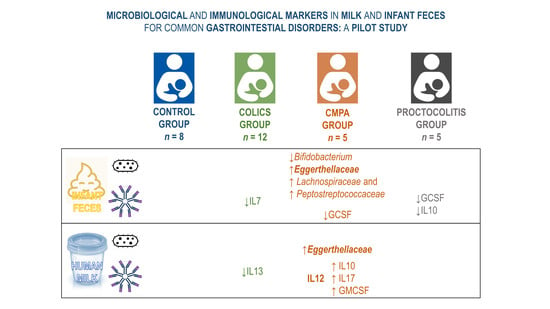
Microbiological and Immunological Markers in Milk and Infant Feces for Common Gastrointestinal Disorders: A Pilot Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Collection of the Samples
2.3. DNA Extraction from the Samples
2.4. PCR Amplification and Sequencing
2.5. Immunological Assays
2.6. Statistical and Bioinformatic Analysis
3. Results
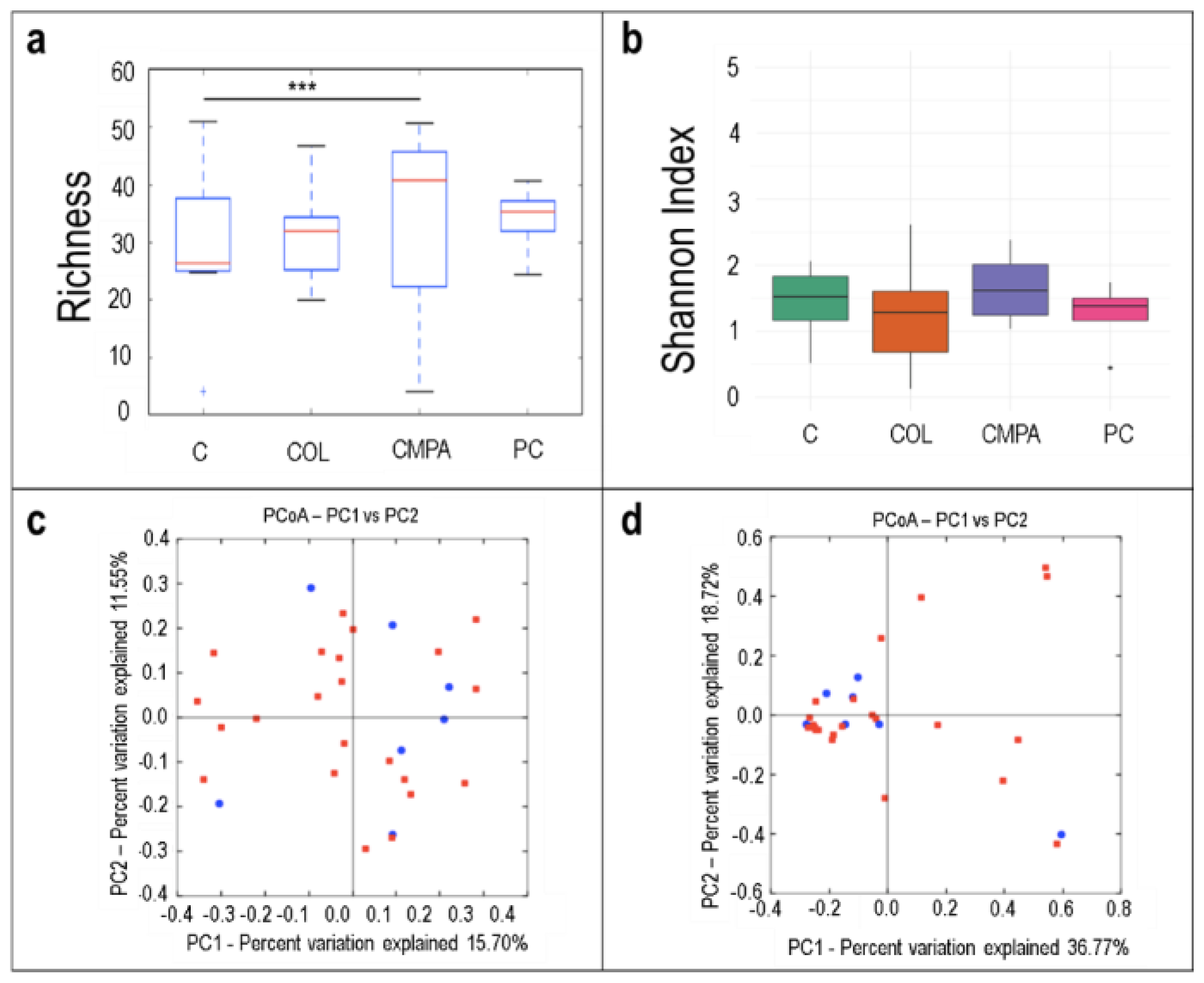
3.1. Metataxonomic Profiling of the Fecal Samples
3.2. Metataxonomic Profiling of the Milk Samples
3.3. Concentration and Frequency of Detection of Immune Factors in the Infant Fecal Samples
3.4. Concentration and Frequency of Detection of Immune Factors in Milk Samples
3.5. Relationship between the Immune Profile of Milk and Infant Fecal Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Benninga, M.A.; Nurko, S.; Faure, C.; Hyman, P.E.; St James Roberts, I.; Schechter, N.L. Childhood functional gastrointestinal disorders: Neonate/toddler. Gastroenterology 2016, 150, 1443–1455.e2. [Google Scholar] [CrossRef]
- Mahon, J.; Lifschitz, C.; Ludwig, T.; Thapar, N.; Glanville, J.; Miqdady, M.; Saps, M.; Quak, S.H.; Lenoir Wijnkoop, I.; Edwards, M.; et al. The costs of functional gastrointestinal disorders and related signs and symptoms in infants: A systematic literature review and cost calculation for England. BMJ Open 2017, 7, e015594. [Google Scholar] [CrossRef]
- Hyman, P.E.; Milla, P.J.; Benninga, M.A.; Davidson, G.P.; Fleisher, D.F.; Taminiau, J. Childhood functional gastrointestinal disorders: Neonate/toddler. Gastroenterology 2006, 130, 1519–1526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caubet, J.C.; Szajewska, H.; Shamir, R.; Nowak-Węgrzyn, A. Non-IgE-mediated gastrointestinal food allergies in children. Pediatr. Allergy Immunol. 2017, 28, 6–17. [Google Scholar] [CrossRef] [PubMed]
- Sung, V.; Hiscock, H.; Tang, M.; Mensah, F.K.; Heine, R.G.; Stock, A.; York, E.; Barr, R.G.; Wake, M. Probiotics to improve outcomes of colic in the community: Protocol for the Baby Biotics randomised controlled trial. BMC Pediatr. 2012, 12, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubois, N.E.; Gregory, K.E. Characterizing the intestinal microbiome in infantile colic: Findings based on an integrative review of the literature. Biol. Res. Nurs. 2016, 18, 307–315. [Google Scholar] [CrossRef]
- Pensabene, L.; Salvatore, S.; D’Auria, E.; Parisi, F.; Concolino, D.; Borrelli, O.; Thapar, N.; Staiano, A.; Vandenplas, Y.; Saps, M. Cow’s milk protein allergy in infancy: A risk factor for functional gastrointestinal disorders in children? Nutrients 2018, 10, 1716. [Google Scholar] [CrossRef] [Green Version]
- Mousan, G.; Kamat, D. Cow’s milk protein allergy. Clin. Pediatr. 2016, 55, 1054–1063. [Google Scholar] [CrossRef]
- Tsabouri, S.; Nicolaou, N.; Douros, K.; Papadopoulou, A.; Priftis, K. Food protein induced proctocolitis: A benign condition with an obscure immunologic mechanism. Endocr. Metab. Immune Disord. Drug Targets 2017, 17, 32–37. [Google Scholar] [CrossRef]
- Vandenplas, Y.; Benninga, M.A.; Broekaert, I.J.; Falconer, J.; Gottrand, F.; Guarino, A.; Lifschitz, C.; Lionetti, P.; Orel, R.; Papadopoulou, A.; et al. Functional gastro-intestinal disorder algorithms focus on early recognition, parental reassurance and nutritional strategies. Acta Paediatr. 2016, 105, 244–252. [Google Scholar] [CrossRef]
- Dong, P.; Feng, J.J.; Yan, D.Y.; Lyu, Y.J.; Xu, X. Early-life gut microbiome and cow’s milk allergy- a prospective case - control 6-month follow-up study. Saudi J. Biol. Sci. 2018, 25, 875–880. [Google Scholar] [CrossRef] [PubMed]
- Chau, K.; Lau, E.; Greenberg, S.; Jacobson, S.; Yazdani-Brojeni, P.; Verma, N.; Koren, G. Probiotics for infantile colic: A randomized, double-blind, placebo-controlled trial investigating Lactobacillus reuteri DSM 17938. J. Pediatr. 2015, 166, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Ling, Z.; Li, Z.; Liu, X.; Cheng, Y.; Luo, Y.; Tong, X.; Yuan, L.; Wang, Y.; Sun, J.; Li, L.; et al. Altered fecal microbiota composition associated with food allergy in infants. Appl. Environ. Microbiol. 2014, 80, 2546–2554. [Google Scholar] [CrossRef] [Green Version]
- Fahey, L.M.; Liacouras, C.A. Eosinophilic gastrointestinal disorders. Pediatr. Clin. N. Am. 2017, 64, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.J.; Marsland, B.J.; Bunyavanich, S.; O’Mahony, L.; Leung, D.Y.; Muraro, A.; Fleisher, T.A. The microbiome in allergic disease: Current understanding and future opportunities-2017 PRACTALL document of the American Academy of Allergy, Asthma and Immunology and the European Academy of Allergy and Clinical Immunology. J. Allergy Clin. Immunol. 2017, 139, 1099–1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández, L.; Langa, S.; Martín, V.; Maldonado, A.; Jiménez, E.; Martín, R.; Rodríguez, J.M. The human milk microbiota: Origin and potential roles in health and disease. Pharmacol. Res. 2013, 69, 1–10. [Google Scholar] [CrossRef]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef] [Green Version]
- Lackey, K.A.; Williams, J.E.; Meehan, C.L.; Zachek, J.A.; Benda, E.D.; Price, W.J.; Foster, J.A.; Sellen, D.W.; Kamau-Mbuthia, E.W.; Kamundia, E.W.; et al. What’s normal? Microbiomes in human milk and infant feces are related to each other but vary geographically: The INSPIRE Study. Front. Nutr. 2019, 6, 45. [Google Scholar] [CrossRef] [Green Version]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Zakrzewski, M.; Proietti, C.; Ellis, J.J.; Hasan, S.; Brion, M.J.; Berger, B.; Krause, L. Calypso: A user-friendly web-server for mining and visualizing microbiome-environment interactions. Bioinformatics 2017, 33, 782–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and "All-species Living Tree Project (LTP)" taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espinosa-Martos, I.; Montilla, A.; de Segura, A.G.; Escuder, D.; Bustos, G.; Pallás, C.; Rodríguez, J.M.; Corzo, N.; Fernández, L. Bacteriological, biochemical, and immunological modifications in human colostrum after Holder pasteurisation. J. Pediatr. Gastroenterol. Nutr. 2013, 56, 560–568. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Harrison, O.J. Homeostatic immunity and the microbiota. Immunity 2017, 46, 562–576. [Google Scholar] [CrossRef]
- Rajani, P.S.; Seppo, A.E.; Jarvinen, K.M. Immunologically active components in human milk and development of atopic disease, with emphasis on food allergy, in the pediatric population. Front. Pediatr. 2018, 6, 218. [Google Scholar] [CrossRef]
- Sagheddu, V.; Patrone, V.; Miragoli, F.; Puglisi, E.; Morelli, L. Infant early gut colonization by Lachnospiraceae: High frequency of Ruminococcus gnavus. Front. Pediatr. 2016, 4, 57. [Google Scholar] [CrossRef] [Green Version]
- Thompson-Chagoyan, O.C.; Vieites, J.M.; Maldonado, J.; Edwards, C.; Gil, A. Changes in fecal microbiota of infants with cow’s milk protein allergy - a Spanish prospective case-control 6-month follow-up study. Pediatr. Allergy Immunol. 2010, 21, e394–e400. [Google Scholar] [CrossRef]
- Werlang, I.; Mueller, N.T.; Pizoni, A.; Wisintainer, H.; Matte, U.; Costa, S.; Ramos, J.; Goldani, M.Z.; Dominguez-Bello, M.G.; Goldani, H. Associations of birth mode with cord blood cytokines, white bloodcells, and newborn intestinal bifidobacteria. PLoS ONE 2018, 13, e0205962. [Google Scholar] [CrossRef]
- Berni Canani, R.; Sangwan, N.; Stefka, A.T.; Nocerino, R.; Paparo, L.; Aitoro, R.; Calignano, A.; Khan, A.A.; Gilbert, J.A.; Nagler, C.R. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J. 2016, 10, 742–750. [Google Scholar] [CrossRef]
- Berni Canani, R.; De Filippis, F.; Nocerino, R.; Paparo, L.; Di Scala, C.; Cosenza, L.; Della Gatta, G.; Calignano, A.; De Caro, C.; Laiola, M.; et al. Gut microbiota composition and butyrate production in children affected by non-IgE-mediated cow’s milk allergy. Sci. Rep. 2018, 8, 12500. [Google Scholar] [CrossRef] [Green Version]
- Diaz, M.; Guadamuro, L.; Espinosa-Martos, I.; Mancabelli, L.; Jimenez, S.; Molinos-Norniella, C.; Perez-Solis, D.; Milani, C.; Rodriguez, J.M.; Ventura, M.; et al. Microbiota and derived parameters in fecal samples of infants with non-IgE cow’s milk protein allergy under a restricted diet. Nutrients 2018, 10, 1481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feehley, T.; Plunkett, C.H.; Bao, R.; Choi Hong, S.M.; Culleen, E.; Belda-Ferre, P.; Campbell, E.; Aitoro, R.; Nocerino, R.; Paparo, L.; et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat. Med. 2019, 25, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Iozzo, P.; Sanguinetti, E. Early dietary patterns and microbiota development: Still a way to go from descriptive interactions to health-relevant solutions. Front. Nutr. 2018, 5, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gardiner, B.J.; Tai, A.Y.; Kotsanas, D.; Francis, M.J.; Roberts, S.A.; Ballard, S.A.; Junckerstorff, R.K.; Korman, T.M. Clinical and microbiological characteristics of Eggerthella lenta bacteremia. J. Clin. Microbiol. 2015, 53, 626–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, S.K.; Woo, P.C.; Fung, A.M.; Chan, K.M.; Woo, G.K.; Yuen, K.Y. Anaerobic, non-sporulating, Gram-positive bacilli bacteraemia characterized by 16S rRNA gene sequencing. J. Med. Microbiol. 2004, 53 Pt 12, 1247–1253. [Google Scholar] [CrossRef]
- Lehtonen, L.; Korvenranta, H.; Eerola, E. Intestinal microflora in colicky and noncolicky infants: Bacterial cultures and gas-liquid chromatography. J. Pediatr. Gastroenterol. Nutr. 1994, 19, 310–314. [Google Scholar] [CrossRef]
- Savino, F.; Cresi, F.; Pautasso, S.; Palumeri, E.; Tullio, V.; Roana, J.; Silvestro, L.; Oggero, R. Intestinal microflora in breastfed colicky and non-colicky infants. Acta Paediatr. 2004, 93, 825–829. [Google Scholar] [CrossRef]
- Savino, F.; Bailo, E.; Oggero, R.; Tullio, V.; Roana, J.; Carlone, N.; Cuffini, A.M.; Silvestro, L. Bacterial counts of intestinal Lactobacillus species in infants with colic. Pediatr. Allergy Immunol. 2005, 16, 72–75. [Google Scholar] [CrossRef]
- Muraro, A.; Halken, S.; Arshad, S.H.; Beyer, K.; Dubois, A.E.J.; Du Toit, G.; Eigenmann, P.A.; Grimshaw, K.E.; Hoest, A.; Lack, G.; et al. EAACI Food Allergy and Anaphylaxis Guidelines Group. EAACI food allergy and anaphylaxis guidelines. Primary prevention of food allergy. Allergy 2014, 69, 590–601. [Google Scholar] [CrossRef]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Delgado Palacio, S.; Arboleya Montes, S.; Mancabelli, L.; et al. The first microbial colonizers of the human gut: Composition, activities, and health implications of the infant gut microbiota. Microbiol. Mol. Biol. Rev. 2017, 81, e00036-17. [Google Scholar] [CrossRef] [Green Version]
- Lodge, C.J.; Tan, D.J.; Lau, M.X.; Dai, X.; Tham, R.; Lowe, A.J.; Bowatte, G.; Allen, K.J.; Dharmage, S.C. Breastfeeding and asthma and allergies: A systematic review and meta-analysis. Acta Paediatr. 2015, 104, 38–53. [Google Scholar] [CrossRef] [PubMed]
- Gomez, M.; Moles, L.; Espinosa-Martos, I.; Bustos, G.; de Vos, W.M.; Fernandez, L.; Rodriguez, J.M.; Fuentes, S.; Jimenez, E. Bacteriological and immunological profiling of meconium and fecal samples from preterm infants: A two-year follow-up study. Nutrients 2017, 9, 1293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eyles, J.L.; Roberts, A.W.; Metcalf, D.; Wicks, I.P. Granulocyte colony-stimulating factor and neutrophils--forgotten mediators of inflammatory disease. Nat. Clin. Pract. Rheumatol. 2006, 2, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Zhao, L.; Wan, Y.Y.; Zhu, B. Mechanism of action of IL-7 and its potential applications and limitations in cancer immunotherapy. Int. J. Mol. Sci. 2015, 16, 10267–10280. [Google Scholar] [CrossRef] [Green Version]
- Iyer, S.S.; Cheng, G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit. Rev. Immunol. 2012, 32, 23–63. [Google Scholar] [CrossRef] [Green Version]
- Waters, R.S.; Perry, J.S.A.; Han, S.; Bielekova, B.; Gedeon, T. The effects of interleukin-2 on immune response regulation. Math. Med. Biol. 2018, 35, 79–119. [Google Scholar] [CrossRef]
- Jarvinen, K.M.; Suarez-Farinas, M.; Savilahti, E.; Sampson, H.A.; Berin, M.C. Immune factors in breast milk related to infant milk allergy are independent of maternal atopy. J. Allergy Clin. Immunol. 2015, 135, 1390–1393.e6. [Google Scholar] [CrossRef] [Green Version]
- Kuwabara, T.; Ishikawa, F.; Kondo, M.; Kakiuchi, T. The role of IL-17 and related cytokines in inflammatory autoimmune diseases. Mediat. Inflamm. 2017, 2017, 3908061. [Google Scholar] [CrossRef] [Green Version]
- Mannon, P.; Reinisch, W. Interleukin 13 and its role in gut defence and inflammation. Gut 2012, 61, 1765–1773. [Google Scholar] [CrossRef] [Green Version]



| Characteristic | Control Group | Colics Group | CMPA Group | Proctocolitis Group |
|---|---|---|---|---|
| n | 8 | 12 | 5 | 5 |
| Gestational age (wks) | 30.38 ± 15.15 | 28.25 ± 16.85 | 31.40 ± 14.82 | 36.00 ± 8.97 |
| Weight (kg) | 4.70 ± 1.03 | 4.30 ± 0.74 | 4.21 ± 0.72 | 5.84 ± 1.40 |
| Length (cm) | 54.84 ± 3.05 | 54.29 ± 3.21 | 54.60 ± 2.58 | 59.60 ± 5.41 |
| Delivery mode, n (%) | ||||
| Vaginal | 7 (88) | 11 (92) | 1 (20) | 3 (60) |
| Cesarean | 1 (12) | 1 (8) | 4 (80) | 2 (40) |
| Gender, n (%) | ||||
| Male | 4 (50) | 5 (42) | 4 (80) | 3 (60) |
| Female | 4 (50) | 7 (58) | 1 (20) | 2 (40) |
| Case definition | Paroxysms of irritability, restlessness or crying at least 3 h/d and at least 3 d/wk, that begin and end without apparent cause in an infant during the first 4 mo with good general condition and good weight gain | Infants with non-IgE-mediated cow’s milk protein allergy | Paroxysms of irritability, restlessness or crying at least 3 h/d and at least 3 d/wk, that begin and end without apparent cause in an infant during the first 4 mo with good general condition and good weight gain | |
| Inclusion criteria | Healthy infants Healthy mother No dietary intervention in the mother | Infants <4 mo Exclusive breastfeeding No dietary interventions in the mother | Infants <6 mo Artificial or mixed breastfeeding (50% of calories contributed by formula) Suggestive symptoms (regurgitation, vomiting, refusal of food, diarrhea, constipation, perianal erythema…) Improvement with cow’s milk protein exclusion diet | Infants <6 mo with clinical symptoms compatible with colitis Exclusive breastfeeding No dietary intervention in the mother Negative stool microbiological study |
| Exclusion criteria | Any diagnosed or suspected pathology | Infants >4 mo Formula-fed infants Presence of signs or symptoms of other pathologies | Infants >6 mo IgE-mediated CMPA Severe atopic dermatitis probably related to cow’s milk proteins (as a single manifestation) Enterocolitis syndrome Exclusive breastfeeding | Infants <6 mo Formula-fed infants Infectious colitis Hircshsprung’s disease Coagulopathy Anal fissure |
| Samples provided * | ||||
| Feces | 7 | 12 | 5 | 5 |
| Milk | 8 | 11 | 3 | 4 |
| Control Group (n = 7) | Colic Group (n = 12) | CMPA Group (n = 5) | Proctocolitis Group (n = 5) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phylum Genus | n (%) | Median (IQR) | n (%) | Median (IQR) | n (%) | Median (IQR) | n (%) | Median (IQR) | p * | p ** |
| Actinobacteria | ||||||||||
| Bifidobacterium | 6 (86) | 64.24 (61.91–75.05) | 12 (100) | 62.10 (39.57–88.63) | 4 (80) | 23.40 (5.27–51.23) | 5 (100) | 54.14 (37.49–77.62) | 0.335 | 0.040 |
| Collinsella | 0 (0) | - | 4 (33) | 4.00 (2.01–14.06) | 2 (40) | 3.41 (3.37–3.45) | 0 (0) | - | 0.178 | 1.000 |
| Bacteroidetes | ||||||||||
| Bacteroides | 4 (57) | 2.78 (0.54–5.52) | 8 (67) | 0.45 (0.22–4.29) | 0 (0) | - | 4 (80) | 1.27 (0.70–3.61) | 0.055 | 0.811 |
| Firmicutes | ||||||||||
| Enterococcus | 5 (71) | 0.97 (0.95–1.37) | 8 (67) | 0.33 (0.20–1.69) | 3 (60) | 0.42 (0.27–1.42) | 5 (100) | 3.28 (2.27–3.29) | 0.552 | 0.300 |
| Lactobacillus | 5 (71) | 1.37 (0.69–1.51) | 8 (67) | 2.10 (0.41–6.31) | 3 (60) | 0.38 (0.29–1.88) | 2 (40) | 16.42 (8.67–24.17) | 0.822 | 0.727 |
| Streptococcus | 7 (100) | 2.00 (0.54–8.79) | 11 (92) | 1.36 (0.46–2.79) | 5 (100) | 1.67 (1.41–2.54) | 5 (100) | 0.99 (0.19–1.17) | 1.000 | 0.395 |
| Erysipelatoclostridium | 5 (71) | 8.39 (1.74–8.78) | 4 (33) | 0.25 (0.18–1.99) | 2 (40) | 0.38 (0.34–0.43) | 1 (20) | 0.25 | 0.333 | 0.012 |
| Veillonella | 5 (71) | 0.70 (0.39–3.78) | 9 (75) | 0.49 (0.34–0.76) | 3 (60) | 1.59 (0.97–12.42) | 5 (100) | 0.89 (0.20–1.19) | 0.579 | 0.665 |
| Proteobacteria | ||||||||||
| Escherichia_Shigella | 5 (71) | 9.56 (0.66–14.73) | 8 (67) | 1.40 (0.47–4.19) | 3 (60) | 2.42 (1.28–40.69) | 5 (100) | 6.56 (0.52–51.11) | 0.552 | 0.871 |
| Klebsiella | 1 (14) | 6.32 | 4 (33) | 6.15 (4.26–6.89) | 1 (20) | 2.81 (2.27–3.34) | 3 (60) | 0.37 (0.33–6.93) | 0.510 | 0.884 |
| Unclassified_Enterobacteriaceae | 3 (43) | 0.95 (0.85–27.89) | 5 (42) | 1.21 (0.38–4.62) | 4 (80) | 4.82 (1.27–19.49) | 3 (60) | 0.55 (0.35–0.76) | 0.579 | 0.488 |
| Control Group (n = 8) | Colic Group (n = 11) | CMPA Group (n = 3) | Proctocolitis Group (n = 4) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phylum Genus | n (%) | Median (IQR) | n (%) | Median (IQR) | n (%) | Median (IQR) | n (%) | Median (IQR) | p* | p ** |
| Actinobacteria | ||||||||||
| Corynebacterium | 7 (88) | 0.88 (0.31–1.76) | 10 (91) | 0.27 (0.19–0.86) | 2 (67) | 1.42 (0.78–2.05) | 2 (50) | 0.51 (0.38–0.64) | 0.238 | 0.735 |
| Kocuria | 2 (25) | 1.18 (1.08–1.29) | 4 (36) | 0.55 (0.22–6.97) | 2 (67) | 0.98 (0.77–1.18) | 2 (50) | 1.38 (0.75–2.01) | 0.702 | 0.865 |
| Firmicutes | ||||||||||
| Bacillus | 4 (57) | 2.78 (0.54–5.52) | 8 (67) | 0.45 (0.22–4.29) | 0 (0) | - | 2 (50) | 3.60 (2.09–5.11) | 0.178 | 0.555 |
| Gemella | 5 (63) | 0.52 (0.31–0.83) | 6 (55) | 1.89 (0.66–4.71) | 1 (33) | 0.11 | 2 (50) | 1.75 (1.43–2.07)) | 0.940 | 0.150 |
| Lactobacillus | 5 (63) | 0.47 (0.30–1.93) | 10 (91) | 0.98 (0.39–1.59) | 2 (67) | 0.86 (0.67–1.04) | 3 (75) | 1.19 (0.74–2.49) | 0.174 | 0.990 |
| Staphylococcus | 8 (100) | 9.63º (1.88–51.35) | 11 (100) | 1.93 (1.06–8.61) | 2 (67) | 6.26 (5.19–7.33) | 4 (100) | 9.44 (3.24–17.64) | 0.115 | 0.652 |
| Streptococcus | 8 (100) | 17.18 (5.08–48.80) | 11 (100) | 23.70 (12.72–36.92) | 2 (67) | 20.48 (16.30–24.66) | 4 (100) | 35.09 (12.29–65.44) | 0.115 | 0.905 |
| Proteobacteria | ||||||||||
| Acinetobacter | 7 (88) | 2.95 (0.59–5.05) | 9 (82) | 1.32 (0.36–1.52) | 3 (100) | 0.43 (0.28–0.56) | 4 (100) | 2.28 (0.22–4.53) | 1.000 | 0.367 |
| Pseudomonas | 4 (50) | 0.37 (0.28–17.02) | 6 (55) | 0.28 (0.18–0.39) | 2 (67) | 0.37 (0.34–0.40) | 3 (75) | 0.96 (0.83–1.11) | 0.886 | 0.303 |
| Unclassified_Enterobacteriaceae | 8 (100) | 9.64 (4.52–19.69) | 10 (91) | 3.89 (2.82–13.42) | 3 (100) | 62.68 (49.53–78.66) | 3 (75) | 1.39 (1.36–31.67) | 0.560 | 0.117 |
| Control Group (n = 7) | Colic Group (n = 12) | CMPA Group (n = 5) | Proctocolitis Group (n = 5) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Immune factor | n (%) | Median (IQR) | n (%) | Median (IQR) | n (%) | Median (IQR) | n (%) | Median (IQR) | p* | p** |
| Innate immunity | ||||||||||
| IL1β (ng/L) | 6 (86) | 533.20 (33.88–1528.44) | 11 (92) | 33.29 (13.25–112.96) | 3 (60) | 12.90 (9.30–102.99) | 4 (100) | 23.80 (13.85–43.85) | 0.300 | 0.593 |
| IL6 (ng/L) | 6 (86) | 10.75 (6.68–13.93) | 12 (100) | 9.32 (5.50–11.08) | 5 (100) | 5.50 (1.00–32.30) | 4 (100) | 5.50 (4.38–8.13) | 0.571 | 0.796 |
| IL12(p70) (ng/L) | 7 (100) | 60.30 (48.24–64.85) | 12 (100) | 37.35 (27.55–48.93) | 5 (100) | 32.70 (14.20–74.00) | 4 (100) | 46.45 (22.35–69.40) | 1.000 | 0.492 |
| IFNγ (ng/L) | 2 (29) | 66.60 (50.50–82.70) | 0 (0) | 1 (20) | 381.70 | 0 (0) | 0.150 | 0.221 | ||
| TNFα (ng/L) | 6 (86) | 48.80 (36.28–89.71) | 8 (67) | 38.85 (19.01–81.20) | 1 (20) | 48.80 | 3 (75) | 48.80 (48.80–81.55) | 0.152 | 0.774 |
| Acquired immunity | ||||||||||
| IL2 (ng/L) | 7 (100) | 54.80 (33.20–62.36) | 12 (100) | 36.80 (20.41–41.30) | 5 (100) | 51.20 (44.00–51.20) | 4 (100) | 58.40 (58.40–60.20) | 1.000 | 0.104 |
| IL4 (ng/L) | 1 (14) | 9.00 | 0 (0) | 0 (0) | 0 (0) | 0.571 | 0.392 | |||
| IL10 (ng/L) | 5 (71) | 34.60 (27.10–42.20) | 10 (83) | 37.15 (7.72–89.73) | 3 (60) | 937.42 (472.71–1105.06) | 3 (75) | 8.00 (5.80–11.50) | 0.811 | 0.256 |
| IL13 (ng/L) | 4 (57) | 11.68 (5.90–18.63) | 4 (33) | 5.10 (3.23–8.58) | 0 (0) | 2 (50) | 15.25 (9.33–21.18) | 0.200 | 0.621 | |
| IL17 (ng/L) | 7 (100) | 66.20 (46.15–120.01) | 12 (100) | 61.28 (36.10–72.58) | 5 (100) | 51.10 (51.10–96.80) | 4 (100) | 26.20 (15.10–48.73) | 1.000 | 0.514 |
| Chemokines | ||||||||||
| IL8 (ng/L) | 5 (71) | 14.30 (9.09–27.00) | 7 (58) | 4.80 (3.00–7.95) | 1 (20) | 4.80 | 3 (75) | 11.10 (7.95–17.45) | 0.351 | 0.407 |
| MCP1 (ng/L) | 3 (43) | 32.50 (28.59–43.15) | 2 (17) | 4.54 (2.64–6.45) | 0 (0) | 1 (25) | 7.00 | 0.377 | 0.145 | |
| MIP1β (ng/L) | 6 (86) | 22.30 (17.58–29.43) | 8 (67) | 39.19 (16.54–76.90) | 1 (20) | 381.60 | 2 (50) | 108.70 (100.55–116.85) | 0.134 | 0.279 |
| Hematopoyetic factors | ||||||||||
| IL5 (ng/L) | 7 (100) | 91.90 (86.33–166.85) | 12 (100) | 59.95 (32.54–110.85) | 5 (100) | 99.90 (84.00–99.90) | 4 (100) | 151.15 (131.40–164.88) | 1.000 | 0.301 |
| IL7 (ng/L) | 5 (71) | 86.30 (74.00–100.37) | 5 (42) | 8.60 (8.60–35.90) | 1 (20) | 35.86 | 1 (25) | 98.40 | 0.352 | 0.152 |
| GCSF (µg/L) | 7 (100) | 400.50 (109.60–575.50) | 12 (100) | 295.15 (119.29–796.05) | 3 (60) | 445.90 (378.00–869.51) | 3 (75) | 87.60 (44.60–396.50) | 0.036 | 0.564 |
| GMCSF (µg/L) | 6 (86) | 12.95 (8.55–18.99) | 8 (67) | 12.20 (3.63–18.60) | 3 (60) | 45.30 (27.30–54.13) | 3 (75) | 15.10 (12.95–17.95) | 0.824 | 0.404 |
| Control Group (n = 7) | Colic Group (n = 12) | CMPA Group (n = 5) | Proctocolitis Group (n = 5) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Immune factor | n (%) | Median (IQR) | n (%) | Median (IQR) | n (%) | Median (IQR) | n (%) | Median (IQR) | p * | p ** |
| Innate immunity | ||||||||||
| IL1β (ng/L) | 6 (86) | 0.73 (0.41–1.18) | 6 (55) | 0.78 (0.19–9.95) | 2 (67) | 0.22 (0.14–0.29) | 4 (100) | 3.85 (2.02–8.28) | 0.339 | 0.075 |
| IL6 (ng/L) | 4 (57) | 5.49 (3.66–9.07) | 5 (45) | 2.21 (2.21–8.25) | 2 (67) | 8.94 (5.58–12.31) | 4 (100) | 2.65 (1.40–4.30) | 0.579 | 0.677 |
| IL12(p70) (ng/L) | 4 (57) | 4.18 (3.50–5.39) | 5 (45) | 5.07 (3.28–7.68) | 2 (67) | 7.89 (7.57–8.21) | 3 (75) | 2.82 (2.11–4.17) | 0.875 | 0.711 |
| IFNγ (ng/L) | 1 (14) | 18.24 | 0 (0) | 0 (0) | 0 (0) | 0.586 | - | |||
| TNFα (ng/L) | 2 (29) | 5.14 (3.79–6.48) | 4 (36) | 3.55 (2.70–5.14) | 2 (67) | 6.24 (5.98–6.51) | 2 (50) | 2.44 (1.89–3.00) | 1.000 | 0.594 |
| Acquired immunity | ||||||||||
| IL2 (ng/L) | 0 (0) | 0 (0) | 2 (67) | 5.69 (4.33–7.06) | 0 (0) | 0.049 | 0.002 | |||
| IL4 (ng/L) | 3 (43) | 0.33 (0.33–0.75) | 1 (9) | 0.14 | 2 (67) | 0.69 (0.61–0.76) | 3 (75) | 0.24 (0.14–0.29) | 0.115 | 0.060 |
| IL10 (ng/L) | 4 (57) | 2.51 (2.38–4.24) | 3 (27) | 2.25 (2.12–10.12) | 3 (100) | 24.00 (13.51–29.46) | 1 (25) | 0.92 | 0.371 | 0.026 |
| IL13 (ng/L) | 6 (86) | 1.42 (1.07–2.08)a | 4 (36) | 0.62 (0.31–0.87)b | 3 (100) | 1.79 (1.08–1.79)ac | 4 (100) | 2.21 (1.45–3.09)ac | 0.126 | 0.003 |
| IL17 (ng/L) | 1 (14) | 6.81 | 0 (0) | 2 (67) | 20.19 (14.15–26.23) | 0 (0) | 0.081 | 0.012 | ||
| Chemokines | ||||||||||
| IL8 (ng/L) | 7 (100) | 73.19 (25.12–129.25) | 11 (100) | 39.95 (24.46–45.51) | 2 (67) | 49.39 (44.76–54.01) | 4 (100) | 142.55 (113.37–177.55) | 0.036 | 0.091 |
| MCP1 (ng/L) | 6 (86) | 79.61 (30.43–145.76) | 8 (73) | 150.45 (86.26–375.50) | 3 (100) | 157.92 (112.39–307.83) | 3 (75) | 39.77 (22.96–46.93) | 0.688 | 0.259 |
| MIP1β (ng/L) | 7 (100) | 14.88 (9.31–44.36) | 11 (100) | 11.96 (8.69–33.27) | 3 (100) | 15.53 (9.87–28.88) | 4 (100) | 26.20 (18.91–38.90) | 0.199 | 0.798 |
| Hematopoyetic factors | ||||||||||
| IL5 (ng/L) | 0 (0) | 0 (0) | 0 (0) | 1 (25) | 1.42 | 0.345 | 0.154 | |||
| IL7 (ng/L) | 2 (29) | 5.04 (3.17–6.90) | 2 (18) | 13.87 (7.84–19.89) | 1 (33) | 15.54 | 1 (25) | 25.24 | 0.919 | 0.949 |
| GCSF (µg/L) | 3 (43) | 17.43 (9.90–17.43) | 0 (0) | 3 (100) | 6.39 (3.56–10.88) | 2 (50) | 16.35 (14.80–17.90) | 0.012 | 0.017 | |
| GMCSF (µg/L) | 3 (43) | 35.74 (29.11–48.91) | 3 (27) | 47.33 (42.44–52.07) | 2 (67) | 582.01 (479.61–684.41) | 2 (50) | 96.66 (53.01–140.30) | 0.779 | 0.337 |
| Immunological Compound | Control Group | Colic Group | CMPA Group | Proctocolitis Group | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Milk (n/N) | Feces (n/N) | p * | Milk (n/N) | Feces (n/N) | p * | Milk (n/N) | Feces (n/N) | p * | Milk (n/N) | Feces (n/N) | p * | |
| Innate immunity | ||||||||||||
| IL1β | 6/7 | 6/7 | 1.000 | 6/11 | 11/12 | 0.069 | 2/3 | 3/5 | 1.000 | 4/4 | 4/4 | 1.000 |
| IL6 | 4/7 | 6/7 | 0.559 | 5/11 | 12/12 | 0.005 | 2/3 | 5/5 | 0.375 | 4/4 | 4/4 | 1.000 |
| IL12 | 4/7 | 7/7 | 0.192 | 5/11 | 12/12 | 0.005 | 2/3 | 5/5 | 0.375 | 3/4 | 4/4 | 1.000 |
| IFNγ | 1/7 | 2/7 | 1.000 | 0/11 | 0/12 | 1.000 | 0/3 | 1/5 | 1.000 | 0/4 | 1/4 | 0.464 |
| TNFα | 2/7 | 6/7 | 0.102 | 4/11 | 8/12 | 0.220 | 2/3 | 1/5 | 0.464 | 2/4 | 3/4 | 1.000 |
| Acquired immunity | ||||||||||||
| IL2 | 0/7 | 7/7 | 0.001 | 0/11 | 12/12 | <0.001 | 2/3 | 5/5 | 0.375 | 0/4 | 4/4 | 0.029 |
| IL4 | 3/7 | 1/7 | 0.559 | 1/11 | 0/12 | 0.478 | 2/3 | 0/5 | 0.107 | 3/4 | 0/4 | 0.143 |
| IL10 | 4/7 | 5/7 | 1.000 | 3/11 | 10/12 | 0.012 | 3/3 | 3/5 | 0.464 | 1/4 | 4/4 | 0.143 |
| IL13 | 6/7 | 4/7 | 0.559 | 4/11 | 4/12 | 1.000 | 3/3 | 0/5 | 0.018 | 4/4 | 3/4 | 1.000 |
| IL17 | 1/7 | 7/7 | 0.004 | 0/11 | 12/12 | <0.001 | 2/3 | 5/5 | 0.375 | 0/4 | 4/4 | 0.029 |
| Chemokines | ||||||||||||
| IL8 | 7/7 | 5/7 | 0.462 | 11/11 | 7/12 | 0.037 | 2/3 | 1/5 | 0.464 | 4/4 | 4/4 | 1.000 |
| MCP1 | 6/7 | 3/7 | 0.266 | 8/11 | 2/12 | 0.012 | 3/3 | 0/5 | 0.018 | 3/4 | 1/4 | 0.486 |
| MIP1β | 7/7 | 6/7 | 1.000 | 11/11 | 8/12 | 0.093 | 3/3 | 1/5 | 0.143 | 4/4 | 2/4 | 0.429 |
| Hematopoyetic factors | ||||||||||||
| IL5 | 0/7 | 7/7 | 0.001 | 0/11 | 12/12 | <0.001 | 0/3 | 5/5 | 0.018 | 1/4 | 4/4 | 0.143 |
| IL7 | 2/7 | 5/7 | 0.286 | 2/11 | 5/12 | 0.371 | 1/3 | 1/5 | 1.000 | 1/4 | 1/4 | 1.000 |
| GCSF | 3/7 | 7/7 | 0.070 | 0/11 | 12/12 | <0.001 | 3/3 | 3/5 | 0.464 | 2/4 | 3/4 | 1.000 |
| GMCSF | 3/7 | 6/7 | 0.266 | 3/11 | 8/12 | 0.100 | 2/3 | 3/5 | 1.000 | 2/4 | 3/4 | 1.000 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aparicio, M.; Alba, C.; Proctocolitis Study Group of CAM Public Health Area 6; Rodríguez, J.M.; Fernández, L. Microbiological and Immunological Markers in Milk and Infant Feces for Common Gastrointestinal Disorders: A Pilot Study. Nutrients 2020, 12, 634. https://doi.org/10.3390/nu12030634
Aparicio M, Alba C, Proctocolitis Study Group of CAM Public Health Area 6, Rodríguez JM, Fernández L. Microbiological and Immunological Markers in Milk and Infant Feces for Common Gastrointestinal Disorders: A Pilot Study. Nutrients. 2020; 12(3):634. https://doi.org/10.3390/nu12030634
Chicago/Turabian StyleAparicio, Marina, Claudio Alba, Proctocolitis Study Group of CAM Public Health Area 6, Juan Miguel Rodríguez, and Leonides Fernández. 2020. "Microbiological and Immunological Markers in Milk and Infant Feces for Common Gastrointestinal Disorders: A Pilot Study" Nutrients 12, no. 3: 634. https://doi.org/10.3390/nu12030634