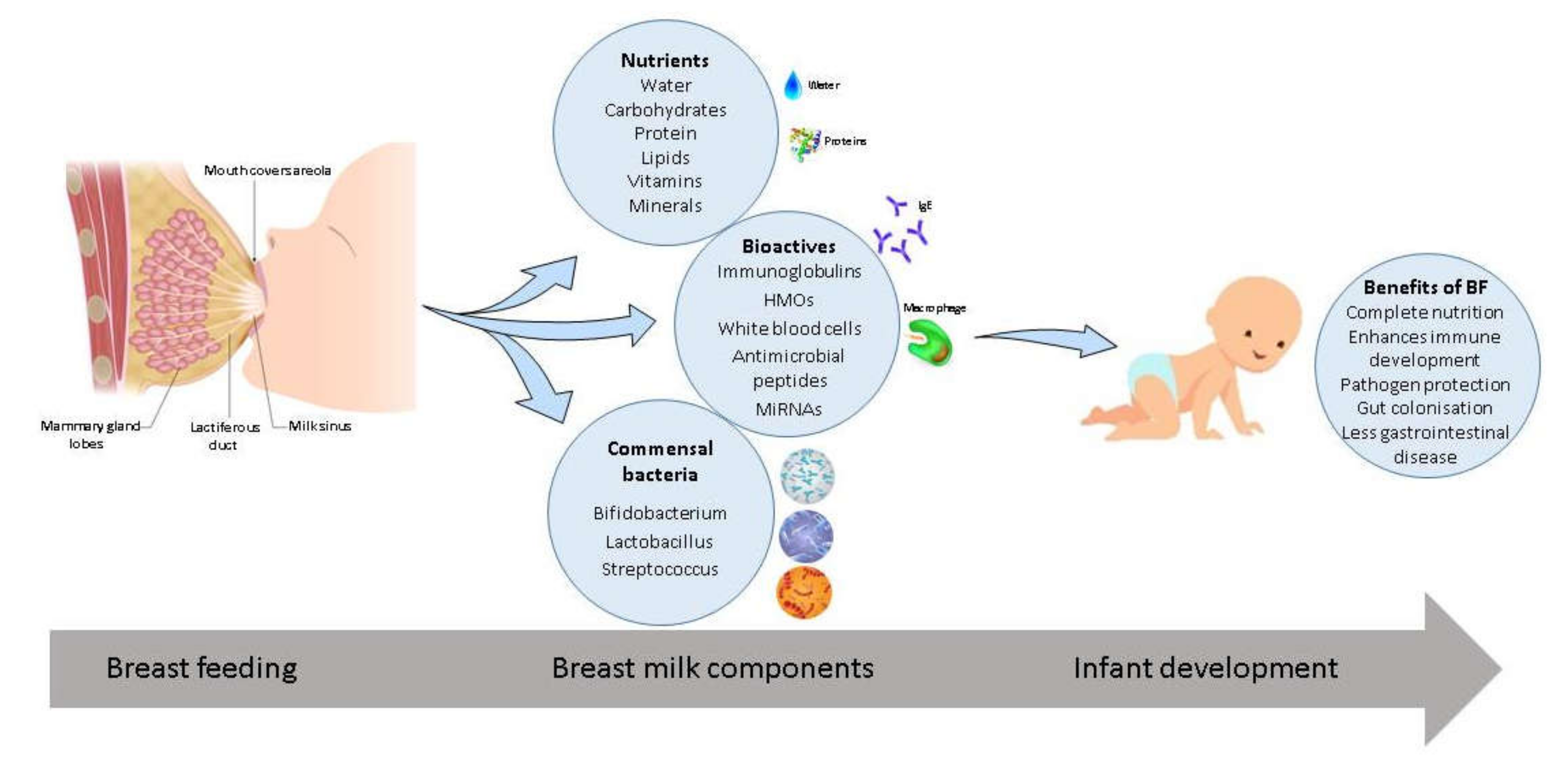
Breast Milk, a Source of Beneficial Microbes and Associated Benefits for Infant Health
Abstract
1. Introduction
2. Mammary Gland Development
2.1. Mammary Gland Development In Utero
2.2. Mammary Gland Development During Puberty and Pregnancy
3. Lactogenesis
4. Milk Synthesis
5. Milk Composition
5.1. Macro and Micronutrient Composition
5.2. Bioactives
5.2.1. Human Milk Oligosaccharides
5.2.2. MicroRNA
5.2.3. Other Bioactive Compounds
5.3. Preterm Milk Composition
6. Infant Formula
7. Breast Milk Microbiome
8. Origins of Milk Microbiome
9. Benefits of Breast Feeding
10. Maternal Implications of Breast Feeding
11. Probiotic Bacteria in Breast Milk
12. Future of Breast Milk Research
Author Contributions
Funding
Conflicts of Interest
References
- Peroni, D.G.; Pescollderungg, L.; Piacentini, G.L.; Rigotti, E.; Maselli, M.; Watschinger, K.; Piazza, M.; Pigozzi, R.; Boner, A.L. Immune regulatory cytokines in the milk of lactating women from farming and urban environments. Pediatr. Allergy Immunol. 2010, 21, 977–982. [Google Scholar] [CrossRef] [PubMed]
- Hermansson, H.; Kumar, H.; Collado, M.C.; Salminen, S.; Isolauri, E.; Rautava, S. Breast milk microbiota is shaped by mode of delivery and intrapartum antibiotic exposure. Front. Nutr. 2019, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Papachatzi, E.; Dimitriou, G.; Dimitropoulos, K.; Vantarakis, A. Pre-pregnancy obesity: Maternal, neonatal and childhood outcomes. J. Neonatal-Perinat. Med. 2013, 6, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Kramer, M.S.; Guo, T.; Platt, R.W.; Sevkovskaya, Z.; Dzikovich, I.; Collet, J.-P.; Shapiro, S.; Chalmers, B.; Hodnett, E.; Vanilovich, I. Infant growth and health outcomes associated with 3 compared with 6 mo of exclusive breastfeeding. Am. J. Clin. Nutr. 2003, 78, 291–295. [Google Scholar] [CrossRef]
- Ladomenou, F.; Moschandreas, J.; Kafatos, A.; Tselentis, Y.; Galanakis, E. Protective effect of exclusive breastfeeding against infections during infancy: A prospective study. Arch. Dis. Child. 2010, 95, 1004–1008. [Google Scholar] [CrossRef]
- Ahern, G.J.; Hennessy, A.; Ryan, C.A.; Ross, R.P.; Stanton, C. Advances in infant formula science. Annu. Rev. Food Sci. Technol. 2019, 10, 75–102. [Google Scholar] [CrossRef]
- Heikkilä, M.P.; Saris, P. Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J. Appl. Microbiol. 2003, 95, 471–478. [Google Scholar] [CrossRef]
- Gavin, A.; Ostovar, K. Microbiological characterization of human milk. J. Food Prot. 1977, 40, 614–616. [Google Scholar] [CrossRef]
- Olivares, M.; Díaz-Ropero, M.; Martín, R.; Rodríguez, J.; Xaus, J. Antimicrobial potential of four Lactobacillus strains isolated from breast milk. J. Appl. Microbiol. 2006, 101, 72–79. [Google Scholar] [CrossRef]
- Jara, S.; Sánchez, M.; Vera, R.; Cofré, J.; Castro, E. The inhibitory activity of Lactobacillus spp. isolated from breast milk on gastrointestinal pathogenic bacteria of nosocomial origin. Anaerobe 2011, 17, 474–477. [Google Scholar] [CrossRef]
- Rajoka, M.S.R.; Mehwish, H.M.; Siddiq, M.; Haobin, Z.; Zhu, J.; Yan, L.; Shao, D.; Xu, X.; Shi, J. Identification, characterization, and probiotic potential of Lactobacillus rhamnosus isolated from human milk. LWT 2017, 84, 271–280. [Google Scholar] [CrossRef]
- Rodríguez, J.M. The origin of human milk bacteria: Is there a bacterial entero-mammary pathway during late pregnancy and lactation? Adv. Nutr. 2014, 5, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Javed, A.; Lteif, A. Development of the human breast. In Seminars in Plastic Surgery. Thieme Medical Publishers: New York, NY, USA; pp. 5–12.
- Howard, B.A.; Gusterson, B.A. Human breast development. J. Mammary Gland Biol. Neoplasia 2000, 5, 119–137. [Google Scholar] [CrossRef]
- Seltzer, V. The breast: Embryology, development, and anatomy. Clin. Obstet. Gynecol. 1994, 37, 879–880. [Google Scholar] [CrossRef] [PubMed]
- Macias, H.; Hinck, L. Mammary gland development. Wiley Interdiscip. Rev. Dev. Biol. 2012, 1, 533–557. [Google Scholar] [CrossRef] [PubMed]
- Watson, C.J.; Khaled, W.T. Mammary development in the embryo and adult: A journey of morphogenesis and commitment. Development 2008, 135, 995–1003. [Google Scholar] [CrossRef]
- Gabriel, A.; Maxwell, G. MedScape. Breast Embryology. Available online: http://emedicine.medscape.com/article/1275146-overview (accessed on 18 July 2016).
- Moore, K.L.; Persaud, T.V.N.; Torchia, M.G. The Developing Human-E-Book: Clinically Oriented Embryology; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Turashvili, G.; Bouchal, J.; Burkadze, G.; Kolar, Z. Mammary gland development and cancer. Cesk Patol 2005, 41, 94–101. [Google Scholar]
- Jayasinghe, Y.; Cha, R.; Horn-Ommen, J.; O’Brien, P.; Simmons, P.S. Establishment of normative data for the amount of breast tissue present in healthy children up to two years of age. J. Pediatr. Adolesc. Gynecol. 2010, 23, 305–311. [Google Scholar] [CrossRef]
- Schmidt, I.M.; Chellakooty, M.; Haavisto, A.-M.; Boisen, K.A.; Damgaard, I.N.; Steendahl, U.; Toppari, J.; Skakkebaek, N.E.; Main, K.M. Gender difference in breast tissue size in infancy: Correlation with serum estradiol. Pediatr. Res. 2002, 52, 682–686. [Google Scholar] [CrossRef]
- McKIERNAN, J.F.; Hull, D. Breast development in the newborn. Arch. Dis. Child. 1981, 56, 525–529. [Google Scholar] [CrossRef]
- Anbazhagan, R.; Bartek, J.; Monaghan, P.; Gusterson, B.A. Growth and development of the human infant breast. Am. J. Anat. 1991, 192, 407–417. [Google Scholar] [CrossRef]
- Naccarato, A.G.; Viacava, P.; Vignati, S.; Fanelli, G.; Bonadio, A.G.; Montruccoli, G.; Bevilacqua, G. Bio-morphological events in the development of the human female mammary gland from fetal age to puberty. Virchows Arch. 2000, 436, 431–438. [Google Scholar] [CrossRef]
- McNally, S.; Martin, F. Molecular regulators of pubertal mammary gland development. Ann. Med. 2011, 43, 212–234. [Google Scholar] [CrossRef] [PubMed]
- Marshall, W.A.; Tanner, J.M. Variations in pattern of pubertal changes in girls. Arch. Dis. Child. 1969, 44, 291. [Google Scholar] [CrossRef]
- Fu, N.Y.; Nolan, E.; Lindeman, G.J.; Visvader, J.E. Stem Cells and the Differentiation Hierarchy in Mammary Gland Development. Physiol. Rev. 2020. [Google Scholar] [CrossRef] [PubMed]
- Brisken, C.; Park, S.; Vass, T.; Lydon, J.P.; O’Malley, B.W.; Weinberg, R.A. A paracrine role for the epithelial progesterone receptor in mammary gland development. Proc. Natl. Acad. Sci. USA 1998, 95, 5076–5081. [Google Scholar] [CrossRef] [PubMed]
- Wiseman, B.S.; Werb, Z. Stromal effects on mammary gland development and breast cancer. Science 2002, 296, 1046–1049. [Google Scholar] [CrossRef]
- Richert, M.M.; Schwertfeger, K.L.; Ryder, J.W.; Anderson, S.M. An atlas of mouse mammary gland development. J. Mammary Gland Biol. Neoplasia 2000, 5, 227–241. [Google Scholar] [CrossRef]
- Taylor-Papadimitriou, J.; Lane, E.; Neville, M. The Mammary Gland: Development, Regulation and Function; Plenum: New York, NY, USA, 1987. [Google Scholar]
- Hoover, K.; Wilson-Clay, B. The Breastfeeding Atlas; LactNews Press: Austin, TX, USA, 2002. [Google Scholar]
- Pang, W.W.; Hartmann, P.E. Initiation of human lactation: Secretory differentiation and secretory activation. J. Mammary Gland Biol. Neoplasia 2007, 12, 211–221. [Google Scholar] [CrossRef]
- Kent, J.C. How breastfeeding works. J. Midwifery Women’s Health 2007, 52, 564–570. [Google Scholar] [CrossRef]
- Truchet, S.; Honvo-Houéto, E. Physiology of milk secretion. Best Pract. Res. Clin. Endocrinol. Metab. 2017, 31, 367–384. [Google Scholar] [CrossRef] [PubMed]
- Kon, S.K.; Cowie, A.T. Milk: The Mammary Gland and Its Secretion; Elsevier: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Pillay, J.; Davis, T.J. Physiology, lactation. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2019. [Google Scholar]
- Neville, M.C.; Morton, J.; Umemura, S. Lactogenesis: The transition from pregnancy to lactation. Pediatr. Clin. North Am. 2001, 48, 35–52. [Google Scholar] [CrossRef]
- Neville, M.C.; Morton, J. Physiology and Endocrine Changes Underlying Human Lactogenesis II. J. Nutr. 2001, 131, 3005S–3008S. [Google Scholar] [CrossRef] [PubMed]
- Kent, J.C.; Mitoulas, L.R.; Cregan, M.D.; Ramsay, D.T.; Doherty, D.A.; Hartmann, P.E. Volume and frequency of breastfeedings and fat content of breast milk throughout the day. Pediatrics 2006, 117, e387–e395. [Google Scholar] [CrossRef]
- Hurst, N.M. Recognizing and treating delayed or failed lactogenesis II. J. Midwifery Women’s Health 2007, 52, 588–594. [Google Scholar] [CrossRef]
- Howie, P.; Houston, M.; Cook, A.; Smart, L.; McArdle, T.; McNeilly, A. How long should a breast feed last? Early Hum. Dev. 1981, 5, 71–77. [Google Scholar] [CrossRef]
- Wilde, C.; Prentice, A.; Peaker, M. Breast-feeding: Matching supply with demand in human lactation. Proc. Nutr. Soc. 1995, 54, 401–406. [Google Scholar] [CrossRef]
- Kent, J.C.; Prime, D.K.; Garbin, C.P. Principles for maintaining or increasing breast milk production. J. Obstet. Gynecol. Neonatal Nurs. 2012, 41, 114–121. [Google Scholar] [CrossRef]
- Hartmann, P.E.; Owens, R.A.; Cox, D.B.; Kent, J.C. Breast development and control of milk synthesis. Food Nutr. Bull. 1996, 17, 1–12. [Google Scholar] [CrossRef]
- Tritos, N.A.; Klibanski, A. Prolactin and its role in human reproduction. In Yen and Jaffe’s Reproductive Endocrinology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 58–74. [Google Scholar]
- Truchet, S.; Ollivier-Bousquet, M. Mammary gland secretion: Hormonal coordination of endocytosis and exocytosis. Animal 2009, 3, 1733–1742. [Google Scholar] [CrossRef]
- Mather, I.H.; Keenan, T.W. Origin and secretion of milk lipids. J. Mammary Gland Biol. Neoplasia 1998, 3, 259–273. [Google Scholar] [CrossRef]
- Mobasheri, A.; Barrett-Jolley, R. Aquaporin water channels in the mammary gland: From physiology to pathophysiology and neoplasia. J. Mammary Gland Biol. Neoplasia 2014, 19, 91–102. [Google Scholar] [CrossRef]
- Shennan, D.; Peaker, M. Transport of milk constituents by the mammary gland. Physiol. Rev. 2000, 80, 925–951. [Google Scholar] [CrossRef]
- Cho, J.-Y.; Leéveilleé, R.E.; Kao, R.; Rousset, B.; Parlow, A.; Burak, W.E., Jr.; Mazzaferri, E.L.; Jhiang, S.M. Hormonal regulation of radioiodide uptake activity and Na+/I−symporter expression in mammary glands. J. Clin. Endocrinol. Metab. 2000, 85, 2936–2943. [Google Scholar] [CrossRef]
- Montalbetti, N.; Dalghi, M.G.; Albrecht, C.; Hediger, M.A. Nutrient transport in the mammary gland: Calcium, trace minerals and water soluble vitamins. J. Mammary Gland Biol. Neoplasia 2014, 19, 73–90. [Google Scholar] [CrossRef]
- Zhao, F.-Q. Biology of glucose transport in the mammary gland. J. Mammary Gland Biol. Neoplasia 2014, 19, 3–17. [Google Scholar] [CrossRef]
- Viña, J.; Puertes, I.R.; Saez, G.T.; Viña, J.R. Role of prolactin in amino acid uptake by the lactating mammary gland of the rat. FEBS Lett. 1981, 126, 250–252. [Google Scholar] [CrossRef]
- McManaman, J.L.; Neville, M.C. Mammary physiology and milk secretion. Adv. Drug Deliv. Rev. 2003, 55, 629–641. [Google Scholar] [CrossRef]
- Kulski, J.; Hartmann, P. Changes in human milk composition during the initiation of lactation. Aust. J. Exp. Biol. Med Sci. 1981, 59, 101–114. [Google Scholar] [CrossRef]
- Kramer, M.S.; Kakuma, R. Optimal duration of exclusive breastfeeding. Cochrane Database Syst. Rev. 2012. [Google Scholar] [CrossRef]
- Casey, C.E.; Neifert, M.R.; Seacat, J.M.; Neville, M.C. Nutrient intake by breast-fed infants during the first five days after birth. Am. J. Dis. Child. 1986, 140, 933–936. [Google Scholar] [CrossRef]
- Castellote, C.; Casillas, R.; Ramírez-Santana, C.; Pérez-Cano, F.J.; Castell, M.; Moretones, M.G.; López-Sabater, M.C.; Franch, À. Premature delivery influences the immunological composition of colostrum and transitional and mature human milk. J. Nutr. 2011, 141, 1181–1187. [Google Scholar] [CrossRef]
- Brown, J.E. Nutrition Through the Life Cycle; Cengage Learning: Boston, MA, USA, 2016. [Google Scholar]
- Pons, S.M.; Bargalló, A.C.; Folgoso, C.C.; Sabater, M.L. Triacylglycerol composition in colostrum, transitional and mature human milk. Eur. J. Clin. Nutr. 2000, 54, 878–882. [Google Scholar] [CrossRef]
- Sundekilde, U.K.; Downey, E.; O’Mahony, J.A.; O’Shea, C.-A.; Ryan, C.A.; Kelly, A.L.; Bertram, H.C. The effect of gestational and lactational age on the human milk metabolome. Nutrients 2016, 8, 304. [Google Scholar] [CrossRef]
- Jenness, R. The composition of human milk. In Seminars in Perinatology; Elsevier: Amsterdam, The Netherlands, 1979; pp. 225–239. [Google Scholar]
- Gao, X.; McMahon, R.J.; Woo, J.G.; Davidson, B.S.; Morrow, A.L.; Zhang, Q. Temporal changes in milk proteomes reveal developing milk functions. J. Proteome Res. 2012, 11, 3897–3907. [Google Scholar] [CrossRef]
- D’Alessandro, A.; Scaloni, A.; Zolla, L. Human milk proteins: An interactomics and updated functional overview. J. Proteome Res. 2010, 9, 3339–3373. [Google Scholar] [CrossRef]
- Saarela, T.; Kokkonen, J.; Koivisto, M. Macronutrient and energy contents of human milk fractions during the first six months of lactation. Acta Paediatr. 2005, 94, 1176–1181. [Google Scholar] [CrossRef]
- Mitoulas, L.R.; Kent, J.C.; Cox, D.B.; Owens, R.A.; Sherriff, J.L.; Hartmann, P.E. Variation in fat, lactose and protein in human milk over 24h and throughout the first year of lactation. Br. J. Nutr. 2002, 88, 29–37. [Google Scholar] [CrossRef]
- Nasser, R.; Stephen, A.M.; Goh, Y.K.; Clandinin, M.T. The effect of a controlled manipulation of maternal dietary fat intake on medium and long chain fatty acids in human breast milk in Saskatoon, Canada. Int. Breastfeed. J. 2010, 5, 3. [Google Scholar] [CrossRef]
- Prentice, A.; Jarjou, L.; Drury, P.J.; Dewit, O.; Crawford, M.A. Breast-milk fatty acids of rural Gambian mothers: Effects of diet and maternal parity. J. Pediatr. Gastroenterol. Nutr. 1989, 8, 486–490. [Google Scholar] [CrossRef]
- Thurl, S.; Munzert, M.; Henker, J.; Boehm, G.; Müller-Werner, B.; Jelinek, J.; Stahl, B. Variation of human milk oligosaccharides in relation to milk groups and lactational periods. Br. J. Nutr. 2010, 104, 1261–1271. [Google Scholar] [CrossRef]
- Lemas, D.J.; Yee, S.; Cacho, N.; Miller, D.; Cardel, M.; Gurka, M.; Janicke, D.; Shenkman, E. Exploring the contribution of maternal antibiotics and breastfeeding to development of the infant microbiome and pediatric obesity. In Seminars in Fetal and Neonatal Medicine; WB Saunders: Philadelphia, PA, USA, 2016; pp. 406–409. [Google Scholar]
- Grapov, D.; Lemay, D.G.; Weber, D.; Phinney, B.S.; Azulay Chertok, I.R.; Gho, D.S.; German, J.B.; Smilowitz, J.T. The human colostrum whey proteome is altered in gestational diabetes mellitus. J. Proteome Res. 2015, 14, 512–520. [Google Scholar] [CrossRef]
- Boix-Amorós, A.; Collado, M.C.; Mira, A. Relationship between milk microbiota, bacterial load, macronutrients, and human cells during lactation. Front. Microbiol. 2016, 7, 492. [Google Scholar] [CrossRef]
- Zambruni, M.; Villalobos, A.; Somasunderam, A.; Westergaard, S.; Nigalye, M.; Turin, C.G.; Zegarra, J.; Bellomo, S.; Mercado, E.; Ochoa, T.J. Maternal and pregnancy-related factors affecting human milk cytokines among Peruvian mothers bearing low-birth-weight neonates. J. Reprod. Immunol. 2017, 120, 20–26. [Google Scholar] [CrossRef]
- Vass, R.A.; Kemeny, A.; Dergez, T.; Ertl, T.; Reglodi, D.; Jungling, A.; Tamas, A. Distribution of bioactive factors in human milk samples. Int. Breastfeed. J. 2019, 14, 9. [Google Scholar] [CrossRef]
- Ballard, O.; Morrow, A.L. Human milk composition: Nutrients and bioactive factors. Pediatr. Clin. 2013, 60, 49–74. [Google Scholar]
- Smilowitz, J.T.; O’sullivan, A.; Barile, D.; German, J.B.; Lönnerdal, B.; Slupsky, C.M. The human milk metabolome reveals diverse oligosaccharide profiles. J. Nutr. 2013, 143, 1709–1718. [Google Scholar] [CrossRef]
- German, J.B.; Freeman, S.L.; Lebrilla, C.B.; Mills, D.A. Human milk oligosaccharides: Evolution, structures and bioselectivity as substrates for intestinal bacteria. In Personalized Nutrition for the Diverse Needs of Infants and Children; Karger Publishers: Basel, Switzerland, 2008; Volume 62, pp. 205–222. [Google Scholar]
- Coppa, G.V.; Gabrielli, O.; Pierani, P.; Catassi, C.; Carlucci, A.; Giorgi, P.L. Changes in carbohydrate composition in human milk over 4 months of lactation. Pediatrics 1993, 91, 637–641. [Google Scholar]
- Weichert, S.; Koromyslova, A.; Singh, B.K.; Hansman, S.; Jennewein, S.; Schroten, H.; Hansman, G.S. Structural basis for norovirus inhibition by human milk oligosaccharides. J. Virol. 2016, 90, 4843–4848. [Google Scholar] [CrossRef]
- Zhang, X.-F.; Tan, M.; Chhabra, M.; Dai, Y.-C.; Meller, J.; Jiang, X. Inhibition of histo-blood group antigen binding as a novel strategy to block norovirus infections. PLoS ONE 2013, 8, e69379. [Google Scholar] [CrossRef][Green Version]
- Morozov, V.; Hansman, G.; Hanisch, F.G.; Schroten, H.; Kunz, C. Human milk oligosaccharides as promising antivirals. Mol. Nutr. Food Res. 2018, 62, 1700679. [Google Scholar] [CrossRef]
- Morrow, A.; Ruiz-Palacios, G.; Altaye, M.; Jiang, X.; Guerrero, M.; Meinzen-Derr, J.; Farkas, T.; Chaturvedi, P.; Pickering, L.; Newburg, D. Human milk oligosaccharide blood group epitopes and innate immune protection against campylobacter and calicivirus diarrhea in breastfed infants. In Protecting Infants through Human Milk; Springer: Berlin/Heidelberg, Germany, 2004; pp. 443–446. [Google Scholar]
- Newburg, D.S.; Walker, W.A. Protection of the neonate by the innate immune system of developing gut and of human milk. Pediatr. Res. 2007, 61, 2–8. [Google Scholar] [CrossRef]
- Ward, R.E.; Ninonuevo, M.; Mills, D.A.; Lebrilla, C.B.; German, J.B. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl. Environ. Microbiol. 2006, 72, 4497–4499. [Google Scholar] [CrossRef]
- Bidart, G.N.; Rodríguez-Díaz, J.; Monedero, V.; Yebra, M.J. A unique gene cluster for the utilization of the mucosal and human milk-associated glycans galacto-N-biose and lacto-N-biose in L actobacillus casei. Mol. Microbiol. 2014, 93, 521–538. [Google Scholar] [CrossRef]
- Triantis, V.; Bode, L.; Van Neerven, R. Immunological effects of human milk oligosaccharides. Front. Pediatr. 2018, 6, 190. [Google Scholar] [CrossRef]
- Marcobal, A.; Sonnenburg, J. Human milk oligosaccharide consumption by intestinal microbiota. Clin. Microbiol. Infect. 2012, 18, 12–15. [Google Scholar] [CrossRef]
- Collado, M.C.; Cernada, M.; Baüerl, C.; Vento, M.; Pérez-Martínez, G. Microbial ecology and host-microbiota interactions during early life stages. Gut Microbes 2012, 3, 352–365. [Google Scholar] [CrossRef]
- Lee, S.A.; Lim, J.Y.; Kim, B.-S.; Cho, S.J.; Kim, N.Y.; Kim, O.B.; Kim, Y. Comparison of the gut microbiota profile in breast-fed and formula-fed Korean infants using pyrosequencing. Nutr. Res. Pract. 2015, 9, 242–248. [Google Scholar] [CrossRef]
- Vandenplas, Y.; Zakharova, I.; Dmitrieva, Y. Oligosaccharides in infant formula: More evidence to validate the role of prebiotics. Br. J. Nutr. 2015, 113, 1339–1344. [Google Scholar] [CrossRef]
- Torres, D.P.; Gonçalves, M.D.P.F.; Teixeira, J.A.; Rodrigues, L.R. Galacto-oligosaccharides: Production, properties, applications, and significance as prebiotics. Compr. Rev. Food Sci. Food Saf. 2010, 9, 438–454. [Google Scholar] [CrossRef]
- Davis, L.M.; Martínez, I.; Walter, J.; Goin, C.; Hutkins, R.W. Barcoded pyrosequencing reveals that consumption of galactooligosaccharides results in a highly specific bifidogenic response in humans. PLoS ONE 2011, 6, e25200. [Google Scholar] [CrossRef]
- Oozeer, R.; van Limpt, K.; Ludwig, T.; Ben Amor, K.; Martin, R.; Wind, R.D.; Boehm, G.; Knol, J. Intestinal microbiology in early life: Specific prebiotics can have similar functionalities as human-milk oligosaccharides. Am. J. Clin. Nutr. 2013, 98, 561S–571S. [Google Scholar] [CrossRef]
- Asakuma, S.; Hatakeyama, E.; Urashima, T.; Yoshida, E.; Katayama, T.; Yamamoto, K.; Kumagai, H.; Ashida, H.; Hirose, J.; Kitaoka, M. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J. Biol. Chem. 2011, 286, 34583–34592. [Google Scholar] [CrossRef]
- Marcobal, A.; Barboza, M.; Froehlich, J.W.; Block, D.E.; German, J.B.; Lebrilla, C.B.; Mills, D.A. Consumption of human milk oligosaccharides by gut-related microbes. J. Agric. Food Chem. 2010, 58, 5334–5340. [Google Scholar] [CrossRef]
- Jantscher-Krenn, E.; Bode, L. Human milk oligosaccharides and their potential benefits for the breast-fed neonate. Minerva Pediatr. 2012, 64, 83–99. [Google Scholar]
- Kitaoka, M. Bifidobacterial enzymes involved in the metabolism of human milk oligosaccharides. Adv. Nutr. 2012, 3, 422S–429S. [Google Scholar] [CrossRef]
- Garrido, D.; Kim, J.H.; German, J.B.; Raybould, H.E.; Mills, D.A. Oligosaccharide binding proteins from Bifidobacterium longum subsp Infantis reveal preference for host glycans. PLoS ONE 2011, 6, e17315. [Google Scholar] [CrossRef]
- Chichlowski, M.; Guillaume De Lartigue, J.; Raybould, H.E.; Mills, D.A. Bifidobacteria isolated from infants and cultured on human milk oligosaccharides affect intestinal epithelial function. J. Pediatr. Gastroenterol. Nutr. 2012, 55, 321. [Google Scholar] [CrossRef]
- Wickramasinghe, S.; Pacheco, A.R.; Lemay, D.G.; Mills, D.A. Bifidobacteria grown on human milk oligosaccharides downregulate the expression of inflammation-related genes in Caco-2 cells. BMC Microbiol. 2015, 15, 172. [Google Scholar] [CrossRef]
- Lawson, M.A.; O’Neill, I.J.; Kujawska, M.; Javvadi, S.G.; Wijeyesekera, A.; Flegg, Z.; Chalklen, L.; Hall, L.J. Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. ISME J. 2020, 14, 635–648. [Google Scholar] [CrossRef]
- Matsuki, T.; Yahagi, K.; Mori, H.; Matsumoto, H.; Hara, T.; Tajima, S.; Ogawa, E.; Kodama, H.; Yamamoto, K.; Yamada, T. A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nat. Commun. 2016, 7, 1–12. [Google Scholar] [CrossRef]
- Egan, M.; Motherway, M.O.C.; Kilcoyne, M.; Kane, M.; Joshi, L.; Ventura, M.; van Sinderen, D. Cross-feeding by Bifidobacterium breve UCC2003 during co-cultivation with Bifidobacterium bifidum PRL2010 in a mucin-based medium. BMC Microbiol. 2014, 14, 282. [Google Scholar] [CrossRef]
- Egan, M.; Motherway, M.O.C.; Ventura, M.; van Sinderen, D. Metabolism of sialic acid by Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 2014, 80, 4414–4426. [Google Scholar]
- Ward, R.E.; Niñonuevo, M.; Mills, D.A.; Lebrilla, C.B.; German, J.B. In vitro fermentability of human milk oligosaccharides by several strains of bifidobacteria. Mol. Nutr. Food Res. 2007, 51, 1398–1405. [Google Scholar]
- Coulet, M.; Phothirath, P.; Allais, L.; Schilter, B. Pre-clinical safety evaluation of the synthetic human milk, nature-identical, oligosaccharide 2′-O-Fucosyllactose (2′ FL). Regul. Toxicol. Pharmacol. 2014, 68, 59–69. [Google Scholar]
- Goehring, K.C.; Marriage, B.J.; Oliver, J.S.; Wilder, J.A.; Barrett, E.G.; Buck, R.H. Similar to those who are breastfed, infants fed a formula containing 2′-fucosyllactose have lower inflammatory cytokines in a randomized controlled trial. J. Nutr. 2016, 146, 2559–2566. [Google Scholar] [CrossRef]
- Ackerman, D.L.; Doster, R.S.; Weitkamp, J.-H.; Aronoff, D.M.; Gaddy, J.A.; Townsend, S.D. Human milk oligosaccharides exhibit antimicrobial and antibiofilm properties against Group B Streptococcus. ACS Infect. Dis. 2017, 3, 595–605. [Google Scholar] [CrossRef]
- Ackerman, D.L.; Craft, K.M.; Doster, R.S.; Weitkamp, J.-H.; Aronoff, D.M.; Gaddy, J.A.; Townsend, S.D. Antimicrobial and antibiofilm activity of human milk oligosaccharides against Streptococcus agalactiae, Staphylococcus aureus, and Acinetobacter baumannii. ACS Infect. Dis. 2017, 4, 315–324. [Google Scholar]
- Chambers, S.A.; Townsend, S.D. Bioorthogonal human milk oligosaccharide probes for antimicrobial target identification within Streptococcus agalactiae. Carbohydr. Res. 2020, 488, 107895. [Google Scholar] [CrossRef]
- Bushati, N.; Cohen, S.M. MicroRNA functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef]
- Alvarez-Garcia, I.; Miska, E.A. MicroRNA functions in animal development and human disease. Development 2005, 132, 4653–4662. [Google Scholar] [CrossRef]
- Wienholds, E.; Plasterk, R.H. MicroRNA function in animal development. FEBS Lett. 2005, 579, 5911–5922. [Google Scholar] [CrossRef]
- Kittelmann, S.; McGregor, A.P. Modulation and evolution of animal development through microRNA regulation of gene expression. Genes 2019, 10, 321. [Google Scholar] [CrossRef]
- Tsukamoto, M.; Iinuma, H.; Matsuda, K.; Yagi, T.; Hashiguchi, Y. Circulating exosomal microRNA-21 as a biomarker in each tumor stage of colorectal cancer. Oncology 2017, 92, 360–370. [Google Scholar] [CrossRef]
- Vu, T.L.; Peng, B.; Zhang, D.X.; Ma, V.; Mathey-Andrews, C.A.; Lam, C.K.; Kiomourtzis, T.; Jin, J.; McReynolds, L.; Huang, L.; et al. Tumor-secreted extracellular vesicles promote the activation of cancer-associated fibroblasts via the transfer of microRNA-125b. J. Extracell. Vesicles 2019, 8, 1599680. [Google Scholar] [CrossRef]
- Kim, S.; Lee, E.; Jung, J.; Lee, J.W.; Kim, H.J.; Kim, J.; Yoo, H.J.; Lee, H.J.; Chae, S.Y.; Jeon, S.-M.; et al. microRNA-155 positively regulates glucose metabolism via PIK3R1-FOXO3a-cMYC axis in breast cancer. Oncogene 2018, 37, 2982–2991. [Google Scholar] [CrossRef]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; How Huang, K.; Jen Lee, M.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef]
- Liao, Y.; Du, X.; Li, J.; Lönnerdal, B. Human milk exosomes and their microRNAs survive digestion in vitro and are taken up by human intestinal cells. Mol. Nutr. Food Res. 2017, 61, 1700082. [Google Scholar] [CrossRef]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as biomarkers and diagnostics. J. Cell. Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef]
- Gallo, A.; Tandon, M.; Alevizos, I.; Illei, G.G. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS ONE 2012, 7, e30679. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, M.; Wang, X.; Li, Q.; Wang, T.; Zhu, Q.; Zhou, X.; Wang, X.; Gao, X.; Li, X. Immune-related microRNAs are abundant in breast milk exosomes. Int. J. Biol. Sci. 2012, 8, 118. [Google Scholar] [CrossRef]
- Alsaweed, M.; Lai, C.T.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. Human milk cells and lipids conserve numerous known and novel miRNAs, some of which are differentially expressed during lactation. PLoS ONE 2016, 11, e0152610. [Google Scholar] [CrossRef]
- Alsaweed, M.; Lai, C.T.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. Human milk miRNAs primarily originate from the mammary gland resulting in unique miRNA profiles of fractionated milk. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef]
- Alsaweed, M.; Hepworth, A.R.; Lefevre, C.; Hartmann, P.E.; Geddes, D.T.; Hassiotou, F. Human milk microRNA and total RNA differ depending on milk fractionation. J. Cell. Biochem. 2015, 116, 2397–2407. [Google Scholar] [CrossRef]
- Carney, M.C.; Tarasiuk, A.; DiAngelo, S.L.; Silveyra, P.; Podany, A.; Birch, L.L.; Paul, I.M.; Kelleher, S.; Hicks, S.D. Metabolism-related microRNAs in maternal breast milk are influenced by premature delivery. Pediatr. Res. 2017, 82, 226–236. [Google Scholar] [CrossRef]
- Shiff, Y.E.; Reif, S.; Marom, R.; Shiff, K.; Reifen, R.; Golan-Gerstl, R. MiRNA-320a is less expressed and miRNA-148a more expressed in preterm human milk compared to term human milk. J. Funct. Foods 2019, 57, 68–74. [Google Scholar] [CrossRef]
- Alsaweed, M.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. MicroRNAs in breastmilk and the lactating breast: Potential immunoprotectors and developmental regulators for the infant and the mother. Int. J. Environ. Res. Public Health 2015, 12, 13981–14020. [Google Scholar] [CrossRef]
- Kosaka, N.; Izumi, H.; Sekine, K.; Ochiya, T. microRNA as a new immune-regulatory agent in breast milk. Silence 2010, 1, 7. [Google Scholar] [CrossRef]
- Chu, M.; Zhao, Y.; Yu, S.; Hao, Y.; Zhang, P.; Feng, Y.; Zhang, H.; Ma, D.; Liu, J.; Cheng, M. MicroRNA-221 may be involved in lipid metabolism in mammary epithelial cells. Int. J. Biochem. Cell Biol. 2018, 97, 118–127. [Google Scholar] [CrossRef]
- Aly, E.; Darwish, A.A.; Lopez-Nicolas, R.; Frontela-Saseta, C.; Ros-Berruezo, G. Bioactive Components of Human Milk: Similarities and Differences between Human Milk and Infant Formula. In Selected Topics in Breastfeeding; IntechOpen: London, UK, 2018. [Google Scholar]
- Vogel, H.J. Lactoferrin, a bird’s eye view. Biochem. Cell Biol. 2012, 90, 233–244. [Google Scholar] [CrossRef]
- Ochoa, T.J.; Chea-Woo, E.; Baiocchi, N.; Pecho, I.; Campos, M.; Prada, A.; Valdiviezo, G.; Lluque, A.; Lai, D.; Cleary, T.G. Randomized double-blind controlled trial of bovine lactoferrin for prevention of diarrhea in children. J. Pediatr. 2013, 162, 349–356. [Google Scholar] [CrossRef]
- Manzoni, P.; Rinaldi, M.; Cattani, S.; Pugni, L.; Romeo, M.G.; Messner, H.; Stolfi, I.; Decembrino, L.; Laforgia, N.; Vagnarelli, F. Bovine lactoferrin supplementation for prevention of late-onset sepsis in very low-birth-weight neonates: A randomized trial. JAMA 2009, 302, 1421–1428. [Google Scholar] [CrossRef]
- Manzoni, P.; Sánchez, R.G.; Meyer, M.; Stolfi, I.; Pugni, L.; Messner, H.; Cattani, S.; Betta, P.M.; Memo, L.; Decembrino, L. Exposure to gastric acid inhibitors increases the risk of infection in preterm very low birth weight infants but concomitant administration of lactoferrin counteracts this effect. J. Pediatr. 2018, 193, 62–67. [Google Scholar] [CrossRef]
- Chang, C.-J.; Chao, J.C.-J. Effect of human milk and epidermal growth factor on growth of human intestinal Caco-2 cells. J. Pediatr. Gastroenterol. Nutr. 2002, 34, 394–401. [Google Scholar] [CrossRef]
- Hirai, C.; Ichiba, H.; Saito, M.; Shintaku, H.; Yamano, T.; Kusuda, S. Trophic effect of multiple growth factors in amniotic fluid or human milk on cultured human fetal small intestinal cells. J. Pediatr. Gastroenterol. Nutr. 2002, 34, 524–528. [Google Scholar] [CrossRef]
- Boesmans, W.; Gomes, P.; Janssens, J.; Tack, J.; Berghe, P.V. Brain-derived neurotrophic factor amplifies neurotransmitter responses and promotes synaptic communication in the enteric nervous system. Gut 2008, 57, 314–322. [Google Scholar] [CrossRef]
- Blum, J.; Baumrucker, C. Colostral and milk insulin-like growth factors and related substances: Mammary gland and neonatal (intestinal and systemic) targets. Domest. Anim. Endocrinol. 2002, 23, 101–110. [Google Scholar] [CrossRef]
- Hurley, W.L.; Theil, P.K. Perspectives on immunoglobulins in colostrum and milk. Nutrients 2011, 3, 442–474. [Google Scholar] [CrossRef]
- Lawrence, R.M.; Lawrence, R.A. Breast milk and infection. Clin. Perinatol. 2004, 31, 501–528. [Google Scholar] [CrossRef]
- Hanson, L.Å.; Korotkova, M. The role of breastfeeding in prevention of neonatal infection. In Seminars in Neonatology; WB Saunders: Philadelphia, PA, USA, 2002. [Google Scholar]
- Lucas, A. Long-term programming effects of early nutrition-Implications for the preterm infant. J. Perinatol. 2005, 25, S2–S6. [Google Scholar] [CrossRef]
- Hylander, M.A.; Strobino, D.M.; Dhanireddy, R. Human milk feedings and infection among very low birth weight infants. Pediatrics 1998, 102, e38. [Google Scholar] [CrossRef]
- Parkinson, J.R.; Hyde, M.J.; Gale, C.; Santhakumaran, S.; Modi, N. Preterm birth and the metabolic syndrome in adult life: A systematic review and meta-analysis. Pediatrics 2013, 131, e1240–e1263. [Google Scholar] [CrossRef]
- Sullivan, S.; Schanler, R.J.; Kim, J.H.; Patel, A.L.; Trawöger, R.; Kiechl-Kohlendorfer, U.; Chan, G.M.; Blanco, C.L.; Abrams, S.; Cotten, C.M. An exclusively human milk-based diet is associated with a lower rate of necrotizing enterocolitis than a diet of human milk and bovine milk-based products. J. Pediatr. 2010, 156, 562–567. [Google Scholar] [CrossRef]
- Patel, P.; Bhatia, J. Human Milk: The Preferred First Food for Premature Infants. J Hum Nutr Food Sci 2016, 4, 1098. [Google Scholar]
- Tudehope, D.I. Human milk and the nutritional needs of preterm infants. J. Pediatr. 2013, 162, S17–S25. [Google Scholar] [CrossRef]
- Bauer, J.; Gerss, J. Longitudinal analysis of macronutrients and minerals in human milk produced by mothers of preterm infants. Clin. Nutr. 2011, 30, 215–220. [Google Scholar] [CrossRef]
- De Figueiredo, C.S.M.; Palhares, D.B.; Melnikov, P.; Moura, A.J.d.C.M.; dos Santos, S.C. Zinc and copper concentrations in human preterm milk. Biol. Trace Elem. Res. 2010, 136, 1–7. [Google Scholar] [CrossRef]
- O’Brien, C.E.; Krebs, N.F.; Westcott, J.L.; Dong, F. Relationships among plasma zinc, plasma prolactin, milk transfer, and milk zinc in lactating women. J. Hum. Lact. 2007, 23, 179–183. [Google Scholar] [CrossRef]
- Dvorak, B.; Fituch, C.C.; Williams, C.S.; Hurst, N.M.; Schanler, R.J. Increased epidermal growth factor levels in human milk of mothers with extremely premature infants. Pediatr. Res. 2003, 54, 15–19. [Google Scholar] [CrossRef]
- Gross, S.J.; Buckley, R.H.; Wakil, S.S.; McAllister, D.C.; David, R.J.; Faix, R.G. Elevated IgA concentration in milk produced by mothers delivered of preterm infants. J. Pediatr. 1981, 99, 389–393. [Google Scholar] [CrossRef]
- Kunz, C.; Kuntz, S.; Rudloff, S.; Moreno, F.; Sanz, M. Food Oligosaccharides: Production, Analysis and Bioactivity; Wiley: Hoboken, NJ, USA, 2014. [Google Scholar]
- Montagne, P.; Cuillière, M.L.; Molé, C.; Béné, M.C.; Faure, G. Immunological and nutritional composition of human milk in relation to prematurity and mothers’ parity during the first 2 weeks of lactation. J. Pediatr. Gastroenterol. Nutr. 1999, 29, 75–80. [Google Scholar] [CrossRef]
- Arora, S.; McJunkin, C.; Wehrer, J.; Kuhn, P. Major factors influencing breastfeeding rates: Mother’s perception of father’s attitude and milk supply. Pediatrics 2000, 106, e67. [Google Scholar] [CrossRef]
- Kozhimannil, K.B.; Jou, J.; Attanasio, L.B.; Joarnt, L.K.; McGovern, P. Medically complex pregnancies and early breastfeeding behaviors: A retrospective analysis. PLoS ONE 2014, 9, e104820. [Google Scholar] [CrossRef]
- Carver, J.D. Advances in nutritional modifications of infant formulas. Am. J. Clin. Nutr. 2003, 77, 1550S–1554S. [Google Scholar] [CrossRef]
- Grant, C.; Rotherham, B.; Sharpe, S.; Scragg, R.; Thompson, J.; Andrews, J.; Wall, C.; Murphy, J.; Lowry, D. Randomized, double-blind comparison of growth in infants receiving goat milk formula versus cow milk infant formula. J. Paediatr. Child Health 2005, 41, 564–568. [Google Scholar] [CrossRef]
- Martin, C.R.; Ling, P.-R.; Blackburn, G.L. Review of infant feeding: Key features of breast milk and infant formula. Nutrients 2016, 8, 279. [Google Scholar] [CrossRef]
- Cook, D.A. Nutrient levels in infant formulas: Technical considerations. J. Nutr. 1989, 119 (Suppl. 12), 1773–1778. [Google Scholar] [CrossRef]
- Thompkinson, D.; Kharb, S. Aspects of infant food formulation. Compr. Rev. Food Sci. Food Saf. 2007, 6, 79–102. [Google Scholar] [CrossRef]
- Koletzko, B.; Baker, S.; Cleghorn, G.; Neto, U.F.; Gopalan, S.; Hernell, O.; Hock, Q.S.; Jirapinyo, P.; Lonnerdal, B.; Pencharz, P. Global standard for the composition of infant formula: Recommendations of an ESPGHAN coordinated international expert group. J. Pediatr. Gastroenterol. Nutr. 2005, 41, 584–599. [Google Scholar] [CrossRef]
- Koletzko, B.; Broekaert, I.; Demmelmair, H.; Franke, J.; Hannibal, I.; Oberle, D.; Schiess, S.; Baumann, B.T.; Verwied-Jorky, S. Protein intake in the first year of life: A risk factor for later obesity? In Early Nutrition and Its Later Consequences: New Opportunities; Springer: Berlin/Heidelberg, Germany, 2005; pp. 69–79. [Google Scholar]
- Høst, A.; Halken, S.; Jacobsen, H.P.; Christensen, A.E.; Herskind, A.M.; Plesner, K. Clinical course of cow’s milk protein allergy/intolerance and atopic diseases in childhood. Pediat. Allergy Immunol. 2002, 13, 23–28. [Google Scholar] [CrossRef]
- Arslanoglu, S.; Moro, G.E.; Boehm, G. Early supplementation of prebiotic oligosaccharides protects formula-fed infants against infections during the first 6 months of life. J. Nutr. 2007, 137, 2420–2424. [Google Scholar] [CrossRef]
- Moro, G.; Arslanoglu, S.; Stahl, B.; Jelinek, J.; Wahn, U.; Boehm, G. A mixture of prebiotic oligosaccharides reduces the incidence of atopic dermatitis during the first six months of age. Arch. Dis. Child. 2006, 91, 814–819. [Google Scholar]
- Grüber, C.; Van Stuijvenberg, M.; Mosca, F.; Moro, G.; Chirico, G.; Braegger, C.P.; Riedler, J.; Boehm, G.; Wahn, U.; Group, M.W. Reduced occurrence of early atopic dermatitis because of immunoactive prebiotics among low-atopy-risk infants. J. Allergy Clin. Immunol. 2010, 126, 791–797. [Google Scholar] [CrossRef]
- Martín, R.; Langa, S.; Reviriego, C.; Jimínez, E.; Marín, M.L.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Human milk is a source of lactic acid bacteria for the infant gut. J. Pediatr. 2003, 143, 754–758. [Google Scholar]
- Ward, T.L.; Hosid, S.; Ioshikhes, I.; Altosaar, I. Human milk metagenome: A functional capacity analysis. BMC Microbiol. 2013, 13, 116. [Google Scholar] [CrossRef]
- Aakko, J.; Kumar, H.; Rautava, S.; Wise, A.; Autran, C.; Bode, L.; Salminen, S. Human milk oligosaccharide categories define the microbiota composition in human colostrum. Benef. Microbes 2017, 8, 563–567. [Google Scholar]
- Murphy, K.; Curley, D.; O’Callaghan, T.F.; O’Shea, C.-A.; Dempsey, E.M.; O’Toole, P.W.; Ross, R.P.; Ryan, C.A.; Stanton, C. The composition of human milk and infant faecal microbiota over the first three months of life: A pilot study. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Lackey, K.A.; Williams, J.E.; Meehan, C.L.; Zachek, J.A.; Benda, E.D.; Price, W.J.; Foster, J.A.; Sellen, D.W.; Kamau-Mbuthia, E.W.; Kamundia, E.W. What’s normal? microbiomes in human milk and infant feces are related to each other but vary geographically: The INSPIRE study. Front. Nutr. 2019, 6, 45. [Google Scholar]
- Pannaraj, P.S.; Li, F.; Cerini, C.; Bender, J.M.; Yang, S.; Rollie, A.; Adisetiyo, H.; Zabih, S.; Lincez, P.J.; Bittinger, K.; et al. Association Between Breast Milk Bacterial Communities and Establishment and Development of the Infant Gut Microbiome. JAMA Pediatr. 2017, 171, 647–654. [Google Scholar] [CrossRef]
- Eidelman, A.I.; Szilagyi, G. Patterns of bacterial colonization of human milk. Obstet. Gynecol. 1979, 53, 550–552. [Google Scholar]
- Osterman, K.L.; Rahm, V.-A. Lactation Mastitis: Bacterial Cultivation of Breast Milk, Symptoms, Treatment, and Outcome. J. Hum. Lact. 2000, 16, 297–302. [Google Scholar] [CrossRef]
- Thomsen, A.C.; Espersen, T.; Maigaard, S. Course and treatment of milk stasis, noninfectious inflammation of the breast, and infectious mastitis in nursing women. Am. J. Obstet. Gynecol. 1984, 149, 492–495. [Google Scholar] [CrossRef]
- Jones, C. Maternal transmission of infectious pathogens in breast milk. J. Paediatr. Child Health 2001, 37, 576–582. [Google Scholar] [CrossRef]
- Kumar, A.; Yadav, M.; Kakkar, S. Human milk as a source of Q-fever infection in breast-fed babies. Indian J. Med. Res. 1981, 73, 510. [Google Scholar]
- Le Thomas, I.; Mariani-Kurkdjian, P.; Collignon, A.; Gravet, A.; Clermont, O.; Brahimi, N.m.; Gaudelus, J.; Aujard, Y.; Navarro, J.; Beaufils, F. Breast milk transmission of a Panton-Valentine leukocidin-producing Staphylococcus aureus strain causing infantile pneumonia. J. Clin. Microbiol. 2001, 39, 728–729. [Google Scholar] [CrossRef][Green Version]
- Ryder, R.W.; Crosby-Ritchie, A.; McDonough, B.; Hall, W.J. Human milk contaminated with Salmonella kottbus: A cause of nosocomial illness in infants. JAMA 1977, 238, 1533–1534. [Google Scholar] [CrossRef]
- Botsford, K.B.; Weinstein, R.A.; Boyer, K.M.; Nathan, C.; Carman, M.; Paton, J.B. Gram-negative bacilli in human milk feedings: Quantitation and clinical consequences for premature infants. J. Pediatr. 1986, 109, 707–710. [Google Scholar] [CrossRef]
- Larson, E.; Zuill, R.; Zier, V.; Berg, B. Storage of human breast milk. Infect. Control Hosp. Epidemiol. 1984, 5, 127–130. [Google Scholar] [CrossRef]
- Sosa, R.; Barness, L. Bacterial Growth in Refrigerated Human Milk. Am. J. Dis. Child. 1987, 141, 111–112. [Google Scholar] [CrossRef]
- Jost, T.; Lacroix, C.; Braegger, C.; Chassard, C. Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br. J. Nutr. 2013, 110, 1253–1262. [Google Scholar] [CrossRef]
- Sinkiewicz, G.; Nordström, E.A. 353 occurrence of Lactobacillus reuteri, lactobacilli and bifidobacteria in human breast milk. Pediatric Res. 2005, 58, 415. [Google Scholar] [CrossRef][Green Version]
- Martín, R.; Jiménez, E.; Heilig, H.; Fernández, L.; Marín, M.L.; Zoetendal, E.G.; Rodríguez, J.M. Isolation of Bifidobacteria from Breast Milk and Assessment of the Bifidobacterial Population by PCR-Denaturing Gradient Gel Electrophoresis and Quantitative Real-Time PCR. Appl. Environ. Microbiol. 2009, 75, 965–969. [Google Scholar] [CrossRef]
- Kansandee, W.; Moonmangmee, D.; Moonmangmee, S.; Itsaranuwat, P. Characterization and Bifidobacterium sp. growth stimulation of exopolysaccharide produced by Enterococcus faecalis EJRM152 isolated from human breast milk. Carbohydr. Polym. 2019, 206, 102–109. [Google Scholar] [CrossRef]
- Martín, R.; Olivares, M.; Marín, M.L.; Fernández, L.; Xaus, J.; Rodríguez, J.M. Probiotic potential of 3 lactobacilli strains isolated from breast milk. J. Hum. Lact. 2005, 21, 8–17. [Google Scholar] [CrossRef]
- Belhadj, F.Z.B.; Boublenza, F.; Karam, N.-E. Stress tolerance in Lactobacillus plantarum LMF6 isolated from human breast milk. South Asian J. Exp. Biol. 2020, 9, 173–184. [Google Scholar]
- Li, N.; Pang, B.; Liu, G.; Zhao, X.; Xu, X.; Jiang, C.; Yang, B.; Liu, Y.; Shi, J. Lactobacillus rhamnosus from human breast milk shows therapeutic function against foodborne infection by multi-drug resistant Escherichia coli in mice. Food Funct. 2020, 11, 435–447. [Google Scholar] [CrossRef]
- Le Doare, K.; Holder, B.; Bassett, A.; Pannaraj, P.S. Mother’s milk: A purposeful contribution to the development of the infant microbiota and immunity. Front. Immunol. 2018, 9, 361. [Google Scholar] [CrossRef]
- Hunt, K.M.; Foster, J.A.; Forney, L.J.; Schütte, U.M.; Beck, D.L.; Abdo, Z.; Fox, L.K.; Williams, J.E.; McGuire, M.K.; McGuire, M.A. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS ONE 2011, 6, e21313. [Google Scholar] [CrossRef]
- Chen, P.-W.; Lin, Y.-L.; Huang, M.-S. Profiles of commensal and opportunistic bacteria in human milk from healthy donors in Taiwan. J. Food Drug Anal. 2018, 26, 1235–1244. [Google Scholar] [CrossRef]
- Li, S.-W.; Watanabe, K.; Hsu, C.-C.; Chao, S.-H.; Yang, Z.-H.; Lin, Y.-J.; Chen, C.-C.; Cao, Y.-M.; Huang, H.-C.; Chang, C.-H. Bacterial composition and diversity in breast milk samples from mothers living in Taiwan and mainland China. Front. Microbiol. 2017, 8, 965. [Google Scholar] [CrossRef]
- Fernández, L.; Browne, P.D.; Aparicio, M.; Alba, C.; Hechler, C.; Beijers, R.; Rodríguez, J.M.; de Weerth, C. Human milk microbiome and maternal postnatal psychosocial distress. Front. Microbiol. 2019, 10, 2333. [Google Scholar]
- Cabrera-Rubio, R.; Collado, M.C.; Laitinen, K.; Salminen, S.; Isolauri, E.; Mira, A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am. J. Clin. Nutr. 2012, 96, 544–551. [Google Scholar] [CrossRef]
- Khodayar-Pardo, P.; Mira-Pascual, L.; Collado, M.C.; Martínez-Costa, C. Impact of lactation stage, gestational age and mode of delivery on breast milk microbiota. J. Perinatol. 2014, 34, 599–605. [Google Scholar] [CrossRef]
- Soto, A.; Martín, V.; Jiménez, E.; Mader, I.; Rodríguez, J.M.; Fernández, L. Lactobacilli and bifidobacteria in human breast milk: Influence of antibiotherapy and other host and clinical factors. J. Pediatr. Gastroenterol. Nutr. 2014, 59, 78. [Google Scholar] [CrossRef]
- Olivares, M.; Albrecht, S.; De Palma, G.; Ferrer, M.D.; Castillejo, G.; Schols, H.A.; Sanz, Y. Human milk composition differs in healthy mothers and mothers with celiac disease. Eur. J. Nutr. 2015, 54, 119–128. [Google Scholar] [CrossRef]
- Kumar, H.; du Toit, E.; Kulkarni, A.; Aakko, J.; Linderborg, K.M.; Zhang, Y.; Nicol, M.P.; Isolauri, E.; Yang, B.; Collado, M.C.; et al. Distinct Patterns in Human Milk Microbiota and Fatty Acid Profiles Across Specific Geographic Locations. Front. Microbiol. 2016, 7, 1619. [Google Scholar] [CrossRef]
- Douglas, C.A.; Ivey, K.L.; Papanicolas, L.E.; Best, K.P.; Muhlhausler, B.S.; Rogers, G.B. DNA extraction approaches substantially influence the assessment of the human breast milk microbiome. Sci. Rep. 2020, 10, 1–10. [Google Scholar]
- Meyer, K.M.; Pace, R.M.; Mohammad, M.; Haymond, M.; Aagaard, K.M. 941: Composition of the breast milk microbiome is influenced by the method of 16S-amplicon sequencing used. Am. J. Obstet. Gynecol. 2019, 220, S607–S608. [Google Scholar] [CrossRef]
- Martín, R.; Jiménez, E.; Olivares, M.; Marín, M.; Fernández, L.; Xaus, J.; Rodríguez, J. Lactobacillus salivarius CECT 5713, a potential probiotic strain isolated from infant feces and breast milk of a mother–child pair. Int. J. Food Microbiol. 2006, 112, 35–43. [Google Scholar] [CrossRef]
- Albesharat, R.; Ehrmann, M.A.; Korakli, M.; Yazaji, S.; Vogel, R.F. Phenotypic and genotypic analyses of lactic acid bacteria in local fermented food, breast milk and faeces of mothers and their babies. Syst. Appl. Microbiol. 2011, 34, 148–155. [Google Scholar] [CrossRef]
- Makino, H.; Kushiro, A.; Ishikawa, E.; Muylaert, D.; Kubota, H.; Sakai, T.; Oishi, K.; Martin, R.; Amor, K.B.; Oozeer, R. Transmission of intestinal Bifidobacterium longum subsp. longum strains from mother to infant, determined by multilocus sequencing typing and amplified fragment length polymorphism. Appl. Environ. Microbiol. 2011, 77, 6788–6793. [Google Scholar] [CrossRef]
- Martín, V.; Maldonado-Barragán, A.; Moles, L.; Rodriguez-Baños, M.; Campo, R.d.; Fernández, L.; Rodríguez, J.M.; Jiménez, E. Sharing of bacterial strains between breast milk and infant feces. J. Hum. Lact. 2012, 28, 36–44. [Google Scholar] [CrossRef]
- Eshaghi, M.; Bibalan, M.H.; Rohani, M.; Esghaei, M.; Douraghi, M.; Talebi, M.; Pourshafie, M.R. Bifidobacterium obtained from mother’s milk and their infant stool; A comparative genotyping and antibacterial analysis. Microb. Pathog. 2017, 111, 94–98. [Google Scholar] [CrossRef]
- Urbaniak, C.; Angelini, M.; Gloor, G.B.; Reid, G. Human milk microbiota profiles in relation to birthing method, gestation and infant gender. Microbiome 2016, 4, 1. [Google Scholar] [CrossRef]
- Biagi, E.; Aceti, A.; Quercia, S.; Beghetti, I.; Rampelli, S.; Turroni, S.; Soverini, M.; Zambrini, A.V.; Faldella, G.; Candela, M.; et al. Microbial Community Dynamics in Mother’s Milk and Infant’s Mouth and Gut in Moderately Preterm Infants. Front. Microbiol. 2018, 9, 2512. [Google Scholar] [CrossRef]
- Dimitriu, P.A.; Iker, B.; Malik, K.; Leung, H.; Mohn, W.; Hillebrand, G.G. New insights into the intrinsic and extrinsic factors that shape the human skin microbiome. MBio 2019, 10, e00839-19. [Google Scholar] [CrossRef]
- Ramsay, D.T.; Kent, J.C.; Owens, R.A.; Hartmann, P.E. Ultrasound imaging of milk ejection in the breast of lactating women. Pediatrics 2004, 113, 361–367. [Google Scholar] [CrossRef]
- Williams, J.E.; Carrothers, J.M.; Lackey, K.A.; Beatty, N.F.; Brooker, S.L.; Peterson, H.K.; Steinkamp, K.M.; York, M.A.; Shafii, B.; Price, W.J. Strong multivariate relations exist among milk, oral, and fecal microbiomes in mother-infant dyads during the first six months postpartum. J. Nutr. 2019, 149, 902–914. [Google Scholar] [CrossRef]
- Geddes, D.T. The use of ultrasound to identify milk ejection in women–tips and pitfalls. Int. Breastfeed. J. 2009, 4, 5. [Google Scholar] [CrossRef]
- Moossavi, S.; Sepehri, S.; Robertson, B.; Bode, L.; Goruk, S.; Field, C.J.; Lix, L.M.; de Souza, R.J.; Becker, A.B.; Mandhane, P.J. Composition and variation of the human milk microbiota are influenced by maternal and early-life factors. Cell Host Microbe 2019, 25, 324–335. [Google Scholar] [CrossRef]
- Moossavi, S.; Azad, M.B. Origins of human milk microbiota: New evidence and arising questions. Gut Microbes 2019, 1–10. [Google Scholar] [CrossRef]
- Riskin, A.; Almog, M.; Peri, R.; Halasz, K.; Srugo, I.; Kessel, A. Changes in immunomodulatory constituents of human milk in response to active infection in the nursing infant. Pediatr. Res. 2012, 71, 220–225. [Google Scholar] [CrossRef]
- Hassiotou, F.; Hepworth, A.R.; Metzger, P.; Tat Lai, C.; Trengove, N.; Hartmann, P.E.; Filgueira, L. Maternal and infant infections stimulate a rapid leukocyte response in breastmilk. Clin. Transl. Immunol. 2013, 2, e3. [Google Scholar] [CrossRef]
- Martén, R.O.; Langa, S.; Reviriego, C.; Jiménez, E.; Martǩn, M.A.L.; Olivares, M.; Boza, J.; Jiménez, J.; Fernández, L.; Xaus, J. The commensal microflora of human milk: New perspectives for food bacteriotherapy and probiotics. Trends Food Sci. Technol. 2004, 15, 121–127. [Google Scholar] [CrossRef]
- Damaceno, Q.S.; Souza, J.P.; Nicoli, J.R.; Paula, R.L.; Assis, G.B.; Figueiredo, H.C.; Azevedo, V.; Martins, F.S. Evaluation of potential probiotics isolated from human milk and colostrum. Probiotics Antimicrob. Proteins 2017, 9, 371–379. [Google Scholar] [CrossRef]
- Rescigno, M.; Urbano, M.; Valzasina, B.; Francolini, M.; Rotta, G.; Bonasio, R.; Granucci, F.; Kraehenbuhl, J.-P.; Ricciardi-Castagnoli, P. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2001, 2, 361–367. [Google Scholar] [CrossRef]
- Langa, S.; Maldonado-Barragán, A.; Delgado, S.; Martín, R.; Martín, V.; Jiménez, E.; Ruíz-Barba, J.L.; Mayo, B.; Connor, R.I.; Suárez, J.E. Characterization of Lactobacillus salivarius CECT 5713, a strain isolated from human milk: From genotype to phenotype. Appl. Microbiol. Biotechnol. 2012, 94, 1279–1287. [Google Scholar] [CrossRef]
- Fernández, L.; Langa, S.; Martín, V.; Maldonado, A.; Jiménez, E.; Martín, R.; Rodríguez, J.M. The human milk microbiota: Origin and potential roles in health and disease. Pharmacol. Res. 2013, 69, 1–10. [Google Scholar] [CrossRef]
- Newburg, D.S. Innate immunity and human milk. J. Nutr. 2005, 135, 1308–1312. [Google Scholar] [CrossRef]
- Macpherson, A.J.; Uhr, T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 2004, 303, 1662–1665. [Google Scholar] [CrossRef]
- Gueimonde, M.; Laitinen, K.; Salminen, S.; Isolauri, E. Breast milk: A source of bifidobacteria for infant gut development and maturation? Neonatology 2007, 92, 64–66. [Google Scholar] [CrossRef]
- Perez, P.F.; Doré, J.; Leclerc, M.; Levenez, F.; Benyacoub, J.; Serrant, P.; Segura-Roggero, I.; Schiffrin, E.J.; Donnet-Hughes, A. Bacterial imprinting of the neonatal immune system: Lessons from maternal cells? Pediatrics 2007, 119, e724–e732. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, E.; Fernández, L.; Maldonado, A.; Martín, R.; Olivares, M.; Xaus, J.; Rodríguez, J. Oral administration of Lactobacillus strains isolated from breast milk as an alternative for the treatment of infectious mastitis during lactation. Appl. Env. Microbiol. 2008, 74, 4650–4655. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, R.; Martín, V.; Maldonado, A.; Jiménez, E.; Fernández, L.; Rodríguez, J.M. Treatment of Infectious Mastitis during Lactation: Antibiotics versus Oral Administration of Lactobacilli Isolated from Breast Milk. Clin. Infect. Dis. 2010, 50, 1551–1558. [Google Scholar] [CrossRef]
- Kordy, K.; Gaufin, T.; Mwangi, M.; Li, F.; Cerini, C.; Lee, D.J.; Adisetiyo, H.; Woodward, C.; Pannaraj, P.S.; Tobin, N.H. Contributions to human breast milk microbiome and enteromammary transfer of Bifidobacterium breve. PLoS ONE 2020, 15, e0219633. [Google Scholar] [CrossRef]
- Elsen, L.V.D.; Garssen, J.; Burcelin, R.; Verhasselt, V. Shaping the gut microbiota by breastfeeding: the gateway to allergy prevention? Front. Pediatr. 2019, 7, 47. [Google Scholar] [CrossRef]
- Eidelman, A.I.; Schanler, R.J. Breastfeeding and the use of human milk. Pediatrics 2012. [Google Scholar] [CrossRef]
- Parikh, N.; Hwang, S.J.; Ingelsson, E.; Benjamin, E.J.; Fox, C.S.; Vasan, R.S.; Murabito, J.M. Breastfeeding in infancy and adult cardiovascular disease risk factors. Am. J. Med. 2009, 122, 656–663. [Google Scholar] [CrossRef]
- Wang, L.; Collins, C.; Ratliff, M.; Xie, B.; Wang, Y. Breastfeeding reduces childhood obesity risks. Child. Obes. 2017, 13, 197–204. [Google Scholar] [CrossRef]
- Klopp, A.; Vehling, L.; Becker, A.B.; Subbarao, P.; Mandhane, P.J.; E Turvey, S.; Lefebvre, D.L.; Sears, M.R.; Azad, M.B.; Daley, D.; et al. Modes of infant feeding and the risk of childhood asthma: A prospective birth cohort study. J. Pediatr. 2017, 190, 192–199. [Google Scholar] [CrossRef]
- Xu, L.; Lochhead, P.; Ko, Y.; Claggett, B.; Leong, R.W.; Ananthakrishnan, A.N. Systematic review with meta-analysis: Breastfeeding and the risk of Crohn’s disease and ulcerative colitis. Aliment. Pharmacol. Ther. 2017, 46, 780–789. [Google Scholar] [CrossRef]
- Dogaru, C.; Nyffenegger, D.; Pescatore, A.M.; Spycher, B.D.; E Kuehni, C. Breastfeeding and childhood asthma: Systematic review and meta-analysis. Am. J. Epidemiology 2014, 179, 1153–1167. [Google Scholar] [CrossRef]
- Horta, B.L.; De Mola, C.L.; Victora, C.G. Long-term consequences of breastfeeding on cholesterol, obesity, systolic blood pressure and type 2 diabetes: A systematic review and meta-analysis. Acta Paediatr. 2015, 104, 30–37. [Google Scholar] [CrossRef]
- Temples, H.S. Breastfeeding reduces risk of Type 2 Diabetes in the (PETS). Nurs. Outlook 2019, 67, 115. [Google Scholar] [CrossRef]
- Michaelsen, F.K.; Lauritzen, L.; Mortensen, E.L. Effects of Breast-feeding on Cognitive Function. In Breast-Feeding: Early Influences on Later Health; Springer: Berlin/Heidelberg, Germany, 2009; pp. 199–215. [Google Scholar]
- Ibrahim, H.S.; El-Ghany, A.; Mohamed, S.; El Shafie, M.T.; Hady, E. Cognitive Functions in Breastfed versus Artificially Fed in Preschool Children. Egypt. J. Hosp. Med. 2019, 77, 5742–5751. [Google Scholar]
- Mackie, R.I.; Sghir, A.; Gaskins, H.R. Developmental microbial ecology of the neonatal gastrointestinal tract. Am. J. Clin. Nutr. 1999, 69, 1035s–1045s. [Google Scholar] [CrossRef]
- Lyons, A.; O’Mahony, D.; O’Brien, F.; Mac Sharry, J.; Sheil, B.; Ceddia, M.; Russell, W.M.; Forsythe, P.; Bienenstock, J.; Kiely, B.; et al. Bacterial strain-specific induction of Foxp3+ T regulatory cells is protective in murine allergy models. Clin. Exp. Allergy 2010, 40, 811–819. [Google Scholar] [CrossRef]
- Nantavisai, K.; Puttikamonkul, S.; Chotelersak, K.; Taweechotipatr, M. In vitro adhesion property and competition against enteropathogens of Lactobacillus strains isolated from Thai infants. Songklanakarin J. Sci. Technol. 2018, 40, 69–74. [Google Scholar]
- Gregory, K.E.; Samuel, B.; Houghteling, P.; Shan, G.; Ausubel, F.; Sadreyev, R.I.; Walker, W.A. Influence of maternal breast milk ingestion on acquisition of the intestinal microbiome in preterm infants. Microbiome 2016, 4, 68. [Google Scholar] [CrossRef]
- Cong, X.; Judge, M.; Xu, W.; Diallo, A.; Janton, S.; Brownell, E.A.; Maas, K.; Graf, J. Influence of infant feeding type on gut microbiome development in hospitalized preterm infants. Nurs. Res. 2017, 66, 123–133. [Google Scholar] [CrossRef]
- Zanella, A.; Silveira, R.C.; Roesch, L.F.W.; Corso, A.L.; Dobbler, P.T.; Mai, V.; Procianoy, R.S. Influence of own mother’s milk and different proportions of formula on intestinal microbiota of very preterm newborns. PLoS ONE 2019, 14, e0217296. [Google Scholar] [CrossRef]
- Inch, S.; Von Xylander, S. Mastitis: Causes and Management; World Health Organization: Geneva, Switzerland, 2000. [Google Scholar]
- Patel, S.; Vaidya, Y.H.; Patel, R.; Pandit, R.J.; Joshi, C.; Kunjadiya, A. Culture independent assessment of human milk microbial community in lactational mastitis. Sci. Rep. 2017, 7, 7804. [Google Scholar] [CrossRef]
- Contreras, G.A.; Rodríguez, J.M. Mastitis: Comparative Etiology and Epidemiology. J. Mammary Gland. Boil. Neoplasia 2011, 16, 339–356. [Google Scholar] [CrossRef]
- Patel, S.; Vaidya, Y.H.; Joshi, C.; Kunjadia, A.P. Culture-dependent assessment of bacterial diversity from human milk with lactational mastitis. Comp. Haematol. Int. 2015, 25, 437–443. [Google Scholar] [CrossRef]
- Moazzez, A. Breast abscess bacteriologic features in the era of community-acquired methicillin-resistant Staphylococcus aureus epidemics. Arch. Surg. 2007, 142, 881. [Google Scholar] [CrossRef]
- Branch-Elliman, W.; Lee, G.M.; Golen, T.H.; Gold, H.S.; Baldini, L.M.; Wright, S.B. Health and economic burden of post-partum Staphylococcus aureus breast abscess. PLoS ONE 2013, 8, e73155. [Google Scholar] [CrossRef]
- Padilha, M.; Iaucci, J.; Cabral, V.; Diniz, E.; Taddei, C.; Saad, S.M.I. Maternal antibiotic prophylaxis affects Bifidobacterium spp. counts in the human milk, during the first week after delivery. Benef. Microbes 2019, 10, 155–163. [Google Scholar]
- Maldonado-Lobón, J.A.; Díaz-López, M.A.; Carputo, R.; Duarte, P.; Díaz-Ropero, M.P.; Valero, A.D.; Sañudo, A.; Sempere, L.; Ruiz-López, M.D.; Bañuelos, O.; et al. Lactobacillus fermentum CECT 5716 reduces Staphylococcus load in the breastmilk of lactating mothers suffering breast pain: A randomized controlled trial. Breastfeed. Med. 2015, 10, 425–432. [Google Scholar] [CrossRef]
- Hurtado, J.A.; Maldonado-Lobón, J.A.; Díaz-Ropero, M.P.; Flores-Rojas, K.; Uberos, J.; Leante, J.L.; Affumicato, L.; Couce, M.L.; Garrido, J.M.; Olivares, M.; et al. Oral administration to nursing women of Lactobacillus fermentum CECT5716 prevents lactational mastitis development: A randomized controlled trial. Breastfeed. Med. 2017, 12, 202–209. [Google Scholar] [CrossRef]
- Bond, D.M.; Morris, J.M.; Nassar, N. Nassar, Study protocol: evaluation of the probiotic Lactobacillus Fermentum CECT5716 for the prevention of mastitis in breastfeeding women: A randomised controlled trial. BMC Pregnancy Childbirth 2017, 17, 148. [Google Scholar] [CrossRef]
- Fernández, L.; Cárdenas, N.; Arroyo, R.; Manzano, S.; Jiménez, E.; Martin, V.; Rodríguez, J.M. Prevention of infectious mastitis by oral administration of Lactobacillus salivarius PS2 during late pregnancy. Clin. Infect. Dis. 2015, 62, 568–573. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Calder, P.C. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Diaz, J.P.; Ruiz-Ojeda, F.J.; Gil-Campos, M. Mechanisms of action of probiotic. Adv. Nutr. 2019, 10, S49–S66. [Google Scholar] [CrossRef]
- Yousefi, B.; Eslami, M.; Ghasemian, A.; Kokhaei, P.; Farrokhi, A.S.; Darabi, N. Probiotics importance and their immunomodulatory properties. J. Cell. Physiol. 2018, 234, 8008–8018. [Google Scholar] [CrossRef]
- Lara-Villoslada, F.; Olivares, M.; Sierra, S.; Rodríguez, J.M.; Boza, J.; Xaus, J. Beneficial effects of probiotic bacteria isolated from breast milk. Br. J. Nutr. 2007, 98, S96–S100. [Google Scholar] [CrossRef]
- Arboleya, S.; Ruas-Madiedo, P.; Margolles, A.; Solís, G.; Salminen, S.; Reyes-Gavilán, C.G.D.L.; Gueimonde, M. Characterization and in vitro properties of potentially probiotic Bifidobacterium strains isolated from breast-milk. Int. J. Food Microbiol. 2011, 149, 28–36. [Google Scholar] [CrossRef]
- Mehanna, N.S.; Tawfik, N.F.; Salem, M.M.; Effat, B.A.; Gad El-Rab, D.A. Assessment of potential probiotic bacteria isolated from breast milk. Middle East J. Sci. Res. 2013, 14, 354–360. [Google Scholar]
- Kozak, K.; Charbonneau, D.L.; Sanozky-Dawes, R.; Klaenhammer, T. Characterization of bacterial isolates from the microbiota of mothers’ breast milk and their infants. Gut Microbes 2016, 6, 341–351. [Google Scholar] [CrossRef]
- Jamyuang, C.; Phoonlapdacha, P.; Chongviriyaphan, N.; Chanput, W.; Nitisinprasert, S.; Nakphaichit, M. Characterization and probiotic properties of Lactobacilli from human breast milk. 3 Biotech 2019, 9, 398. [Google Scholar] [CrossRef]
- Halloran, K.; Underwood, M.A. Underwood, Probiotic mechanisms of action. Early Hum. Dev. 2019, 135, 58–65. [Google Scholar] [CrossRef]
- Díaz-Ropero, M.; Martin, R.; Sierra, S.; Lara-Villoslada, F.; Rodríguez, J.M.; Xaus, J.; Olivares, M. Two Lactobacillus strains, isolated from breast milk, differently modulate the immune response. J. Appl. Microbiol. 2007, 102, 337–343. [Google Scholar] [CrossRef]
- Solís, G.; Reyes-Gavilan, C.D.L.; Fernández, N.; Margolles, A.; Gueimonde, M.; Reyes-Gavilán, C.G.D.L. Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe 2010, 16, 307–310. [Google Scholar] [CrossRef]
- Rajoka, M.S.R.; Zhao, H.; Mehwish, H.M.; Li, N.; Lu, Y.; Lian, Z.; Shao, D.; Jin, M.; Li, Q.; Zhao, L.; et al. Anti-tumor potential of cell free culture supernatant of Lactobacillus rhamnosus strains isolated from human breast milk. Food Res. Int. 2019, 123, 286–297. [Google Scholar] [CrossRef]
- Gunyakti, A.; Özüsağlam, M.A. Lactobacillus gasseri from human milk with probiotic potential and some technological properties. LWT 2019, 109, 261–269. [Google Scholar] [CrossRef]
- Mu, Q.; Tavella, V.J.; Luo, X. Role of Lactobacillus reuteri in Human Health and Diseases. Front. Microbiol. 2018, 9, 757. [Google Scholar] [CrossRef]
- Kosek, M.N.; Peñataro-Yori, P.; Paredes-Olortegui, M.; Lefante, J.; Ramal-Asayag, C.; Zamora-Babilonia, M.; Oberhelman, R.A. Safety of Lactobacillus Reuteri DSM 17938 in Healthy Children 2–5 Years of Age. Pediatr. Infect. Disease J. 2019, 38, e178–e180. [Google Scholar] [CrossRef]
- Fatheree, N.Y.; Liu, Y.; Taylor, C.M.; Hoang, T.K.; Cai, C.; Rahbar, M.H.; Hessabi, M.; Ferris, M.; McMurtry, V.; Wong, C.; et al. Lactobacillus reuteri for infants with colic: A double-blind, placebo-controlled, randomized clinical trial. J. Pediatr. 2017, 191, 170–178. [Google Scholar] [CrossRef]
- Sung, V.; D’Amico, F.; Cabana, M.D.; Chau, K.; Koren, G.; Savino, F.; Szajewska, H.; Deshpande, G.; Dupont, C.; Indrio, F.; et al. Lactobacillus reuteri to treat infant colic: A meta-analysis. Pediatrics 2018, 141, e20171811. [Google Scholar] [CrossRef]
- Rodenas, C.L.G.; Lepage, M.; Ngom-Bru, C.; Fotiou, A.; Papagaroufalis, K.; Berger, B. Effect of formula containing Lactobacillus reuteri DSM 17938 on fecal microbiota of infants born by cesarean-section. J. Pediatr. Gastroenterol. Nutr. 2016, 63, 681–687. [Google Scholar] [CrossRef]
- Francavilla, R.; Lionetti, E.; Castellaneta, S.; Ciruzzi, F.; Indrio, F.; Masciale, A.; Fontana, F.; La Rosa, M.M.; Cavallo, L.; Francavilla, A. Randomised clinical trial: L actobacillus reuteri DSM 17938 vs. placebo in children with acute diarrhoea-a double-blind study. Aliment. Pharmacol. Ther. 2012, 36, 363–369. [Google Scholar]
- Dinleyici, E.C.; Dalgic, N.; Guven, S.; Metin, O.; Yasa, O.; Kurugol, Z.; Turel, O.; Tanir, G.; SamiYazar, A.; Sancar, M.; et al. Lactobacillus reuteri DSM 17938 shortens acute infectious diarrhea in a pediatric outpatient setting. Jornal Pediatria 2015, 91, 392–396. [Google Scholar] [CrossRef]
- Dinleyici, E.C.; Vandenplas, Y.; PROBAGE Study Group. Lactobacillus reuteri DSM 17938 effectively reduces the duration of acute diarrhoea in hospitalised children. Acta Paediatr. 2014, 103, e300–e305. [Google Scholar]
- Urbanska, M.; Gieruszczak-Białek, D.; Szajewska, H. Systematic review with meta-analysis: Lactobacillus reuteri DSM 17938 for diarrhoeal diseases in children. Aliment. Pharmacol. Ther. 2016, 43, 1025–1034. [Google Scholar] [CrossRef]
- Gutiérrez-Castrellón, P.; López-Velázquez, G.; Garcia, M.L.D.; Jimenez-Gutierrez, C.; Mancilla-Ramirez, J.; Estevez-Jimenez, J.; Parra, M. Diarrhea in preschool children and Lactobacillus reuteri: A randomized controlled trial. Pediatrics 2014, 133, e904–e909. [Google Scholar] [CrossRef]
- Khalkhali, S.; Mojgani, N. In vitro and in vivo safety analysis of Enterococcus faecium 2C isolated from human breast milk. Microb. Pathog. 2018, 116, 73–77. [Google Scholar] [CrossRef]
- Bagci, U.; Togay, S.O.; Temiz, A.; Ay, M. Probiotic characteristics of bacteriocin-producing Enterococcus faecium strains isolated from human milk and colostrum. Folia Microbiol. 2019, 64, 735–750. [Google Scholar] [CrossRef]
- Fouhy, F.; Deane, J.; Rea, M.C.; O’Sullivan, O.; Ross, P.; O’Callaghan, G.; Plant, B.J.; Stanton, C. The effects of freezing on faecal microbiota as determined using MiSeq sequencing and culture-based investigations. PLoS ONE 2015, 10, e0119355. [Google Scholar] [CrossRef]
- Lackey, K.A.; Williams, J.E.; Price, W.J.; Carrothers, J.M.; Brooker, S.L.; Shafii, B.; McGuire, M.A.; McGuire, M. Comparison of commercially-available preservatives for maintaining the integrity of bacterial DNA in human milk. J. Microbiol. Methods 2017, 141, 73–81. [Google Scholar] [CrossRef]
- Fouhy, F.; Clooney, A.G.; Stanton, C.; Claesson, M.J.; Cotter, P. 16S rRNA gene sequencing of mock microbial populations-impact of DNA extraction method, primer choice and sequencing platform. BMC Microbiol. 2016, 16, 123. [Google Scholar] [CrossRef]
- Martín, R.; Bermúdez-Humarán, L.G.; Langella, P. Searching for the bacterial effector: The example of the multi-skilled commensal bacterium Faecalibacterium prausnitzii. Front. Microbiol. 2018, 9, 346. [Google Scholar] [CrossRef]
- Cani, P.D.; De Vos, W.M. Next-generation beneficial microbes: the case of Akkermansia muciniphila. Front. Microbiol. 2017, 8, 1765. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lyons, K.E.; Ryan, C.A.; Dempsey, E.M.; Ross, R.P.; Stanton, C. Breast Milk, a Source of Beneficial Microbes and Associated Benefits for Infant Health. Nutrients 2020, 12, 1039. https://doi.org/10.3390/nu12041039
Lyons KE, Ryan CA, Dempsey EM, Ross RP, Stanton C. Breast Milk, a Source of Beneficial Microbes and Associated Benefits for Infant Health. Nutrients. 2020; 12(4):1039. https://doi.org/10.3390/nu12041039
Chicago/Turabian StyleLyons, Katríona E., C. Anthony Ryan, Eugene M. Dempsey, R. Paul Ross, and Catherine Stanton. 2020. "Breast Milk, a Source of Beneficial Microbes and Associated Benefits for Infant Health" Nutrients 12, no. 4: 1039. https://doi.org/10.3390/nu12041039
APA StyleLyons, K. E., Ryan, C. A., Dempsey, E. M., Ross, R. P., & Stanton, C. (2020). Breast Milk, a Source of Beneficial Microbes and Associated Benefits for Infant Health. Nutrients, 12(4), 1039. https://doi.org/10.3390/nu12041039