y.; Zhang, P.; Wu, L.; Li, M.; Lu, Y.; Chen, H.; et al. Identification of HNF-4α as a key transcription factor to promote ChREBP expression in response to glucose. Sci. Rep. 2016, 6, 23944. [Google Scholar] [CrossRef] [Green Version]
- Aaronson, D.S.; Horvath, C.M. A road map for those who don’t know JAK-STAT. Science 2002, 296, 1653–1655. [Google Scholar] [CrossRef]
- Shi, H.; Kokoeva, M.V.; Inouye, K.; Tzameli, I.; Yin, H.; Flier, J.S. TLR4 links innate immunity and fatty acid-induced insulin resistance. J. Clin. Investig. 2006, 116, 3015–3025. [Google Scholar] [CrossRef]
- Phillips, C.M.; Goumidi, L.; Bertrais, S.; Field, M.R.; Peloso, G.M.; Shen, J.; McManus, R.; Hercberg, S.; Lairon, D.; Planells, R.; et al. Dietary saturated fat modulates the association between STAT3 polymorphisms and abdominal obesity in adults. J. Nutr. 2009, 139, 2011–2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, S.; Tong, Q.; Liu, B.; Huang, W.; Tian, Y.; Fu, X. Targeting STAT3 in Cancer Immunotherapy. Mol. Cancer 2020, 19, 145. [Google Scholar] [CrossRef] [PubMed]
- Kwan, H.Y.; Liu, B.; Huang, C.; Fatima, S.; Su, T.; Zhao, X.; Ho, A.H.M.; Han, Q.; Hu, X.; Gong, R.-H.; et al. Signal transducer and activator of transcription-3 drives the high-fat diet-associated prostate cancer growth. Cell Death Dis. 2019, 10, 637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, Y.-S.; Maeda, N. PPARgamma: A critical determinant of body fat distribution in humans and mice. Trends Cardiovasc. Med. 2005, 15, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Evans, R.M.; Barish, G.D.; Wang, Y.-X. PPARs and the complex journey to obesity. Nat. Med. 2004, 10, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthy, M.V.; Lodhi, I.J.; Yin, L.; Malapaka, R.R.V.; Xu, H.E.; Turk, J.; Semenkovich, C.F. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell 2009, 138, 476–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.-X.; Lee, C.-H.; Tiep, S.; Yu, R.T.; Ham, J.; Kang, H.; Evans, R.M. Peroxisome-proliferator-activated receptor delta activates fat metabolism to prevent obesity. Cell 2003, 113, 159–170. [Google Scholar] [CrossRef] [Green Version]
- Herceg, Z. Epigenetics and cancer: Towards an evaluation of the impact of environmental and dietary factors. Mutagenesis 2007, 22, 91–103. [Google Scholar] [CrossRef] [Green Version]
- Jump, D.B. Fatty acid regulation of gene transcription. Crit. Rev. Clin. Lab. Sci. 2004, 41, 41–78. [Google Scholar] [CrossRef]
- Yue, L.; Ye, F.; Gui, C.; Luo, H.; Cai, J.; Shen, J.; Chen, K.; Shen, X.; Jiang, H. Ligand-binding regulation of LXR/RXR and LXR/PPAR heterodimerizations: SPR technology-based kinetic analysis correlated with molecular dynamics simulation. Protein Sci. 2005, 14, 812–822. [Google Scholar] [CrossRef] [Green Version]
- Mangelsdorf, D.J.; Thummel, C.; Beato, M.; Herrlich, P.; Schütz, G.; Umesono, K.; Blumberg, B.; Kastner, P.; Mark, M.; Chambon, P.; et al. The nuclear receptor superfamily: The second decade. Cell 1995, 83, 835–839. [Google Scholar] [CrossRef] [Green Version]
- Horton, J.D.; Bashmakov, Y.; Shimomura, I.; Shimano, H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc. Natl. Acad. Sci. USA 1998, 95, 5987–5992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimano, H.; Horton, J.D.; Shimomura, I.; Hammer, R.E.; Brown, M.S.; Goldstein, J.L. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J. Clin. Investig. 1997, 99, 846–854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zelcer, N.; Tontonoz, P. Liver X receptors as integrators of metabolic and inflammatory signaling. J. Clin. Investig. 2006, 116, 607–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martini, C.; Pallottini, V. Cholesterol: From feeding to gene regulation. Genes Nutr. 2007, 2, 181–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barone, R.; Rizzo, R.; Tabbì, G.; Malaguarnera, M.; Frye, R.E.; Bastin, J. Nuclear Peroxisome Proliferator-Activated Receptors (PPARs) as Therapeutic Targets of Resveratrol for Autism Spectrum Disorder. Int. J. Mol. Sci. 2019, 20, 1878. [Google Scholar] [CrossRef] [Green Version]
- Cheang, W.S.; Wong, W.T.; Wang, L.; Cheng, C.K.; Lau, C.W.; Ma, R.C.W.; Xu, A.; Wang, N.; Huang, Y.; Tian, X.Y. Resveratrol ameliorates endothelial dysfunction in diabetic and obese mice through sirtuin 1 and peroxisome proliferator-activated receptor δ. Pharmacol. Res. 2019, 139, 384–394. [Google Scholar] [CrossRef] [PubMed]
- Hoang, M.-H.; Jia, Y.; Lee, J.H.; Kim, Y.; Lee, S.-J. Kaempferol reduces hepatic triglyceride accumulation by inhibiting Akt. J. Food Biochem. 2019, 43, e13034. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Moneim, A.; El-Twab, S.M.A.; Yousef, A.I.; Reheim, E.S.A.; Ashour, M.B. Modulation of hyperglycemia and dyslipidemia in experimental type 2 diabetes by gallic acid and p-coumaric acid: The role of adipocytokines and PPARγ. Biomed. Pharmacother. 2018, 105, 1091–1097. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.X.; Li, Y.B.; Zhao, R.P. Epigallocatechin Gallate Attenuates β-Amyloid Generation and Oxidative Stress Involvement of PPARγ in N2a/APP695 Cells. Neurochem. Res. 2017, 42, 468–480. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Miao, B.; Hu, K.-Q.; Fu, X.; Wang, X.-D. Apo-10’-lycopenoic acid inhibits cancer cell migration and angiogenesis and induces peroxisome proliferator-activated receptor γ. J. Nutr. Biochem. 2018, 56, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Song, E.A.; Lim, J.W.; Kim, H. Docosahexaenoic acid inhibits IL-6 expression via PPARγ-mediated expression of catalase in cerulein-stimulated pancreatic acinar cells. Int. J. Biochem. Cell Biol. 2017, 88, 60–68. [Google Scholar] [CrossRef]
- Hwang, J.-K.; Yu, H.-N.; Noh, E.-M.; Kim, J.-M.; Hong, O.-Y.; Youn, H.J.; Jung, S.H.; Kwon, K.-B.; Kim, J.-S.; Lee, Y.-R. DHA blocks TPA-induced cell invasion by inhibiting MMP-9 expression via suppression of the PPAR-γ/NF-κB pathway in MCF-7 cells. Oncol. Lett. 2017, 13, 243–249. [Google Scholar] [CrossRef] [Green Version]
- Xin, F.-Z.; Zhao, Z.-H.; Zhang, R.-N.; Pan, Q.; Gong, Z.-Z.; Sun, C.; Fan, J.-G. Folic acid attenuates high-fat diet-induced steatohepatitis via deacetylase SIRT1-dependent restoration of PPARα. World J. Gastroenterol. 2020, 26, 2203–2220. [Google Scholar] [CrossRef]
- Guo, T.; Wang, Y.; Guo, Y.; Wu, S.; Chen, W.; Liu, N.; Wang, Y.; Geng, D. 1, 25-D(3) Protects From Cerebral Ischemia by Maintaining BBB Permeability via PPAR-γ Activation. Front. Cell. Neurosci. 2018, 12, 480. [Google Scholar] [CrossRef]
- Hoseini, R.; Damirchi, A.; Babaei, P. Vitamin D increases PPARγ expression and promotes beneficial effects of physical activity in metabolic syndrome. Nutrition 2016, 36, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Liu, D.; Zeng, R.; Xian, T.; Lu, Y.; Zeng, G.; Sun, Z.; Huang, B.; Huang, Q. Epigallocatechin-3-gallate inhibits adipogenesis through down-regulation of PPARγ and FAS expression mediated by PI3K-AKT signaling in 3T3-L1 cells. Eur. J. Pharmacol. 2017, 795, 134–142. [Google Scholar] [CrossRef]
- Hawley, C.; Mankins, C.; Byrd, S.K. Effects of the Bioflavonoid Quercetin on TLR4 Expression and NFKb activation in A375 Melanoma Cells. FASEB J. 2020, 34 (Suppl. 1), 1. [Google Scholar] [CrossRef]
- Zhao, M.; Ma, J.; Zhu, H.Y.; Zhang, X.H.; Du, Z.Y.; Xu, Y.J.; Yu, X.D. Apigenin inhibits proliferation and induces apoptosis in human multiple myeloma cells through targeting the trinity of CK2, Cdc37 and Hsp90. Mol. Cancer 2011, 10, 104. [Google Scholar] [CrossRef] [Green Version]
- De Bittencourt Pasquali, M.A.; Gelain, D.P.; Zeidán-Chuliá, F.; Pires, A.S.; Gasparotto, J.; Terra, S.R.; Moreira, J.C.F. Vitamin A (retinol) downregulates the receptor for advanced glycation endproducts (RAGE) by oxidant-dependent activation of p38 MAPK and NF-kB in human lung cancer A549 cells. Cell. Signal. 2013, 25, 939–954. [Google Scholar] [CrossRef]
- Wongsirisin, P.; Yodkeeree, S.; Yamada, S.; Limtrakul, P. Curcumin inhibition of the effects of Tip α induced cytokine expression in gastric cancer patients. PharmaNutrition 2018, 6, 24, 100–106. [Google Scholar] [CrossRef]
- Berrak, O.; Akkoc, Y.; Arisan, E.D.; Coker-Gurkan, A.; Obakan-Yerlikaya, P.; Palavan-Unsal, N. The inhibition of PI3K and NFkappaB promoted curcumin-induced cell cycle arrest at G2/M via altering polyamine metabolism in Bcl-2 overexpressing MCF-7 breast cancer cells. Biomed. Pharmacother. 2016, 77, 150–160. [Google Scholar] [CrossRef]
- Fan, Z.; Yao, J.; Li, Y.; Hu, X.; Shao, H.; Tian, X. Anti-inflammatory and antioxidant effects of curcumin on acute lung injury in a rodent model of intestinal ischemia reperfusion by inhibiting the pathway of NF-Kb. Int. J. Clin. Exp. Pathol. 2015, 8, 3451–3459. [Google Scholar]
- Mishra, A.; Kumar, R.; Tyagi, A.; Kohaar, I.; Hedau, S.; Bharti, A.C.; Sarker, S.; Dey, D.; Saluja, D.; Das, B. Curcumin modulates cellular AP-1, NF-kB, and HPV16 E6 proteins in oral cancer. Ecancermedicalscience 2015, 9, 525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Suwanwela, N.C.; Patumraj, S. Curcumin by down-regulating NF-kB and elevating Nrf2, reduces brain edema and neurological dysfunction after cerebral I/R. Microvasc. Res. 2016, 106, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Rasheduzzaman, M.; Jeong, J.-K.; Park, S.-Y. Resveratrol sensitizes lung cancer cell to TRAIL by p53 independent and suppression of Akt/NF-κB signaling. Life Sci. 2018, 208, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Chekalina, N.; Burmak, Y.; Petrov, Y.; Borisova, Z.; Manusha, Y.; Kazakov, Y.; Kaidashev, I. Quercetin reduces the transcriptional activity of NF-kB in stable coronary artery disease. Indian Heart J. 2018, 70, 593–597. [Google Scholar] [CrossRef]
- Chen, T.; Zhang, X.; Zhu, G.; Liu, H.; Chen, J.; Wang, Y.; He, X. Quercetin inhibits TNF-α induced HUVECs apoptosis and inflammation via downregulating NF-kB and AP-1 signaling pathway in vitro. Medicine 2020, 99, e22241. [Google Scholar] [CrossRef]
- Zhu, M.; Zhou, X.; Zhao, J. Quercetin prevents alcohol-induced liver injury through targeting of PI3K/Akt/nuclear factor-κB and STAT3 signaling pathway. Exp. Ther. Med. 2017, 14, 6169–6175. [Google Scholar] [CrossRef] [Green Version]
- Tong, J.; Shen, Y.; Zhang, Z.; Hu, Y.; Zhang, X.; Han, L. Apigenin inhibits epithelial-mesenchymal transition of human colon cancer cells through NF-κB/Snail signaling pathway. Biosci. Rep. 2019, 39, BSR20190452. [Google Scholar] [CrossRef] [Green Version]
- Xia, Y.; Yuan, M.; Li, S.; Thuan, U.T.; Nguyen, T.T.; Kang, T.W.; Liao, W.; Lian, S.; Jung, Y.D. Apigenin Suppresses the IL-1β-Induced Expression of the Urokinase-Type Plasminogen Activator Receptor by Inhibiting MAPK-Mediated AP-1 and NF-κB Signaling in Human Bladder Cancer T24 Cells. J. Agric. Food Chem. 2018, 66, 7663–7673. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.-S.; Choi, H.-S.; Kim, S.-R.; Choi, Y.K.; Woo, S.-M.; Shin, I.; Woo, J.-K.; Park, S.-Y.; Shin, Y.C.; Ko, S.-K. Apigenin induces apoptosis via extrinsic pathway, inducing p53 and inhibiting STAT3 and NFκB signaling in HER2-overexpressing breast cancer cells. Mol. Cell. Biochem. 2012, 366, 319–334. [Google Scholar] [CrossRef]
- Ai, X.-Y.; Qin, Y.; Liu, H.-J.; Cui, Z.-H.; Li, M.; Yang, J.-H.; Zhong, W.-L.; Liu, Y.-R.; Chen, S.; Sun, T.; et al. Apigenin inhibits colonic inflammation and tumorigenesis by suppressing STAT3-NF-κB signaling. Oncotarget 2017, 8, 100216–100226. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Yao, X.; Sun, B.; Jiang, W.; Liao, C.; Dai, X.; Chen, Y.; Chen, J.; Ding, R. Pretreatment with kaempferol attenuates microglia-mediate neuroinflammation by inhibiting MAPKs–NF–κB signaling pathway and pyroptosis after secondary spinal cord injury. Free Radic. Biol. Med. 2021, 168, 142–154. [Google Scholar] [CrossRef]
- Chen, H.-Y.; Jiang, Y.-W.; Kuo, C.-L.; Way, T.-D.; Chou, Y.-C.; Chang, Y.-S.; Chung, J.-G. Chrysin inhibit human melanoma A375.S2 cell migration and invasion via affecting MAPK signaling and NF-κB signaling pathway in vitro. Environ. Toxicol. 2019, 34, 434–442. [Google Scholar] [CrossRef]
- Liang, Y.; Feng, G.; Wu, L.; Zhong, S.; Gao, X.; Tong, Y.; Cui, W.; Qin, Y.; Xu, W.; Xiao, X.; et al. Caffeic acid phenethyl ester suppressed growth and metastasis of nasopharyngeal carcinoma cells by inactivating the NF-κB pathway. Drug Des. Dev. Ther. 2019, 13, 1335–1345. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Li, F.; Huang, Y.; Zhou, T.; Chen, S.; Li, G.; Shi, J.; Dong, N.; Xu, K. Caffeic Acid Phenethyl Ester Ameliorates Calcification by Inhibiting Activation of the AKT/NF-κB/NLRP3 Inflammasome Pathway in Human Aortic Valve Interstitial Cells. Front. Pharmacol. 2020, 11, 826. [Google Scholar] [CrossRef]
- Li, L.; Sun, W.; Wu, T.; Lu, R.; Shi, B. Caffeic acid phenethyl ester attenuates lipopolysaccharide-stimulated proinflammatory responses in human gingival fibroblasts via NF-κB and PI3K/Akt signaling pathway. Eur. J. Pharmacol. 2017, 794, 61–68. [Google Scholar] [CrossRef]
- Jia, Y.; Jiang, S.; Chen, C.; Lu, G.; Xie, Y.; Sun, X.; Huang, L. Caffeic acid phenethyl ester attenuates nuclear factor-κB-mediated inflammatory responses in Müller cells and protects against retinal ganglion cell death. Mol. Med. Rep. 2019, 19, 4863–4871. [Google Scholar] [PubMed] [Green Version]
- Cheng, H.; Zhang, Y.; Lu, W.; Gao, X.; Xu, C.; Bao, H. Caffeic acid phenethyl ester attenuates neuropathic pain by suppressing the p38/NF-κB signal pathway in microglia. J. Pain Res. 2018, 11, 2709–2719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.-L.; Han, N.-Z.; Liu, S.-S. Caffeic acid phenethyl ester inhibits the progression of ovarian cancer by regulating NF-κB signaling. Biomed. Pharmacother. 2018, 99, 825–831. [Google Scholar] [CrossRef] [PubMed]
- Fratantonio, D.; Speciale, A.; Canali, R.; Natarelli, L.; Ferrari, D.; Saija, A.; Virgili, F.; Cimino, F. Low nanomolar caffeic acid attenuates high glucose-induced endothelial dysfunction in primary human umbilical-vein endothelial cells by affecting NF-κB and Nrf2 pathways. BioFactors 2017, 43, 54–62. [Google Scholar] [CrossRef]
- Qu, Z.; Jia, L.; Xie, T.; Zhen, J.; Si, P.; Cui, Z.; Xue, Y.; Sun, C.; Wang, W. (−)-Epigallocatechin-3-Gallate Protects Against Lithium-Pilocarpine-Induced Epilepsy by Inhibiting the Toll-Like Receptor 4 (TLR4)/Nuclear Factor-κB (NF-κB) Signaling Pathway. Med Sci. Monit. Int. Med J. Exp. Clin. Res. 2019, 25, 1749–1758. [Google Scholar] [CrossRef]
- Zhang, L.; Xie, J.; Gan, R.; Wu, Z.; Luo, H.; Chen, X.; Lu, Y.; Wu, L.; Zheng, D. Synergistic inhibition of lung cancer cells by EGCG and NF-κB inhibitor BAY11-7082. J. Cancer 2019, 10, 6543–6556. [Google Scholar] [CrossRef]
- Xu, C.; Shen, G.; Chen, C.; Gelinas, C.; Kong, A.N. Suppression of NF-kappaB and NF-kappaB-regulated gene expression by sulforaphane and PEITC through IkappaBalpha, IKK pathway in human prostate cancer PC-3 cells. Oncogene 2005, 24, 4486–4495. [Google Scholar] [CrossRef] [Green Version]
- Jeong, Y.; Lim, J.; Kim, H. Lycopene Inhibits Reactive Oxygen Species-Mediated NF-κB Signaling and Induces Apoptosis in Pancreatic Cancer Cells. Nutrients 2019, 11, 762. [Google Scholar] [CrossRef] [Green Version]
- Assar, E.; Vidalle, M.; Chopra, M.; Hafizi, S. Lycopene acts through inhibition of IkB kinase to suppress NF-kB signaling in human prostate and breast cancer cells. Tumor Biol. 2016, 37, 9375–9385. [Google Scholar] [CrossRef] [Green Version]
- Enguita, M.; Razquin, N.; Pamplona, R.; Quiroga, J.; Prieto, J.; Fortes, P. The cirrhotic liver is depleted of docosahexaenoic acid (DHA), a key modulator of NF-κB and TGFβ pathways in hepatic stellate cells. Cell Death Dis. 2019, 10, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, M.; Lim, J.W.; Kim, H. Docoxahexaenoic Acid Induces Apoptosis of Pancreatic Cancer Cells by Suppressing Activation of STAT3 and NF-κB. Nutrients 2018, 10, 1621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagherieh, M.; Kheirollahi, A.; Zamani-Garmsiri, F.; Emamgholipour, S.; Meshkani, R. Folic acid ameliorates palmitate-induced inflammation through decreasing homocysteine and inhibiting NF-κB pathway in HepG2 cells. Arch. Physiol. Biochem. 2021. [Google Scholar] [CrossRef] [PubMed]
- Gasparian, A.V.; Yao, Y.J.; Lü, J.; Yemelyanov, A.Y.; Lyakh, L.A.; Slaga, T.J.; Budunova, I.V. Selenium Compounds Inhibit IκB Kinase (IKK) and Nuclear Factor-κB (NF-κB) in Prostate Cancer Cells 1 Supported by Department of Defense Prostate Cancer Research Program DAMD17-01-1-0015. Mol. Cancer Ther. 2002, 1, 1079. [Google Scholar] [PubMed]
- Christensen, M.; Nartey, E.; Hada, A.; Legg, R.; Barzee, B. High Selenium Reduces NF-κB-Regulated Gene Expression in Uninduced Human Prostate Cancer Cells. Nutr. Cancer 2007, 58, 197–204. [Google Scholar] [CrossRef]
- Chen, Y.-C.; Sosnoski, D.; Gandhi, U.; Novinger, L.; Prabhu, K.S.; Mastro, A. Selenium modifies the osteoblast inflammatory stress response to bone metastatic breast cancer. Carcinogenesis 2009, 30, 1941–1948. [Google Scholar] [CrossRef] [Green Version]
- Faure, P.; Ramon, O.; Favier, A.; Halimi, S. Selenium supplementation decreases nuclear factor-kappa B activity in peripheral blood mononuclear cells from type 2 diabetic patients. Eur. J. Clin. Investig. 2004, 34, 475–481. [Google Scholar] [CrossRef]
- Crispen, P.L.; Uzzo, R.G.; Golovine, K.; Makhov, P.; Pollack, A.; Horwitz, E.M.; Greenberg, R.E.; Kolenko, V.M. Vitamin E succinate inhibits NF-κB and prevents the development of a metastatic phenotype in prostate cancer cells: Implications for chemoprevention. Prostate 2007, 67, 582–590. [Google Scholar] [CrossRef]
- Wang, G.; Song, X.; Zhao, L.; Li, Z.; Liu, B. Resveratrol Prevents Diabetic Cardiomyopathy by Increasing Nrf2 Expression and Transcriptional Activity. BioMed Res. Int. 2018, 2018, 2150218. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Zhao, X.; Zhu, H.; Wang, J.; Ma, J.; Gu, M. Apigenin Protects Against Renal Tubular Epithelial Cell Injury and Oxidative Stress by High Glucose via Regulation of NF-E2-Related Factor 2 (Nrf2) Pathway. Med Sci. Monit. Int. Med J. Exp. Clin. Res. 2019, 25, 5280–5288. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, J.; Zhao, G.; Lin, M.; Lang, Y.; Zhang, D.; Feng, D.; Tu, C. Apigenin protects human melanocytes against oxidative damage by activation of the Nrf2 pathway. Cell Stress Chaperones 2020, 25, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.; Piao, M.; Hyun, Y.; Zhen, A.X.; Cho, S.; Ahn, M.; Yi, J.; Hyun, J. Luteolin promotes apoptotic cell death via upregulation of Nrf2 expression by DNA demethylase and the interaction of Nrf2 with p53 in human colon cancer cells. Exp. Mol. Med. 2019, 51, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, Q.; Wu, R.; Xiao, X.; Yang, C.; Yang, A.; Wang, C.; Lin, L.; Kong, A.-N. The dietary flavone luteolin epigenetically activates the Nrf2 pathway and blocks cell transformation in human colorectal cancer HCT116 cells. J. Cell. Biochem. 2018, 119, 9573–9582. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liu, B.-F.; Xie, F.-J.; Yang, W.-L.; Cao, N. Luteolin induces mitochondrial apoptosis in HT29 cells by inhibiting the Nrf2/ARE signaling pathway. Exp. Ther. Med. 2020, 19, 2179–2187. [Google Scholar] [CrossRef] [Green Version]
- Sampath, C.; Rashid, M.R.; Sang, S.; Ahmedna, M. Green tea epigallocatechin 3-gallate alleviates hyperglycemia and reduces advanced glycation end products via nrf2 pathway in mice with high fat diet-induced obesity. Biomed. Pharmacother. 2017, 87, 73–81. [Google Scholar] [CrossRef]
- Enkhbat, T.; Nishi, M.; Yoshikawa, K.; Jun, H.; Tokunaga, T.; Takasu, C.; Kashihara, H.; Ishikawa, D.; Tominaga, M.; Shimada, M. Epigallocatechin-3-gallate Enhances Radiation Sensitivity in Colorectal Cancer Cells Through Nrf2 Activation and Autophagy. Anticancer Res. 2018, 38, 6247. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Pérez, J.; Martínez-Rosas, M.; Conde-Castañón, C.A.; Toscano-Garibay, J.D.; Ruiz-Pérez, N.J.; Flores, P.L.; Mera Jiménez, E.; Flores-Estrada, J. Epigallocatechin 3-Gallate Has a Neuroprotective Effect in Retinas of Rabbits with Ischemia/Reperfusion through the Activation of Nrf2/HO-1. Int. J. Mol. Sci. 2020, 21, 3716. [Google Scholar] [CrossRef]
- Zhou, J.-W.; Wang, M.; Sun, N.-X.; Qing, Y.; Yin, T.-F.; Li, C.; Wu, D. Sulforaphane-induced epigenetic regulation of Nrf2 expression by DNA methyltransferase in human Caco-2 cells. Oncol. Lett. 2019, 18, 2639–2647. [Google Scholar] [CrossRef] [Green Version]
- Zhao, F.; Zhang, J.-L.; Chang, N. Epigenetic modification of Nrf2 by sulforaphane increases the antioxidative and anti-inflammatory capacity in a cellular model of Alzheimer’s disease. Eur. J. Pharmacol. 2018, 824, 1–10. [Google Scholar] [CrossRef]
- Zhu, W.; Ding, Y.; Kong, W.; Li, T.; Chen, H. Docosahexaenoic Acid (DHA) Provides Neuroprotection in Traumatic Brain Injury Models via Activating Nrf2-ARE Signaling. Inflammation 2018, 41, 1182–1193. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Xiu, P.; Li, F.; Xin, C.; Li, K. Vitamin A Supplementation Alleviates Extrahepatic Cholestasis Liver Injury through Nrf2 Activation. Oxid. Med. Cell. Longev. 2014, 2014, 273692. [Google Scholar] [CrossRef] [Green Version]
- Fang, J.; Yin, H.; Yang, Z.; Tan, M.; Wang, F.; Chen, K.; Zuo, Z.; Shu, G.; Cui, H.; Ouyang, P.; et al. Vitamin E protects against cadmium-induced sub-chronic liver injury associated with the inhibition of oxidative stress and activation of Nrf2 pathway. Ecotoxicol. Environ. Saf. 2021, 208, 111610. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, S.; Ghorbani, M.; Sabzichi, M.; Ramezani, F.; Hamishehkar, H.; Samadi, N. Targeted hyaluronic acid-based lipid nanoparticle for apigenin delivery to induce Nrf2-dependent apoptosis in lung cancer cells. J. Drug Deliv. Sci. Technol. 2019, 49, 268–276. [Google Scholar] [CrossRef]
- Li, L.; Luo, W.; Qian, Y.; Zhu, W.; Qian, J.; Li, J.; Jin, Y.; Xu, X.; Liang, G. Luteolin protects against diabetic cardiomyopathy by inhibiting NF-κB-mediated inflammation and activating the Nrf2-mediated antioxidant responses. Phytomedicine 2019, 59, 152774. [Google Scholar] [CrossRef] [PubMed]
- Fouzder, C.; Mukhuty, A.; Kundu, R. Kaempferol inhibits Nrf2 signalling pathway via downregulation of Nrf2 mRNA and induces apoptosis in NSCLC cells. Arch. Biochem. Biophys. 2021, 697, 108700. [Google Scholar] [CrossRef] [PubMed]
- Sabzichi, M.; Mohammadian, J.; Bazzaz, R.; Pirouzpanah, M.; Shaaker, M.; Hamishehkar, H.; Chavoshi, H.; Salehi, R.; Samadi, N. Chrysin loaded nanostructured lipid carriers (NLCs) triggers apoptosis in MCF-7 cancer cells by inhibiting the Nrf2 pathway. Process Biochem. 2017, 60, 84–91. [Google Scholar] [CrossRef]
- Wang, J.; Wang, H.; Sun, K.; Wang, X.; Pan, H.; Zhu, J.; Ji, X.; Li, X. Chrysin suppresses proliferation, migration, and invasion in glioblastoma cell lines via mediating the ERK/Nrf2 signaling pathway. Drug Des. Dev. Ther. 2018, 12, 721–733. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Li, X.; Wei, J.a.; Chen, H.; Lu, Y.; Li, L.; Han, L.; Lu, C. Gallic acid inhibits the expression of keratin 16 and keratin 17 through Nrf2 in psoriasis-like skin disease. Int. Immunopharmacol. 2018, 65, 84–95. [Google Scholar] [CrossRef]
- Radan, M.; Dianat, M.; Badavi, M.; Mard, S.A.; Bayati, V.; Goudarzi, G. In vivo and in vitro evidence for the involvement of Nrf2-antioxidant response element signaling pathway in the inflammation and oxidative stress induced by particulate matter (PM10): The effective role of gallic acid. Free Radic. Res. 2019, 53, 210–225. [Google Scholar] [CrossRef]
- Dworski, R.; Han, W.; Blackwell, T.S.; Hoskins, A.; Freeman, M.L. Vitamin E prevents NRF2 suppression by allergens in asthmatic alveolar macrophages in vivo. Free Radic. Biol. Med. 2011, 51, 516–521. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Nie, P.; Lu, X.; Li, C.; Dong, X.; Yang, F.; Luo, P.; Li, B. Nrf2 participates in the anti-apoptotic role of zinc in Type 2 diabetic nephropathy through Wnt/β-catenin signaling pathway. J. Nutr. Biochem. 2020, 84, 108451. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, L.; Zhang, T.; Wu, W.; Liu, J.; Wang, X.; Wan, Y.; Geng, H.; Sun, X.; Qian, W.; et al. Curcumin Negatively Regulates Cigarette Smoke-Induced Renal Cell Carcinoma Epithelial-Mesenchymal Transition Through the ERK5/AP-1 Pathway. OncoTargets Ther. 2020, 13, 9689–9700. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, J.; Zhao, L.; Geng, H.; Ma, J.; Zhang, Z.; Yu, D.; Zhong, C. Curcumin reverses benzidine-induced epithelial-mesenchymal transition via suppression of ERK5/AP-1 in SV-40 immortalized human urothelial cells. Int. J. Oncol. 2017, 50, 1321–1329. [Google Scholar] [CrossRef] [Green Version]
- Pang, J.-H.S.; Yen, J.-H.; Wu, H.-T.; Huang, S.-T. Gallic Acid Inhibited Matrix Invasion and AP-1/ETS-1-Mediated MMP-1 Transcription in Human Nasopharyngeal Carcinoma Cells. Int. J. Mol. Sci. 2017, 18, 1354. [Google Scholar] [CrossRef] [Green Version]
- Chuang, C.-H.; Huang, C.-S.; Hu, M.-L. Vitamin E and rutin synergistically inhibit expression of vascular endothelial growth factor through down-regulation of binding activity of activator protein-1 in human promyelocytic leukemia (HL-60) cells. Chem. Biol. Interact. 2009, 183, 434–441. [Google Scholar] [CrossRef]
- Uzzo, R.; Crispen, P.; Golovine, K.; Makhov, P.; Horwitz, E.; Kolenko, V. Diverse effects of zinc on NF-κB and AP-1 transcription factors: Implications for prostate cancer progression. Carcinogenesis 2006, 27, 1980–1990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, K.-H.; Wu, H.-H.; Lin, R.-C.; Lin, Y.-C.; Lu, P.W.-A.; Yang, S.-F.; Yang, J.-S. Curcumin Analogue L48H37 Suppresses Human Osteosarcoma U2OS and MG-63 Cells’ Migration and Invasion in Culture by Inhibition of uPA via the JAK/STAT Signaling Pathway. Molecules 2020, 26, 30. [Google Scholar] [CrossRef] [PubMed]
- Petiti, J.; Rosso, V.; Lo Iacono, M.; Panuzzo, C.; Calabrese, C.; Signorino, E.; Pironi, L.; Cartellà, A.; Bracco, E.; Pergolizzi, B.; et al. Curcumin induces apoptosis in JAK2-mutated cells by the inhibition of JAK2/STAT and mTORC1 pathways. J. Cell. Mol. Med. 2019, 23, 4349–4357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, L.; Jiang, D. Resveratrol eliminates cancer stem cells of osteosarcoma by STAT3 pathway inhibition. PLoS ONE 2018, 13, e0205918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Wang, G.; Jin, G.; Yao, K.; Zhao, Z.; Bie, L.; Guo, Y.; Li, N.; Deng, W.; Chen, X.; et al. Resveratrol suppresses colon cancer growth by targeting the AKT/STAT3 signaling pathway. Int. J. Mol. Med. 2019, 43, 630–640. [Google Scholar] [CrossRef] [Green Version]
- Zhong, L.-X.; Nie, J.-H.; Liu, J.; Lin, L.-Z. Correlation of ARHI upregulation with growth suppression and STAT3 inactivation in resveratrol-treated ovarian cancer cells. Cancer Biomark. 2018, 21, 787–795. [Google Scholar] [CrossRef]
- Sun, X.; Xu, Q.; Zeng, L.; Xie, L.; Zhao, Q.; Xu, H.; Wang, X.; Jiang, N.; Fu, P.; Sang, M. Resveratrol suppresses the growth and metastatic potential of cervical cancer by inhibiting STAT3(Tyr705) phosphorylation. Cancer Med. 2020, 9, 8685–8700. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Li, J.; Liu, T.; Li, S.; Feng, J.; Yu, Q.; Zhang, J.; Chen, J.; Zhou, Y.; Ji, J.; et al. Quercetin shows anti-tumor effect in hepatocellular carcinoma LM3 cells by abrogating JAK2/STAT3 signaling pathway. Cancer Med. 2019, 8, 4806–4820. [Google Scholar] [CrossRef]
- Omar, H.; Obaya, E.; Sabry, D.; Abdelkader, A.; Maher, M.; Mekawy, D. Apigenin inhibits proliferation of hepatocellular carcinoma cell by upregulation of cleaved caspases-3/8 and downregulation of pSTAT-3/pJAK-1/pJAK-2. Gene Rep. 2020, 21, 100964. [Google Scholar] [CrossRef]
- Seo, H.-S.; Ku, J.M.; Choi, H.S.; Woo, J.-K.; Lee, B.H.; Kim, D.S.; Song, H.J.; Jang, B.-H.; Shin, Y.C.; Ko, S.-G. Apigenin overcomes drug resistance by blocking the signal transducer and activator of transcription 3 signaling in breast cancer cells. Oncol. Rep. 2017, 38, 715–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeda, Y.; Takahashi, H.; Nakai, N.; Yanagita, T.; Ando, N.; Okubo, T.; Saito, K.; Shiga, K.; Hirokawa, T.; Hara, M.; et al. Apigenin induces apoptosis by suppressing Bcl-xl and Mcl-1 simultaneously via signal transducer and activator of transcription 3 signaling in colon cancer. Int. J. Oncol. 2018, 52, 1661–1673. [Google Scholar] [CrossRef]
- Su, T.; Huang, C.; Yang, C.; Jiang, T.; Su, J.; Chen, M.; Fatima, S.; Gong, R.; Hu, X.; Bian, Z.; et al. Apigenin inhibits STAT3/CD36 signaling axis and reduces visceral obesity. Pharmacol. Res. 2020, 152, 104586. [Google Scholar] [CrossRef]
- Song, S.; Su, Z.; Xu, H.; Niu, M.; Chen, X.; Min, H.; Zhang, B.; Sun, G.; Xie, S.; Wang, H.; et al. Correction: Luteolin selectively kills STAT3 highly activated gastric cancer cells through enhancing the binding of STAT3 to SHP-1. Cell Death Dis. 2018, 9, 787. [Google Scholar] [CrossRef] [Green Version]
- Kato, H.; Naiki-Ito, A.; Suzuki, S.; Inaguma, S.; Komura, M.; Nakao, K.; Naiki, T.; Kachi, K.; Kato, A.; Matsuo, Y.; et al. DPYD, down-regulated by the potentially chemopreventive agent luteolin, interacts with STAT3 in pancreatic cancer. Carcinogenesis 2021, 42(7), 940–950. [Google Scholar] [CrossRef]
- Cummins, C.B.; Wang, X.; Nunez Lopez, O.; Graham, G.; Tie, H.-Y.; Zhou, J.; Radhakrishnan, R.S. Luteolin-Mediated Inhibition of Hepatic Stellate Cell Activation via Suppression of the STAT3 Pathway. Int. J. Mol. Sci. 2018, 19, 1567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonoki, H.; Tanimae, A.; Endo, S.; Matsunaga, T.; Furuta, T.; Ichihara, K.; Ikari, A. Kaempherol and Luteolin Decrease Claudin-2 Expression Mediated by Inhibition of STAT3 in Lung Adenocarcinoma A549 Cells. Nutrients 2017, 9, 597. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; He, L.; Liu, J.; Zhou, L. Luteolin Attenuates Diabetic Nephropathy through Suppressing Inflammatory Response and Oxidative Stress by Inhibiting STAT3 Pathway. Exp. Clin. Endocrinol. Diabetes 2020. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Tong, Y.; Ying, J.; Lei, Z.; Wan, L.; Zhu, X.; Ye, F.; Mao, P.; Wu, X.; Pan, R.; et al. Chrysin induces cell growth arrest, apoptosis, and ER stress and inhibits the activation of STAT3 through the generation of ROS in bladder cancer cells. Oncol. Lett. 2018, 15, 9117–9125. [Google Scholar] [CrossRef]
- Zhang, T.; Ma, L.; Wu, P.; Li, W.; Li, T.; Gu, R.; Dan, X.; Li, Z.; Fan, X.; Xiao, Z. Gallic acid has anticancer activity and enhances the anticancer effects of cisplatin in non-small cell lung cancer A549 cells via the JAK/STAT3 signaling pathway. Oncol. Rep. 2019, 41, 1779–1788. [Google Scholar] [CrossRef] [Green Version]
- Tasaki, S.; Horiguchi, A.; Asano, T.; Ito, K.; Asano, T.; Asakura, H. Docosahexaenoic acid inhibits the phosphorylation of STAT3 and the growth and invasion of renal cancer cells. Exp. Ther. Med. 2017, 14, 1146–1152. [Google Scholar] [CrossRef] [Green Version]
- D’Eliseo, D.; Di Renzo, L.; Santoni, A.; Velotti, F. Docosahexaenoic acid (DHA) promotes immunogenic apoptosis in human multiple myeloma cells, induces autophagy and inhibits STAT3 in both tumor and dendritic cells. Genes cancer 2017, 8, 426–437. [Google Scholar] [CrossRef] [Green Version]
- Miao, Z.; Yu, F.; Ren, Y.; Yang, J. d,l-Sulforaphane Induces ROS-Dependent Apoptosis in Human Gliomablastoma Cells by Inactivating STAT3 Signaling Pathway. Int. J. Mol. Sci. 2017, 18, 72. [Google Scholar] [CrossRef] [Green Version]
- Fu, H.; Wang, C.; Yang, D.; Wei, Z.; Xu, J.; Hu, Z.; Zhang, Y.; Wang, W.; Yan, R.; Cai, Q. Curcumin regulates proliferation, autophagy, and apoptosis in gastric cancer cells by affecting PI3K and P53 signaling. J. Cell. Physiol. 2018, 233, 4634–4642. [Google Scholar] [CrossRef]
- Sidhar, H.; Giri, R.K. Induction of Bex genes by curcumin is associated with apoptosis and activation of p53 in N2a neuroblastoma cells. Sci. Rep. 2017, 7, 41420. [Google Scholar] [CrossRef]
- Xu, S.; Yang, Z.; Fan, Y.; Guan, B.; Jia, J.; Gao, Y.; Wang, K.; Wu, K.; Wang, X.; Zheng, P.; et al. Curcumin enhances temsirolimus-induced apoptosis in human renal carcinoma cells through upregulation of YAP/p53. Oncol. Lett. 2016, 12, 4999–5006. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.K.; Banerjee, S.; Acosta, E.P.; Lillard, J.W.; Singh, R. Resveratrol induces cell cycle arrest and apoptosis with docetaxel in prostate cancer cells via a p53/ p21WAF1/CIP1 and p27KIP1 pathway. Oncotarget 2017, 8, 17216–17228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Gao, Z.; Zhang, X. Resveratrol Induces Apoptosis in Murine Prostate Cancer Cells via Hypoxia-Inducible Factor 1-alpha (HIF-1α)/Reactive Oxygen Species (ROS)/P53 Signaling. Med Sci. Monit. Int. Med J. Exp. Clin. Res. 2018, 24, 8970–8976. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wu, X.; Lv, J.; Sun, H.; Zhou, F. Resveratrol induces p53 in colorectal cancer through SET7/9. Oncol. Lett. 2019, 17, 3783–3789. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Yin, X.; Sui, S. Resveratrol inhibited the progression of human hepatocellular carcinoma by inducing autophagy via regulating p53 and the phosphoinositide 3-kinase/protein kinase B pathway. Oncol. Rep. 2018, 40, 2758–2765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, P.A.; Bhattacharya, S.; Elmayan, A.; Darjatmoko, S.R.; Thuro, B.A.; Yan, M.B.; van Ginkel, P.R.; Polans, A.S.; Kuo, J.S. Resveratrol targeting of AKT and p53 in glioblastoma and glioblastoma stem-like cells to suppress growth and infiltration. J. Neurosurg. 2017, 126, 1448–1460. [Google Scholar] [CrossRef] [Green Version]
- Liontas, A.; Yeger, H. Curcumin and resveratrol induce apoptosis and nuclear translocation and activation of p53 in human neuroblastoma. Anticancer Res. 2004, 24, 87–98. [Google Scholar]
- Shih, A.; Davis, F.B.; Lin, H.Y.; Davis, P.J. Resveratrol induces apoptosis in thyroid cancer cell lines via a MAPK- and p53-dependent mechanism. J. Clin. Endocrinol. Metab. 2002, 87, 1223–1232. [Google Scholar] [CrossRef]
- Kuo, P.L.; Lin, C.C. Green tea constituent (-)-epigallocatechin-3-gallate inhibits Hep G2 cell proliferation and induces apoptosis through p53-dependent and Fas-mediated pathways. J. Biomed. Sci. 2003, 10, 219–227. [Google Scholar] [PubMed]
- Karagül, M.İ.; Aktaş, S.; Yetkin, D.; Bayrak, G.; Çelikcan, D. P53, Bcl2 and Bax Expression and Apoptosis in Perifosine and Vitamin D-Treated Endometrial Cancer Cell Line (HEC1A). Proceedings 2018, 2, 1564. [Google Scholar] [CrossRef] [Green Version]
- Yu, T.; Wang, Z.; You, X.; Zhou, H.; He, W.; Li, B.; Xia, J.; Zhu, H.; Zhao, Y.; Yu, G.; et al. Resveratrol promotes osteogenesis and alleviates osteoporosis by inhibiting p53. Aging 2020, 12, 10359–10369. [Google Scholar] [CrossRef]
- Ferraz da Costa, D.C.; Campos, N.P.C.; Santos, R.A.; Guedes-da-Silva, F.H.; Martins-Dinis, M.M.D.C.; Zanphorlin, L.; Ramos, C.; Rangel, L.P.; Silva, J.L. Resveratrol prevents p53 aggregation in vitro and in breast cancer cells. Oncotarget 2018, 9, 29112–29122. [Google Scholar] [CrossRef] [Green Version]
- Diao, Q.X.; Zhang, J.Z.; Zhao, T.; Xue, F.; Gao, F.; Ma, S.M.; Wang, Y. Vitamin E promotes breast cancer cell proliferation by reducing ROS production and p53 expression. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 2710–2717. [Google Scholar]
- Hellsten, S.V.; Tripathi, R.; Ceder, M.M.; Fredriksson, R. Nutritional Stress Induced by Amino Acid Starvation Results in Changes for Slc38 Transporters in Immortalized Hypothalamic Neuronal Cells and Primary Cortex Cells. Front. Mol. Biosci. 2018, 5, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anthony, T.G.; McDaniel, B.J.; Byerley, R.L.; McGrath, B.C.; Cavener, D.R.; McNurlan, M.A.; Wek, R.C. Preservation of liver protein synthesis during dietary leucine deprivation occurs at the expense of skeletal muscle mass in mice deleted for eIF2 kinase GCN2. J. Biol. Chem. 2004, 279, 36553–36561. [Google Scholar] [CrossRef] [Green Version]
- Costa-Mattioli, M.; Walter, P. The integrated stress response: From mechanism to disease. Science 2020, 368, eaat5314. [Google Scholar] [CrossRef] [PubMed]
- Bogorad, A.M.; Lin, K.Y.; Marintchev, A. eIF2B Mechanisms of Action and Regulation: A Thermodynamic View. Biochemistry 2018, 57, 1426–1435. [Google Scholar] [CrossRef] [PubMed]
- Shan, J.; Ord, D.; Ord, T.; Kilberg, M.S. Elevated ATF4 expression, in the absence of other signals, is sufficient for transcriptional induction via CCAAT enhancer-binding protein-activating transcription factor response elements. J. Biol. Chem. 2009, 284, 21241–21248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kilberg, M.S.; Balasubramanian, M.; Fu, L.; Shan, J. The transcription factor network associated with the amino acid response in mammalian cells. Adv. Nutr. 2012, 3, 295–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sensing, Signaling and Cell Adaptation, 1st ed. eBook. Volume 3. Available online: https://www.elsevier.com/books/sensing-signaling-and-cell-adaptation/storey/978-0-444-51147-8 (accessed on 13 October 2021).
- Nutrient Metabolism, 2nd ed. eBook. Available online: https://www.elsevier.com/books/nutrient-metabolism/kohlmeier/978-0-12-387784-0 (accessed on 13 October 2021).
- Meeran, S.M.; Katiyar, S.K. Cell cycle control as a basis for cancer chemoprevention through dietary agents. Front. Biosci. J. Virtual Libr. 2008, 13, 2191–2202. [Google Scholar] [CrossRef] [Green Version]
- Anand, P.; Thomas, S.G.; Kunnumakkara, A.B.; Sundaram, C.; Harikumar, K.B.; Sung, B.; Tharakan, S.T.; Misra, K.; Priyadarsini, I.K.; Rajasekharan, K.N.; et al. Biological activities of curcumin and its analogues (Congeners) made by man and Mother Nature. Biochem. Pharmacol. 2008, 76, 1590–1611. [Google Scholar] [CrossRef]
- Shankar, S.; Ganapathy, S.; Chen, Q.; Srivastava, R.K. Curcumin sensitizes TRAIL-resistant xenografts: Molecular mechanisms of apoptosis, metastasis and angiogenesis. Mol. Cancer 2008, 7, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, R.-X.; Ong, C.-N.; Shen, H.-M. Protein kinase C inhibition and x-linked inhibitor of apoptosis protein degradation contribute to the sensitization effect of luteolin on tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis in cancer cells. Cancer Res. 2005, 65, 7815–7823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shishodia, S.; Aggarwal, B.B. Guggulsterone inhibits NF-kappaB and IkappaBalpha kinase activation, suppresses expression of anti-apoptotic gene products, and enhances apoptosis. J. Biol. Chem. 2004, 279, 47148–47158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, V.; Belloir, C.; Siess, M.H.; Le Bon, A.M. Inhibition of carcinogen-induced DNA damage in rat liver and colon by garlic powders with varying alliin content. Nutr. Cancer 2006, 55, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. Polyunsaturated fatty acids and inflammation. Biochem. Soc. Trans. 2005, 33, 423–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, K.H.; Barve, A.; Yu, S.; Huang, M.T.; Kong, A.N. Cancer chemoprevention by phytochemicals: Potential molecular targets, biomarkers and animal models. Acta Pharmacol. Sin. 2007, 28, 1409–1421. [Google Scholar] [CrossRef]
- Yang, F.; Oz, H.S.; Barve, S.; de Villiers, W.J.; McClain, C.J.; Varilek, G.W. The green tea polyphenol (-)-epigallocatechin-3-gallate blocks nuclear factor-kappa B activation by inhibiting I kappa B kinase activity in the intestinal epithelial cell line IEC-6. Mol. Pharmacol. 2001, 60, 528–533. [Google Scholar]
- Shen, G.; Jeong, W.-S.; Hu, R.; Kong, A.-N.T. Regulation of Nrf2, NF-kappaB, and AP-1 signaling pathways by chemopreventive agents. Antioxid. Redox Signal. 2005, 7, 1648–1663. [Google Scholar] [CrossRef]
- Hess, J.; Angel, P.; Schorpp-Kistner, M. AP-1 subunits: Quarrel and harmony among siblings. J. Cell Sci. 2004, 117, 5965–5973. [Google Scholar] [CrossRef] [Green Version]
- Jeong, W.-S.; Kim, I.-W.; Hu, R.; Kong, A.-N.T. Modulation of AP-1 by natural chemopreventive compounds in human colon HT-29 cancer cell line. Pharm. Res. 2004, 21, 649–660. [Google Scholar] [CrossRef] [PubMed]
- Granado-Serrano, A.B.; Martín, M.A.; Haegeman, G.; Goya, L.; Bravo, L.; Ramos, S. Epicatechin induces NF-kappaB, activator protein-1 (AP-1) and nuclear transcription factor erythroid 2p45-related factor-2 (Nrf2) via phosphatidylinositol-3-kinase/protein kinase B (PI3K/AKT) and extracellular regulated kinase (ERK) signalling in HepG2 cells. Br. J. Nutr. 2010, 103, 168–179. [Google Scholar] [PubMed] [Green Version]
- Zhou, C.; Verma, S.; Blumberg, B. The steroid and xenobiotic receptor (SXR), beyond xenobiotic metabolism. Nucl. Recept. Signal. 2009, 7, e001. [Google Scholar] [CrossRef] [Green Version]
- Alkhalaf, M. Resveratrol-induced apoptosis is associated with activation of p53 and inhibition of protein translation in T47D human breast cancer cells. Pharmacology 2007, 80, 134–143. [Google Scholar] [CrossRef]
- Singh, R.P.; Tyagi, A.; Sharma, G.; Mohan, S.; Agarwal, R. Oral silibinin inhibits in vivo human bladder tumor xenograft growth involving down-regulation of survivin. Clin. Cancer Res. 2008, 14, 300–308. [Google Scholar] [CrossRef] [Green Version]
- Tian, B.; Wang, Z.; Zhao, Y.; Wang, D.; Li, Y.; Ma, L.; Li, X.; Li, J.; Xiao, N.; Tian, J.; et al. Effects of curcumin on bladder cancer cells and development of urothelial tumors in a rat bladder carcinogenesis model. Cancer Lett. 2008, 264, 299–308. [Google Scholar] [CrossRef]
- Block, G.; Patterson, B.; Subar, A. Fruit, vegetables, and cancer prevention: A review of the epidemiological evidence. Nutr. Cancer 1992, 18, 1–29. [Google Scholar] [CrossRef]
- Li, Y.H.; Niu, Y.B.; Sun, Y.; Zhang, F.; Liu, C.X.; Fan, L.; Mei, Q.B. Role of phytochemicals in colorectal cancer prevention. World J. Gastroenterol. 2015, 21, 9262–9272. [Google Scholar] [CrossRef] [PubMed]
- Reddy, L.; Odhav, B.; Bhoola, K.D. Natural products for cancer prevention: A global perspective. Pharmacol. Ther. 2003, 99, 1–13. [Google Scholar] [CrossRef]
- Vieira, A.R.; Abar, L.; Vingeliene, S.; Chan, D.S.; Aune, D.; Navarro-Rosenblatt, D.; Stevens, C.; Greenwood, D.; Norat, T. Fruits, vegetables and lung cancer risk: A systematic review and meta-analysis. Ann. Oncol. 2016, 27, 81–96. [Google Scholar] [CrossRef] [PubMed]
- Dzialo, M.; Mierziak, J.; Korzun, U.; Preisner, M.; Szopa, J.; Kulma, A. The Potential of Plant Phenolics in Prevention and Therapy of Skin Disorders. Int. J. Mol. Sci. 2016, 17, 160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harb, H.; Irvine, J.; Amarasekera, M.; Hii, C.S.; Kesper, D.A.; Ma, Y.; D’Vaz, N.; Renz, H.; Potaczek, D.P.; Prescott, S.L.; et al. The role of PKCζ in cord blood T-cell maturation towards Th1 cytokine profile and its epigenetic regulation by fish oil. Biosci. Rep. 2017, 37, BSR20160485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harb, H.; Alashkar Alhamwe, B.; Garn, H.; Renz, H.; Potaczek, D.P. Recent developments in epigenetics of pediatric asthma. Curr. Opin. Pediatr. 2016, 28, 754–763. [Google Scholar] [CrossRef] [PubMed]
- Dunstan, J.A.; Mori, T.A.; Barden, A.; Beilin, L.J.; Taylor, A.L.; Holt, P.G.; Prescott, S.L. Fish oil supplementation in pregnancy modifies neonatal allergen-specific immune responses and clinical outcomes in infants at high risk of atopy: A randomized, controlled trial. J. Allergy Clin. Immunol. 2003, 112, 1178–1184. [Google Scholar] [CrossRef] [PubMed]
- Acevedo, N.; Alashkar Alhamwe, B.; Caraballo, L.; Ding, M.; Ferrante, A.; Garn, H.; Garssen, J.; Hii, C.S.; Irvine, J.; Llinás-Caballero, K.; et al. Perinatal and Early-Life Nutrition, Epigenetics, and Allergy. Nutrients 2021, 13, 724. [Google Scholar] [CrossRef] [PubMed]
- Alaskhar Alhamwe, B.; Khalaila, R.; Wolf, J.; von Bülow, V.; Harb, H.; Alhamdan, F.; Hii, C.S.; Prescott, S.L.; Ferrante, A.; Renz, H.; et al. Histone modifications and their role in epigenetics of atopy and allergic diseases. Allergy Asthma Clin. Immunol. 2018, 14, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acevedo, N.; Frumento, P.; Harb, H.; Alashkar Alhamwe, B.; Johansson, C.; Eick, L.; Alm, J.; Renz, H.; Scheynius, A.; Potaczek, D.P. Histone Acetylation of Immune Regulatory Genes in Human Placenta in Association with Maternal Intake of Olive Oil and Fish Consumption. Int. J. Mol. Sci. 2019, 20, 1060. [Google Scholar] [CrossRef] [Green Version]
- Fenech, M.; El-Sohemy, A.; Cahill, L.; Ferguson, L.R.; French, T.A.; Tai, E.S.; Milner, J.; Koh, W.P.; Xie, L.; Zucker, M.; et al. Nutrigenetics and nutrigenomics: Viewpoints on the current status and applications in nutrition research and practice. J. Nutrigenet. Nutrigenomics 2011, 4, 69–89. [Google Scholar] [CrossRef] [Green Version]
- Fiedor, J.; Burda, K. Potential role of carotenoids as antioxidants in human health and disease. Nutrients 2014, 6, 466–488. [Google Scholar] [CrossRef] [Green Version]
- Fiedor, J.; Fiedor, L.; Haessner, R.; Scheer, H. Cyclic endoperoxides of beta-carotene, potential pro-oxidants, as products of chemical quenching of singlet oxygen. Biochim. Biophys. Acta 2005, 1709, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Alpha-Tocopherol Beta Carotene Cancer Prevention Study Group. The effect of vitamin E and beta carotene on the incidence of lung cancer and other cancers in male smokers. N. Engl. J. Med. 1994, 330, 1029–1035. [Google Scholar] [CrossRef] [PubMed]
- Satia, J.A.; Littman, A.; Slatore, C.G.; Galanko, J.A.; White, E. Long-term use of beta-carotene, retinol, lycopene, and lutein supplements and lung cancer risk: Results from the VITamins And Lifestyle (VITAL) study. Am. J. Epidemiol. 2009, 169, 815–828. [Google Scholar] [CrossRef]
- Divella, R.; Daniele, A.; Savino, E.; Paradiso, A. Anticancer Effects of Nutraceuticals in the Mediterranean Diet: An Epigenetic Diet Model. Cancer Genom. Proteom. 2020, 17, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Murphy, H.C. The Use of Whole Animals Versus Isolated Organs or Cell Culture in Research. Trans. Neb. Acad. Sci. Affil. Soc. 1991, 18, 4. [Google Scholar]
- Morand, C.; De Roos, B.; Garcia-Conesa, M.T.; Gibney, E.R.; Landberg, R.; Manach, C.; Milenkovic, D.; Rodriguez-Mateos, A.; Van de Wiele, T.; Tomas-Barberan, F. Why interindividual variation in response to consumption of plant food bioactives matters for future personalised nutrition. Proc. Nutr. Soc. 2020, 79, 225–235. [Google Scholar] [CrossRef]
- Maki, K.C.; Slavin, J.L.; Rains, T.M.; Kris-Etherton, P.M. Limitations of Observational Evidence: Implications for Evidence-Based Dietary Recommendations. Adv. Nutr. 2014, 5, 7–15. [Google Scholar] [CrossRef] [Green Version]
- Pokimica, B.; García-Conesa, M.-T. Critical Evaluation of Gene Expression Changes in Human Tissues in Response to Supplementation with Dietary Bioactive Compounds: Moving Towards Better-Quality Studies. Nutrients 2018, 10, 807. [Google Scholar] [CrossRef] [Green Version]
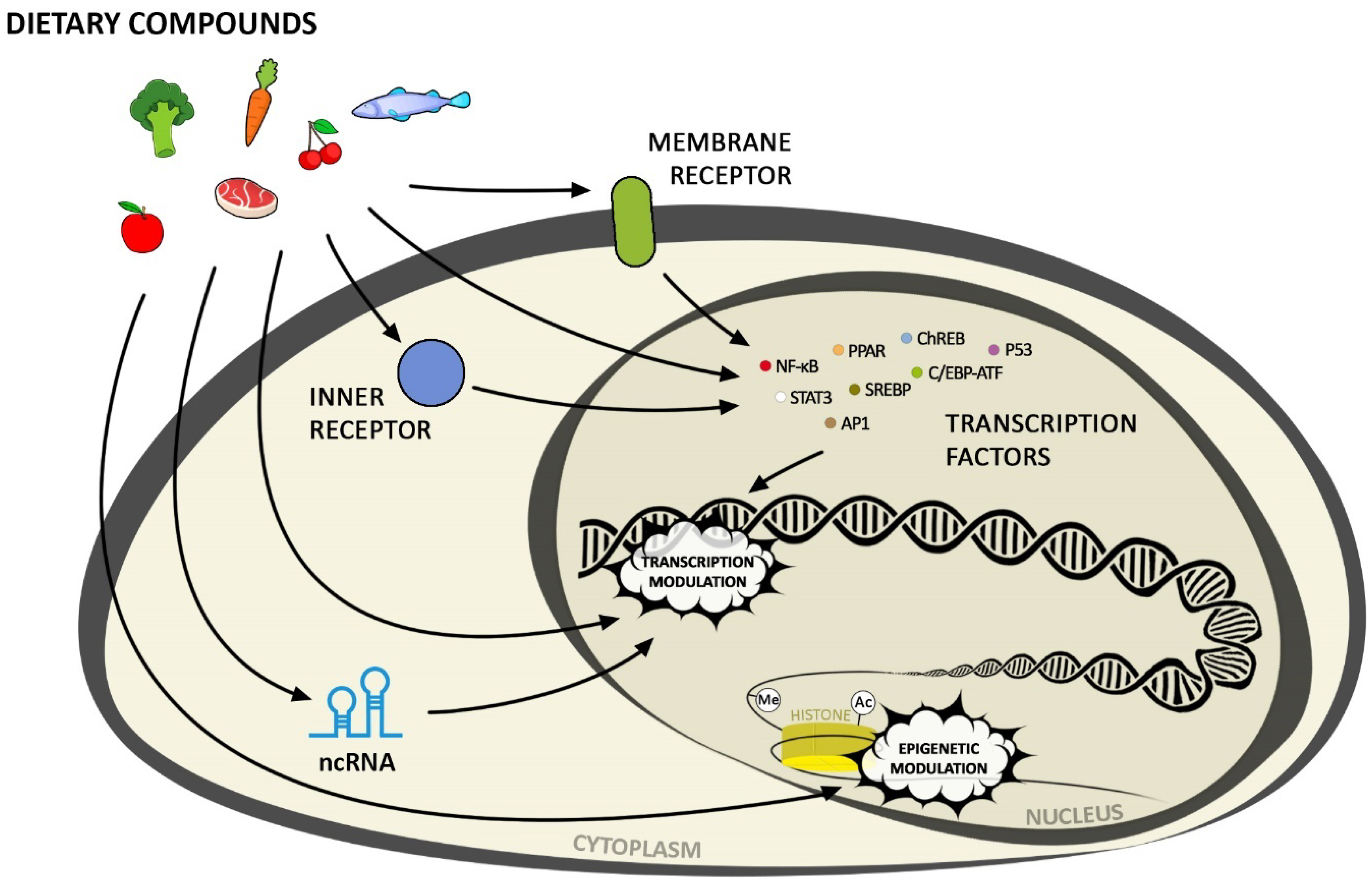
| Mechanisms | Bioactive Component | Disorders | References |
|---|---|---|---|
| upregulation of DNMT | omega-3 fatty acids: DHA, EPA | colorectal cancer | [25,26] |
| folic acid | colorectal cancer, breast cancer | [27,28] | |
| methionine | lung cancer | [29] | |
| vitamin A | congenital heart defects | [30] | |
| DNA methyltransferase inhibition | kaempferol | bladder cancer | [31] |
| gallic acid | lung cancer and oral cancer | [32] | |
| epigallocatechin-3-gallate | breast cancer, diabetic kidney disease | [33,34] | |
| β-caroten | colorectal cancer | [35] | |
| sulforaphane | breast cancer, cardiomyopathy | [36,37] | |
| omega-3 fatty acids: EPA | hepatocarcinoma | [38] | |
| vitamin A | congenital heart defects | [30] | |
| histone deacetylase inhibition | resveratrol | breast cancer, renal cell carcinoma, colorectal cancer | [39,40,41,42] |
| apigenin | prostate cancer, lung cancer | [43,44] | |
| luteolin | lung cancer, leukemia | [45,46] | |
| chrysin | melanoma | [47] | |
| cinnamic acid derivatives | colon and cervical cancer | [48] | |
| gallic acid | prostate cancer, cardiovascular diseases | [49] | |
| epigallocatechin-3-gallate | cardiac diastolic dysfunction, prostate cancer, acute promyelocytic leukemia | [50,51,52] | |
| sulforaphane | Alzheimer’s disease, melanoma, colon cancer, cardiomyopathy | [37,53,54,55] | |
| omega-3 fatty acids: EPA | hepatocarcinoma | [38] | |
| vitamin D | breast cancer | [56] | |
| telomerase inhibition, telomere shortening | epigallocatechin-3-gallate | glioblastoma | [57] |
| Mechanisms | Bioactive Component | Disorders | References |
|---|---|---|---|
| ↓ miR-143 and miR-124 | curcumin | osteoarthritis | [111] |
| ↑ miR-99a | retinoblastoma | [112] | |
| ↑ miR-34a, miR-503, miR-424 | resveratrol | breast cancer | [113] |
| ↑ miRNA-200 | pancreatic ductal adenocarcinoma | [114] | |
| ↑ miR-122-5p | breast cancer | [115] | |
| ↑ miR-200c | colorectal cancer | [116] | |
| ↓ miR-155, miR-34a ↑ miR-21, miR-181, miR-186 | type 2 diabetes, hypertensive patients with coronary artery disease | [117] | |
| ↓ miR-221 | melanoma | [118] | |
| ↑ miR-29b | quercetin | diabetic retinopathy | [119] |
| ↑ miR-146a | breast cancer | [120] | |
| ↓ miR-206 | osteoporosis | [121] | |
| ↓ miR-21 | breast cancer | [122] | |
| ↓ miR-22 | oral lichen planus | [123] | |
| ↓ miR-216a | peripheral arterial disease | [124] | |
| ↓ miR-21 | hepatic steatosis and fibrosis | [125] | |
| ↓ miR-15a and miR-16 | hepatocellular carcinoma | [126] | |
| ↓ miR-16 | oral cancer | [127] | |
| ↑ hsa-miR-24, hsa-miR-6769b-3p, hsa-miR-6836-3p, hsa-miR-199a-3p, hsa-miR-663a, hsa-miR-4739, hsa-miR-6892-3p, hsa-miR-7107-5p, hsa-miR-1273g-3p, hsa-miR-1343, and hsa-miR-6089; ↓ hsa-miR-181a-5p and hsa-miR-148a-3p | apigenin | hepatocellular carcinoma | [128] |
| ↑ miR-34a-5p | lung cancer | [129] | |
| ↑ miR-152-5p | cervical carcinoma | [130] | |
| ↑ miRNA-215-5p | colorectal cancer | [131] | |
| ↑ miR-34a-5p | luteolin | lung cancer | [132] |
| ↑ miR-203 | breast cancer | [133] | |
| ↑ miR-6809-5p | hepatocellular carcinoma | [134] | |
| ↑ miRNA-34a | gastric cancer | [135] | |
| ↓ miR-21 and ↑ miR-16 and -34a | breast cancer | [136] | |
| ↓ microRNA-132 | Bronchopneumonia | [137] | |
| ↓ miRNA-301-3p | pancreatic cancer | [138] | |
| ↑ microRNA-340 | kaempferol | lung cancer | [139] |
| ↑ miR-339-5p | colon cancer | [140] | |
| ↓ microRNA-21 | liver cancer | [141] | |
| ↓ miR-146a | osteoarthritis | [142] | |
| ↑ miR-203 | hypertension | [143] | |
| ↑ miR-132 and miR-502c | chrysin | breast cancer | [144] |
| ↑ miR-9 and Let-7 | gastric cancer | [145] | |
| ↓ miR-18a, miR-21, and miR-221 genes | gastric cancer | [146] | |
| ↓ microRNA (miR)-92a | atherosclerosis | [147] | |
| ↓ miR-636 | caffeic acid | diabetic nephropathy | [148] |
| ↑ miR-221 | epigallocatechin-3-gallate | hepatic fibrosis | [149] |
| ↑ miR-548m | hepatitis C | [150] | |
| ↑ microRNA-let-7b | melanoma | [151] | |
| ↑ miR-520a-3p | prostate cancer | [152] | |
| ↑ miR-384 | ischemic heart disease | [153] | |
| ↓ miR-25 | breast cancer | [154] | |
| ↑ miR-9-3 | sulforaphane | lung cancer | [155] |
| ↑ miR135b-5p | pancreatic cancer | [156] | |
| ↓ miRNA-423-5p | liver fibrosis | [157] | |
| ↓ miR30a-3p | pancreatic cancer | [158] | |
| ↓ miR-155 | acute myeloid leukemia | [159] | |
| ↓ miR-21 | colon cancer | ||
| ↑ miRNA-124-3p | nasopharyngeal cancer | [160] | |
| ↓ miR-23b, miR-92b, miR-381, and miR-382 | breast cancer | [36] | |
| ↑ miR-29a-3p and miR-200a | Carotenoids: lycopene, β-carotene, lutein, astaxanthin | colorectal cancer | [161] |
| ↑ miR-320d, miR-1246 and miRNA-1290 | neuroblastoma | [162] | |
| ↑ miR-let-7f-1 | prostate cancer | [163] | |
| ↑ miR-99a | omega-3 fatty acids: DHA, EPA | breast cancer | [164] |
| ↑ miR-138-5p | lung cancer | [165] | |
| ↑ miR-34a | multiple myeloma | [166] | |
| ↑ miR-194 and ↓ miR-106b | breast cancer | [167] | |
| ↓ miR-21 | breast cancer | [168] | |
| ↓ microRNA-155 | carotid restenosis | [169] | |
| ↓ microRNA-20a | gastric cancer | [170] | |
| ↓ miR-21 | Parkinson’s disease | [171] | |
| ↓ miRNA-146a and -155 | vitamin D | obesity | [172] |
| ↑ miR-100 and -125b | prostate cancer | [173] | |
| ↑ miR-10a | vitamin E | breast cancer | [174] |
| ↑ miR-374, miR-16, miR-199a-5p, miR-195, and miR-30e; ↓ miR-3571, miR-675, and miR-450a | selenium | cardiac dysfunction | [175] |
| ↓ miR-21, miR-31, and miR-223; ↓ miR-375 | zinc | esophageal cancer | [176] |
| Mechanisms | Bioactive Component | Disorders | References |
|---|---|---|---|
| ↓ BRAF-activated long noncoding RNA (BANCR) | luteolin | thyroid carcinoma | [182] |
| long non-coding RNA | epigallocatechin-3-gallate | lung cancer | [183] |
| ↓ lnc RNA LINC00511 | gastric cancer | [184] | |
| ↓ lncRNAs H19 | sulforaphane | pancreatic ductal adenocarcinoma | [185] |
| ↓ lncRNA LUCAT1 | vitamin D | oral squamous cell carcinoma | [186] |
| ↓ lncRNA CCAT2 | ovarian cancer | [187] | |
| ↑ lncRNA MEG3 | colorectal cancer | [188] |
| Mechanisms | Bioactive Component | Disorders | References |
|---|---|---|---|
| PPAR activation | resveratrol | autism spectrum disorder, obesity and insulin resistance | [211,212] |
| kaempferol | hyperlipidemia | [213] | |
| gallic acid and p-coumaric acid | type 2 diabetes | [214] | |
| epigallocatechin-3-gallate | Alzheimer’s disease | [215] | |
| lycopene | liver and lung cancer | [216] | |
| omega-3 fatty acids: DHA | pancreatic acinar, breast cancer, Parkinson’s disease | [171,217,218] | |
| folic acid | non-alcoholic steatohepatitis | [219] | |
| vitamin D | cerebral ischemia, metabolic syndrome | [220,221] | |
| downregulation of PPARγ | epigallocatechin-3-gallate | obesity | [222] |
| NF-κB activation | quercetin | melanoma | [223] |
| apigenin | multiple myeloma | [224] | |
| vitamin A | lung cancer | [225] | |
| NF-κB inhibition | curcumin | gastric cancer, breast cancer, acute lung injury, oral cancer, cerebral ischemia/reperfusion (I/R) injury | [226,227,228,229,230] |
| resveratrol | lung cancer, melanoma | [118,231] | |
| quercetin | coronary artery disease, coronary heart disease, alcohol-induced liver injury | [232,233,234] | |
| apigenin | colon cancer, bladder cancer, breast cancer, inflammatory bowel disease (IBD) and colitis-associated cancer (CAC) | [235,236,237,238] | |
| keampferol | spinal cord injury, hypertension | [143,239] | |
| chrysin | melanoma | [240] | |
| caffeic acid phenethyl ester | nasopharyngeal carcinoma, calcific aortic valve disease, periodontal diseases, glaucoma, neuropathic pain, ovarian cancer | [241,242,243,244,245,246] | |
| caffeic acid | hyperglycemia | [247] | |
| epigallocatechin-3-gallate | temporal lobe epilepsy, lung cancer | [248,249] | |
| sulforaphane | prostate cancer | [250] | |
| lycopene | pancreatic cancer, prostate and breast cancer | [251,252] | |
| omega-3 fatty acids: DHA | liver cirrhosis, breast cancer, pancreatic cancer | [218,253,254] | |
| folic acid | steatohepatitis | [255] | |
| selenium | prostate cancer, breast cancer, type 2 diabetes | [256,257,258,259] | |
| vitamin D | obesity | [172] | |
| vitamin E | prostate cancer | [260] | |
| Nrf2 activation | curcumin | cerebral ischemia/reperfusion (I/R) injury | [230] |
| resveratrol | diabetic cardiomyopathy | [261] | |
| apigenin | vitiligo, diabetic nephropathy | [262,263] | |
| luteolin | colon cancer, colorectal cancer, diabetic cardiomyopathy | [264,265,266] | |
| epigallocatechin-3-gallate | hyperglycemia, obesity, colorectal cancer, retinal ischemia-reperfusion | [267,268,269] | |
| sulforaphane | colon cancer, Alzheimer’s disease, cardiomyopathy | [37,270,271] | |
| omega-3 fatty acids: DHA | traumatic brain injury | [272] | |
| vitamin A | cholestasis | [273] | |
| vitamin E | chronic liver injury | [274] | |
| Nrf2 inhibition | apigenin | lung cancer | [275] |
| luteolin | colon cancer | [276] | |
| keampferol | non-small cell lung cancer | [277] | |
| chrysin | breast cancer, glioblastoma | [278,279] | |
| gallic acid | psoriasis-like skin disease, respiratory diseases | [280,281] | |
| vitamin E | asthma | [282] | |
| zinc | diabetic nephropathy | [283] | |
| AP-1 inhibition | curcumin | renal cell carcinoma, bladder cancer, oral cancer | [229,284,285] |
| gallic acid | nasopharyngeal cancer | [286] | |
| quercetin | coronary heart disease | [233] | |
| apigenin | bladder cancer | [236] | |
| vitamin E | leukemia | [287] | |
| zinc | prostate cancer | [288] | |
| STAT3 inhibition | curcumin | osteosarcoma, myeloproliferative neoplasms, retinoblastoma | [112,289,290] |
| resveratrol | osteosarcoma, colon cancer, ovarian cancer, cervical cancer | [291,292,293,294] | |
| quercetin | hepatocellular carcinoma, alcohol-induced liver injury | [234,295] | |
| apigenin | hepatocellular carcinoma, breast cancer, colon cancer, visceral obesity, inflammatory bowel disease (IBD) and colitis-associated cancer (CAC) | [237,238,296,297,298,299] | |
| luteolin | gastric cancer, pancreatic cancer, hepatic fibrosis, lung adenocarcinoma | [300,301,302,303] | |
| keampferol | diabetic nephropathy | [304] | |
| chrysin | bladder cancer | [305] | |
| gallic acid | non-small cell lung cancer | [306] | |
| omega-3 fatty acids: DHA | renal cancer, multiple myeloma, pancreatic cancer | [254,307,308] | |
| sulforaphane | nasopharyngeal cancer, glioblastoma multiforme | [160,309] | |
| activation of p53 | curcumin | gastric cancer, neuroblastoma, renal cell carcinoma | [310,311,312] |
| resveratrol | prostate cancer, colon cancer, hepatocellular carcinoma, glioblastoma multiform, neuroblastoma, thyroid cancer | [313,314,315,316,317,318,319] | |
| epigallocatechin-3-gallate | liver cancer | [320] | |
| vitamin D | endometrial cancer | [321] | |
| inhibition of p53 | resveratrol | osteoporosis, breast cancer | [322,323] |
| vitamin E | breast cancer | [324] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mierziak, J.; Kostyn, K.; Boba, A.; Czemplik, M.; Kulma, A.; Wojtasik, W. Influence of the Bioactive Diet Components on the Gene Expression Regulation. Nutrients 2021, 13, 3673. https://doi.org/10.3390/nu13113673
Mierziak J, Kostyn K, Boba A, Czemplik M, Kulma A, Wojtasik W. Influence of the Bioactive Diet Components on the Gene Expression Regulation. Nutrients. 2021; 13(11):3673. https://doi.org/10.3390/nu13113673
Chicago/Turabian StyleMierziak, Justyna, Kamil Kostyn, Aleksandra Boba, Magdalena Czemplik, Anna Kulma, and Wioleta Wojtasik. 2021. "Influence of the Bioactive Diet Components on the Gene Expression Regulation" Nutrients 13, no. 11: 3673. https://doi.org/10.3390/nu13113673






