Is Machine Learning a Better Way to Identify COVID-19 Patients Who Might Benefit from Hydroxychloroquine Treatment?—The IDENTIFY Trial
Abstract
:1. Introduction
2. Experimental Section
2.1. Patient Enrollment
2.2. Data Processing
2.3. Treatment
2.4. Covariates
2.5. End Point
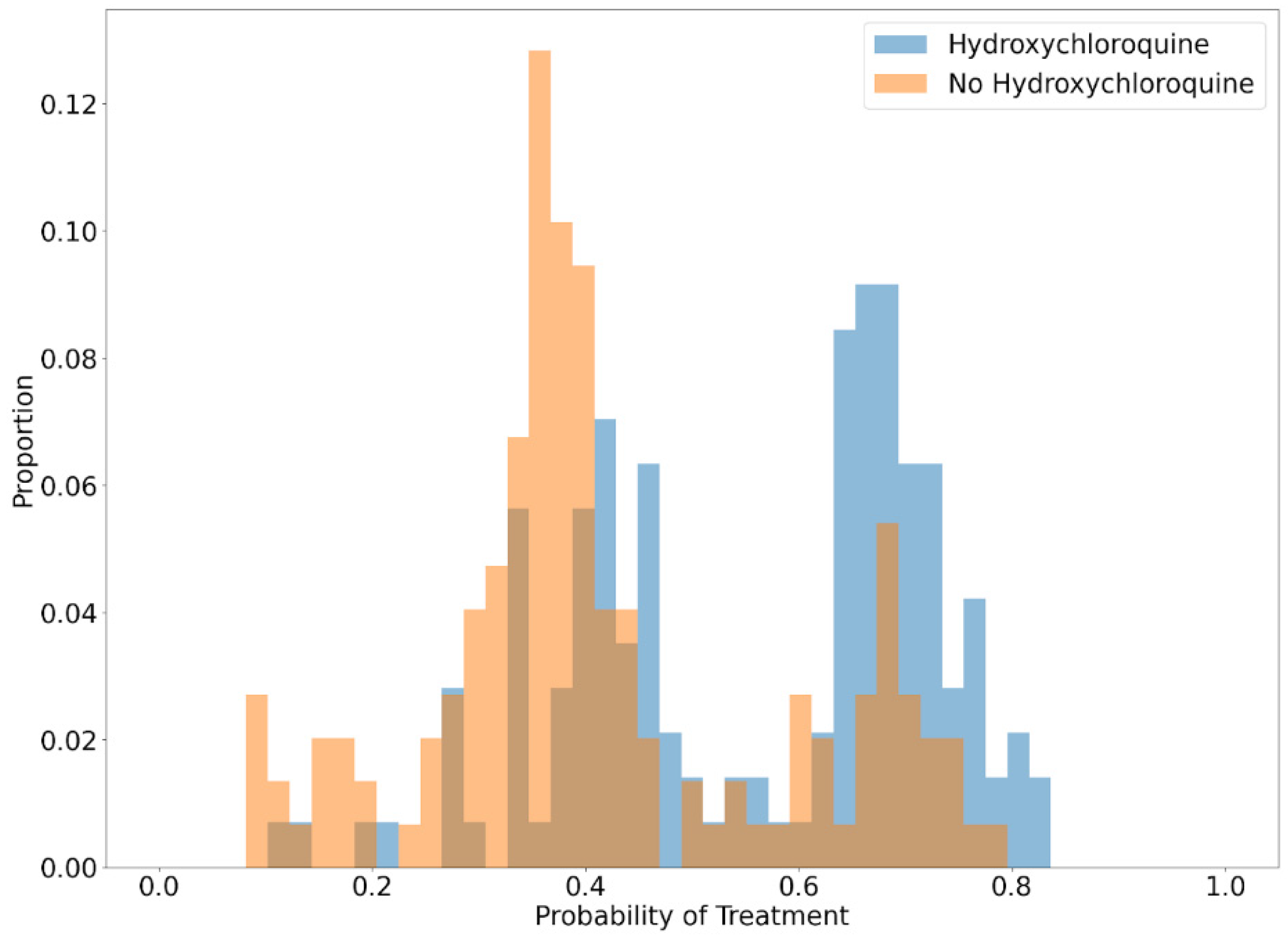
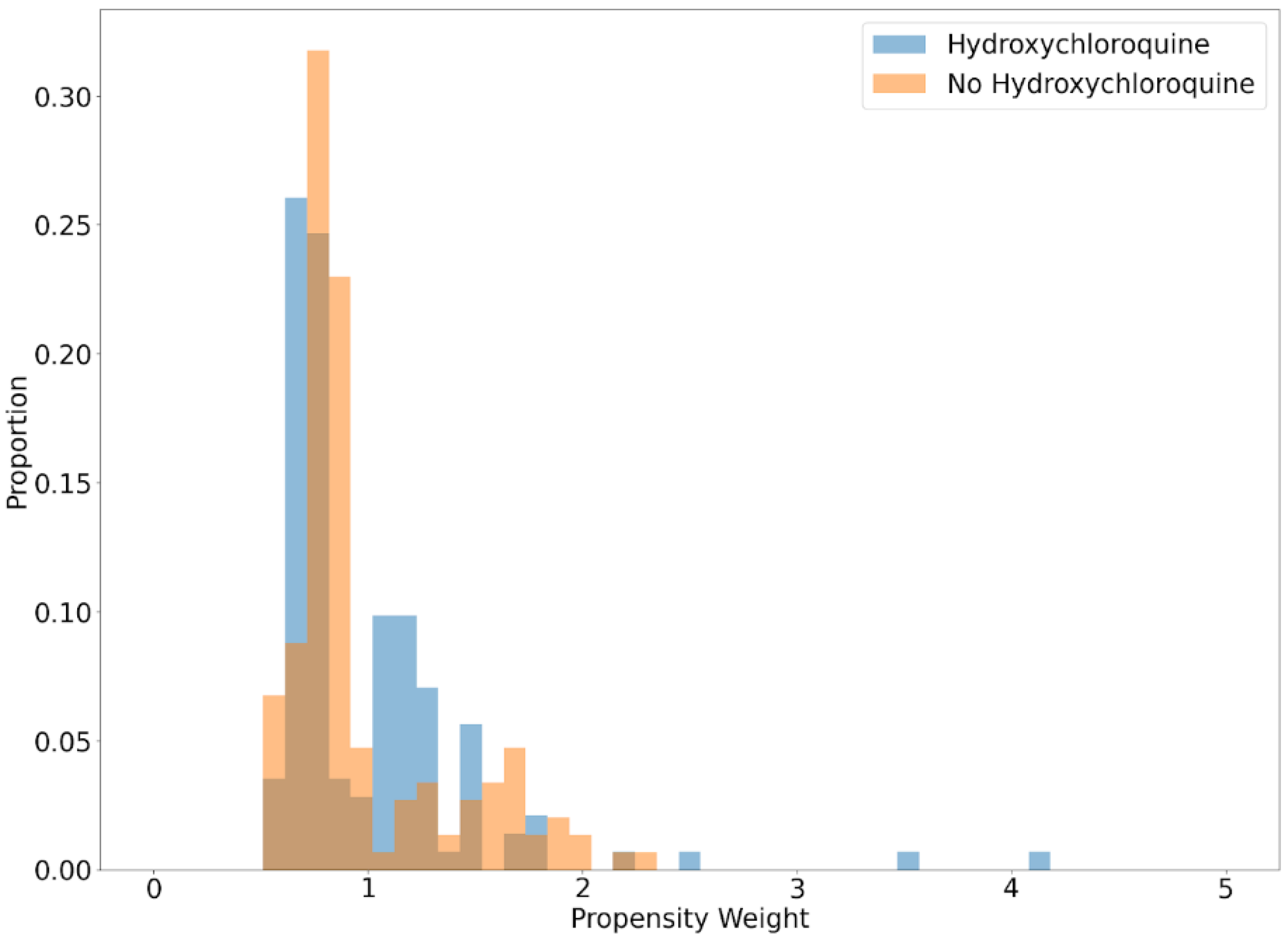
2.6. Statistical Analysis
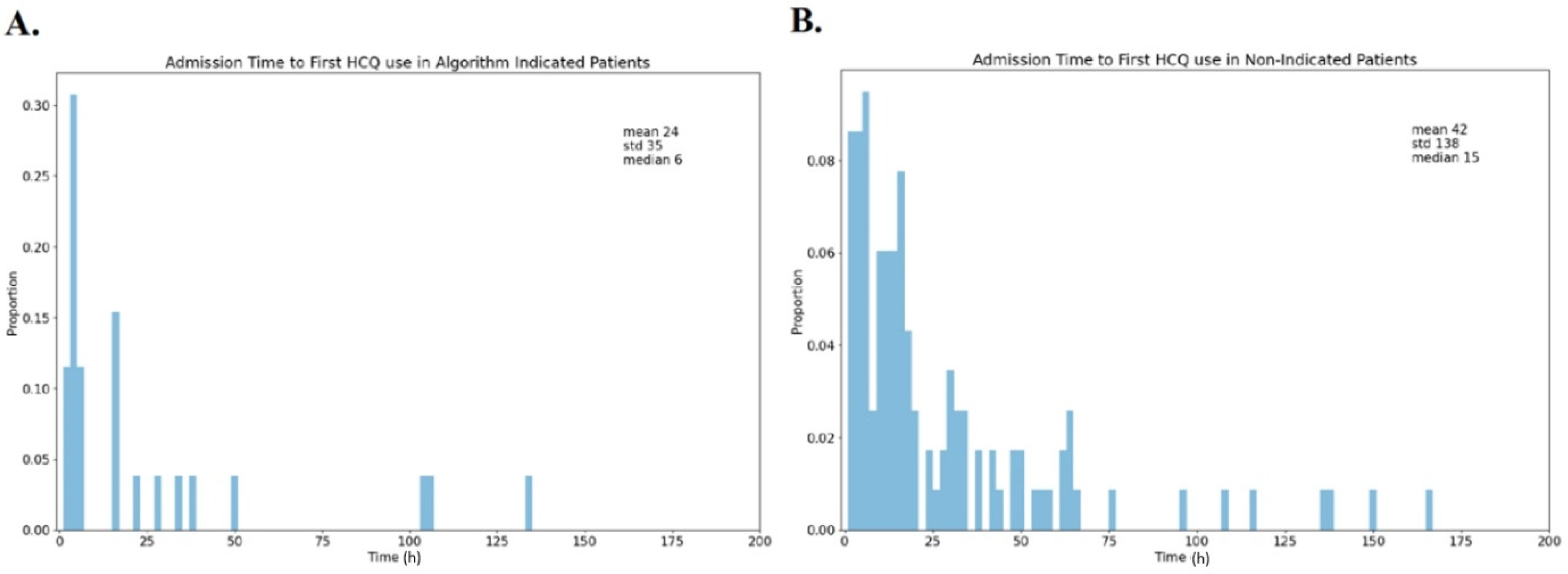
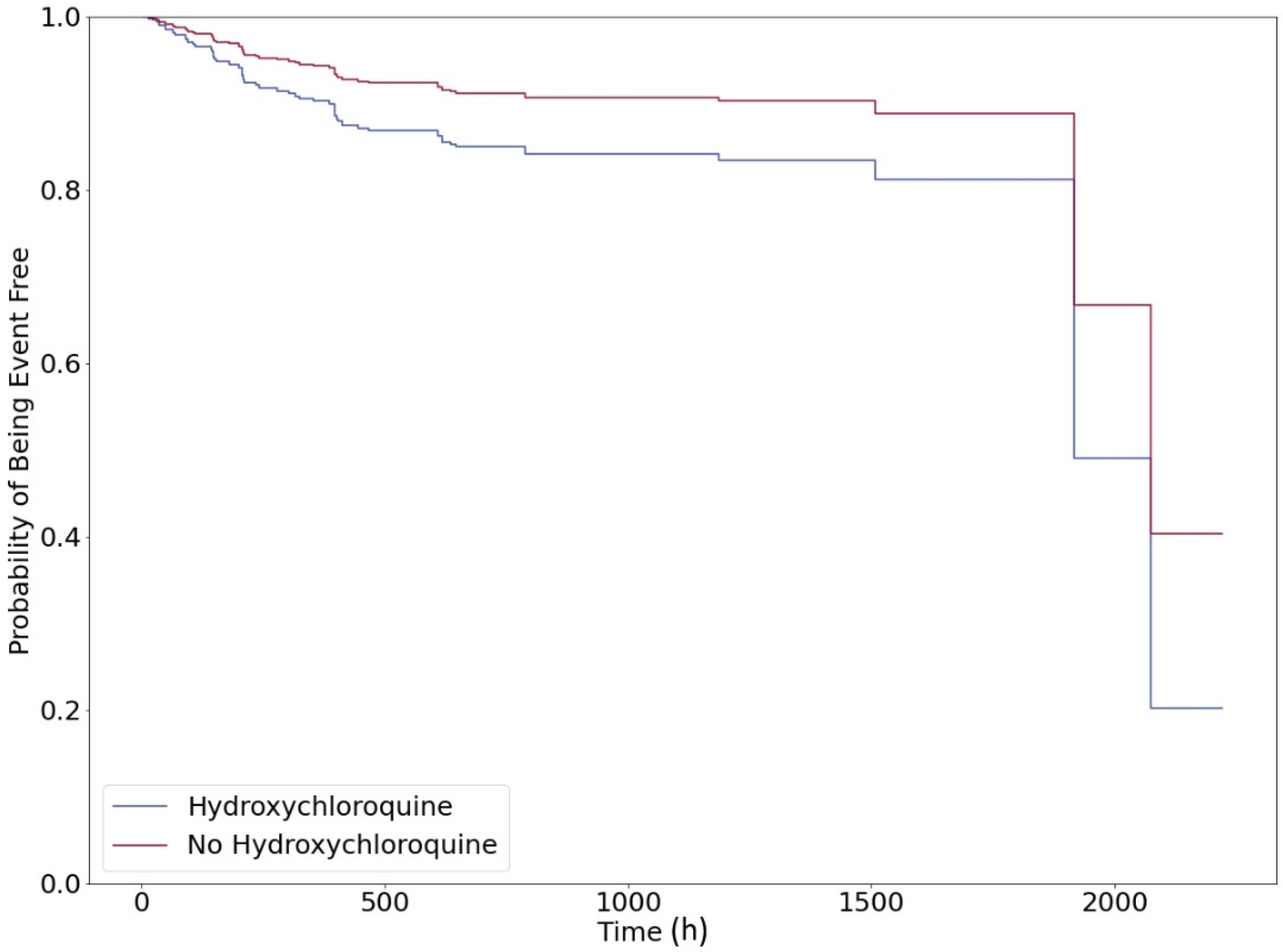
3. Results
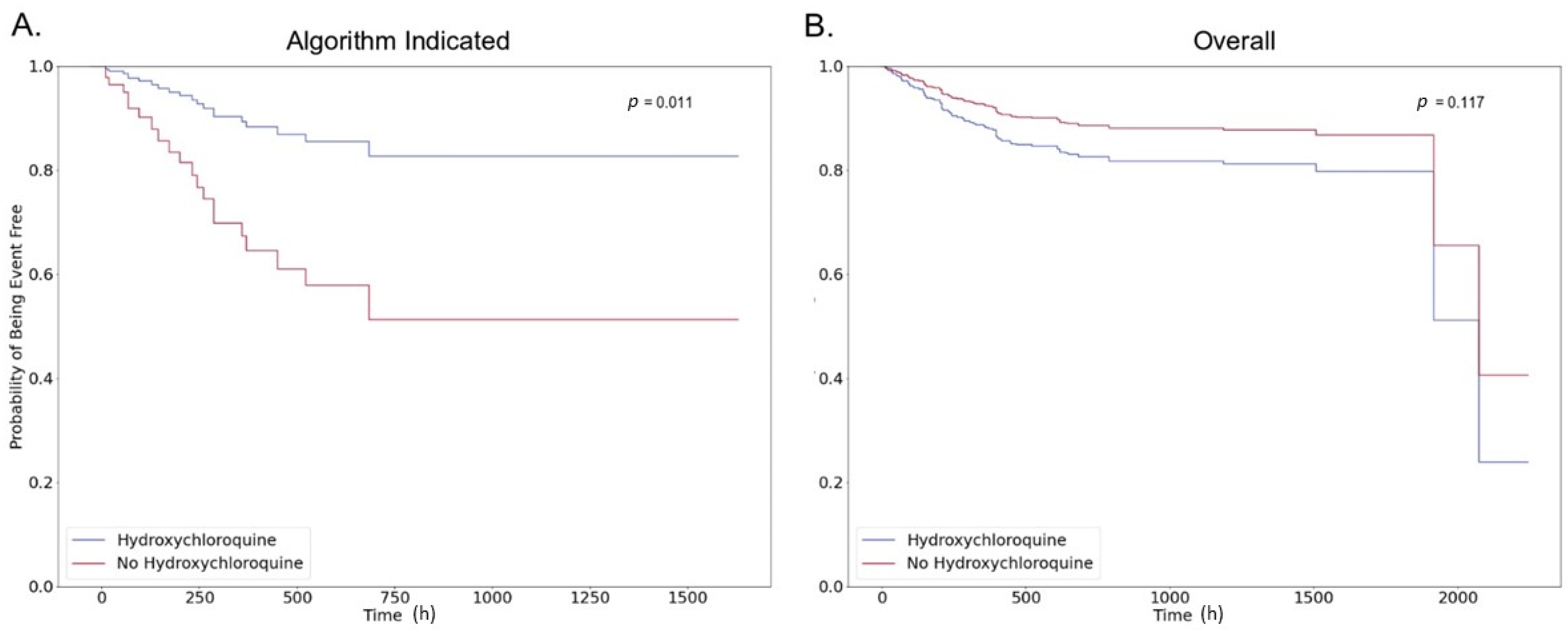
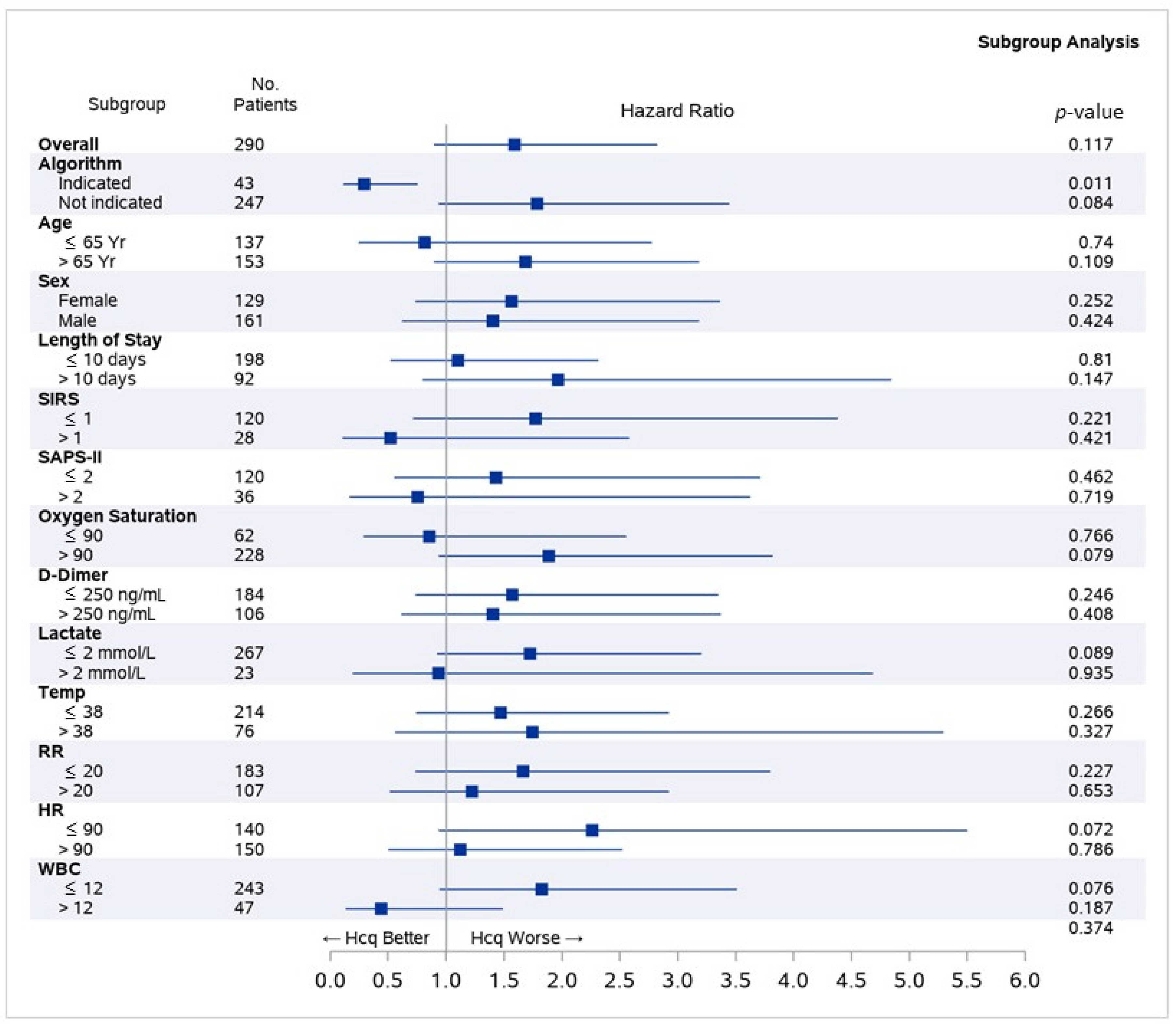
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
- Patient was admitted to the hospital as an inpatient
- Patient tested positive for COVID 19
- Patient had electronic health record data collected within four hours of receiving a COVID-19 test
| Demographics | Full Study Population | Treated with Hydroxychloroquine | Not Treated with Hydroxychloroquine | Indicated for Treatment by Algorithm | |
|---|---|---|---|---|---|
| Age | Age < 30 | 10 (3.4%) | 9 (6.3%) | 1 (0.7%) | 4 (9.3%) |
| 30–39 | 49 (16.9%) | 23 (16.2%) | 26 (17.6%) | 6 (14.0%) | |
| 50–59 | 34 (11.7%) | 21 (14.8%) | 13 (8.8%) | 3 (7.0%) | |
| 60–69 | 63 (21.7%) | 28 (19.7%) | 35 (23.6%) | 10 (23.3%) | |
| 70–79 | 70 (24.1%) | 35 (24.6%) | 35 (23.6%) | 11 (25.6%) | |
| Age > 80 | 64 (22.1%) | 26 (18.3%) | 38 (25.7%) | 9 (20.9%) | |
| Gender | Female | 129 (44.5%) | 59 (41.5%) | 70 (47.3%) | 17 (39.5%) |
| Diagnoses | Initial O2 Sat | 93.52 (5.52) | 92.96 (5.45) | 94.07 (5.52) | 89.16 (7.3) |
| Sepsis | 15 (5.2%) | 10 (7.0%) | 5 (3.4%) | 6 (14.0%) | |
| ARDS | 37 (12.8%) | 21 (14.8%) | 16 (10.8%) | 9 (20.9%) | |
| Pneumonia | 40 (13.8%) | 30 (21.1%) | 10 (6.8%) | 12 (27.9%) | |
| AKI | 26 (9.0%) | 13 (9.2%) | 13 (8.8%) | 5 (11.6%) | |
| Arrhythmia | 1 (0.3%) | 0 (0.0%) | 1 (0.7%) | 1 (2.3%) | |
| Medications | Remdesivir | 16 (5.5%) | 5 (3.5%) | 11 (7.4%) | 3 (7.0%) |
| Macrolide | 130 (44.8%) | 85 (59.9%) | 45 (30.4%) | 22 (51.2%) | |
| Hydroxy-chloroquine | 142 (49.0%) | 142 (100.0%) | 0 (0.0%) | 26 (60.5%) | |
| ARB | 22 (7.6%) | 7 (4.9%) | 15 (10.1%) | 2 (4.7%) | |
| ACEI | 26 (9.0%) | 16 (11.3%) | 10 (6.8%) | 1 (2.3%) | |
| NSAID | 72 (24.8%) | 35 (24.6%) | 37 (25.0%) | 9 (20.9%) | |
| Steroids | 85 (29.3%) | 52 (36.6%) | 33 (22.3%) | 16 (37.2%) | |
| History | Cardio | 41 (14.1%) | 11 (7.7%) | 30 (20.3%) | 2 (4.7%) |
| Renal | 5 (1.7%) | 4 (2.8%) | 1 (0.7%) | 0 (0.0%) | |
| Hepatic | 5 (1.7%) | 3 (2.1%) | 2 (1.4%) | 0 (0.0%) | |
| Diabetes | 27 (9.3%) | 9 (6.3%) | 18 (12.2%) | 1 (2.3%) | |
| Organ Transplant | 1 (0.3%) | 1 (0.7%) | 0 (0.0%) | 0 (0.0%) | |
| HIV | 1 (0.3%) | 0 (0.0%) | 1 (0.7%) | 0 (0.0%) | |
| Psych | 21 (7.2%) | 8 (5.6%) | 13 (8.8%) | 0 (0.0%) | |
| COPD | 5 (1.7%) | 2 (1.4%) | 3 (2.0%) | 0 (0.0%) | |
| Cancer | 32 (11.0%) | 15 (10.6%) | 17 (11.5%) | 1 (2.3%) | |
| ETOH | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | |
| PNA | 63 (21.7%) | 31 (21.8%) | 32 (21.6%) | 9 (20.9%) |
| Algorithm Indicated | Not Algorithm Indicated | |||
|---|---|---|---|---|
| Treated | Untreated | Treated | Untreated | |
| Hospital Length of Stay | 374.6 (288.1) | 147.2 (170.7) | 256.2 (268.7) | 229.1 (344.9) |
| Mechanical Ventilation | 14 (53.8%) | 6 (35.3%) | 29 (25.0) | 19 (14.5%) |
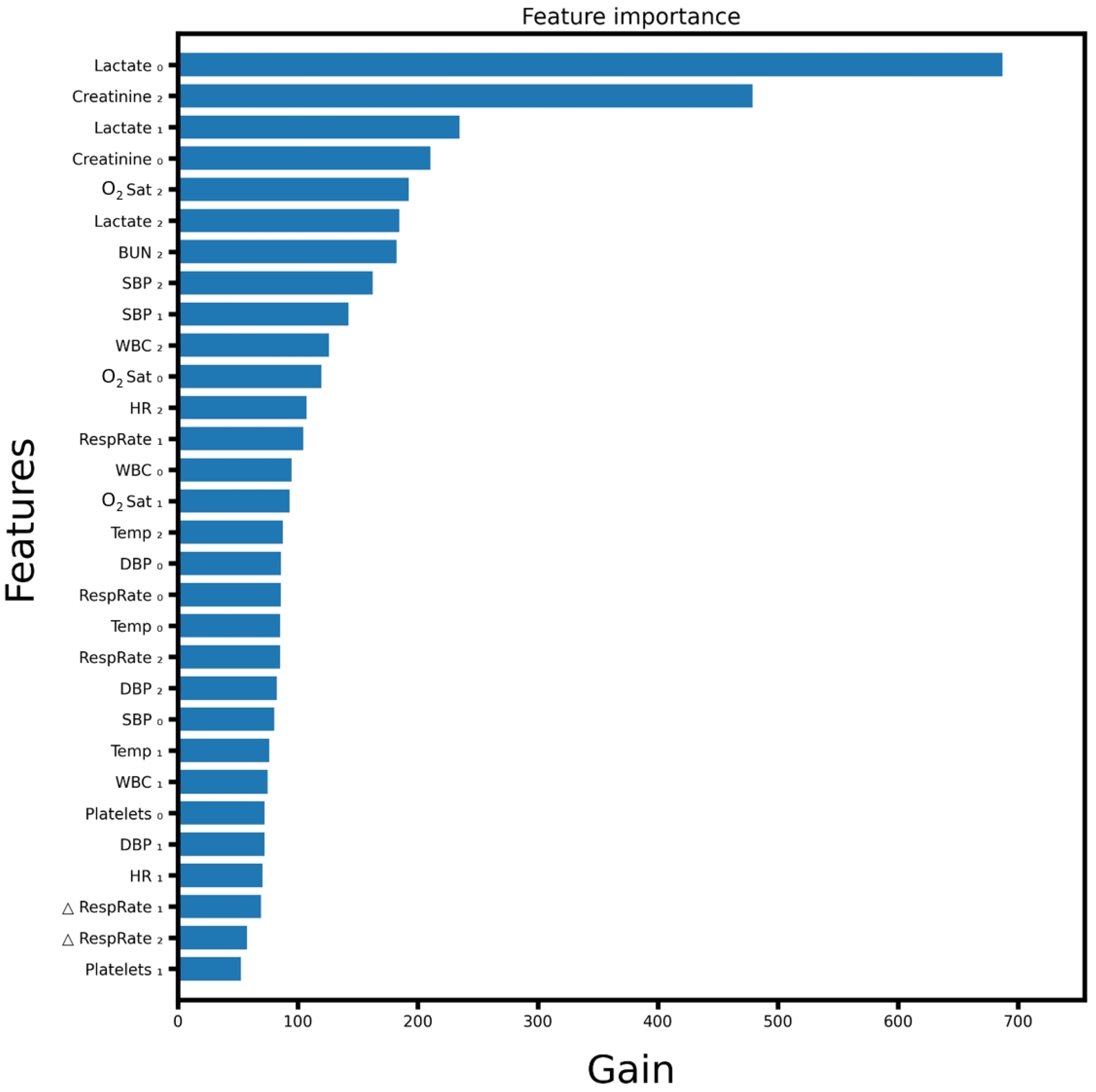
References
- Lai, C.-C.; Shih, T.-P.; Ko, W.-C.; Tang, H.-J.; Hsueh, P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents 2020, 55, 105924. [Google Scholar] [CrossRef] [PubMed]
- Devaux, C.A.; Rolain, J.-M.; Colson, P.; Raoult, D. New insights on the antiviral effects of chloroquine against coronavirus: What to expect for COVID-19? Int. J. Antimicrob. Agents 2020, 55, 105938. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Wang, Y.; Wen, D.; Liu, W.; Wang, J.; Fan, G.; Ruan, L.; Song, B.; Cai, Y.; Wei, M.; et al. A Trial of Lopinavir–Ritonavir in Adults Hospitalized with Severe Covid-19. New Engl. J. Med. 2020, 382, 1787–1799. [Google Scholar] [CrossRef] [PubMed]
- Borba, M.G.S.; Val, F.F.A.; Sampaio, V.S.; Alexandre, M.A.A.; Melo, G.C.; Brito, M.; Mourão, M.P.G.; Brito-Sousa, J.D.; Baía-da-Silva, D.; Guerra, M.V.F.; et al. Effect of High vs Low Doses of Chloroquine Diphosphate as Adjunctive Therapy for Patients Hospitalized with Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection: A Randomized Clinical Trial. JAMA Netw. Open 2020, 24, e208857. [Google Scholar] [CrossRef] [Green Version]
- Gautret, P.; Lagier, J.C.; Parola, P.; Hoang, V.T.; Meddeb, L.; Mailhe, M.; Doudier, B.; Courjon, J.; Giordanengo, V.; Vieira, V.E.; et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: Results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents 2020, 56, 105949. [Google Scholar] [CrossRef]
- Cortegiani, A.; Ingoglia, G.; Ippolito, M.; Giarratano, A.; Einav, S. A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J. Crit. Care 2020, 57, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Geleris, J.; Sun, Y.; Platt, J.; Zucker, J.; Baldwin, M.; Hripcsak, G.; Labella, A.; Manson, D.K.; Kubin, C.; Barr, R.G.; et al. Observational Study of Hydroxychloroquine in Hospitalized Patients with Covid-19. N. Engl. J. Med. 2020, 382, 2411–2418. [Google Scholar] [CrossRef] [PubMed]
- Biot, C.; Daher, W.; Chavain, N.; Fandeur, T.; Khalife, J.; Dive, D.; De Clercq, E. Design and Synthesis of Hydroxyferroquine Derivatives with Antimalarial and Antiviral Activities. J. Med. Chem. 2006, 49, 2845–2849. [Google Scholar] [CrossRef] [Green Version]
- Yao, X.; Ye, F.; Zhang, M.; Cui, C.; Huang, B.; Niu, P.; Liu, X.; Zhao, L.; Dong, E.; Song, C.; et al. In Vitro Antiviral Activity and Projection of Optimized Dosing Design of Hydroxychloroquine for the Treatment of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis. 2020, 71, 732–739. [Google Scholar] [CrossRef] [Green Version]
- Molina, J.; Delaugerre, C.; Le Goff, J.; Mela-Lima, B.; Ponscarme, D.; Goldwirt, L.; De Castro, N. No evidence of rapid antiviral clearance or clinical benefit with the combination of hydroxychloroquine and azithromycin in patients with severe COVID-19 infection. Méd. Mal. Infect. 2020, 50, 384. [Google Scholar] [CrossRef]
- Horby, P.; Mafham, M.M.; Linsell, L.; Bell, J.L.; Staplin, N.; Emberson, J.; Wiselka, M.; Ustianowski, A.; Elmahi, E.; Prudon, B.; et al. Effect of Hydroxychloroquine in Hospitalized Patients with COVID-19: Preliminary results from a multi-centre, randomized, controlled trial. medRxiv 2020. pre-print. [Google Scholar] [CrossRef]
- Sarma, P.; Kaur, H.; Kumar, H.; Mahendru, D.; Avti, P.; Bhattacharyya, A.; Prajapat, M.; Shekhar, N.; Kumar, S.; Singh, R.; et al. Virological and clinical cure in COVID-19 patients treated with hydroxychloroquine: A systematic review and meta-analysis. J. Med Virol. 2020, 92, 776–785. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Singh, A.; Singh, R.; Misra, A. Hydroxychloroquine in patients with COVID-19: A Systematic Review and meta-analysis. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Ayerbe, L.; Risco-Risco, C.; Ayis, S. The association of treatment with hydroxychloroquine and hospital mortality in COVID-19 patients. Intern. Emerg. Med. 2020, 15, 1501–1506. [Google Scholar] [CrossRef]
- Arshad, S.; Kilgore, P.; Chaudhry, Z.S.; Jacobsen, G.; Wang, D.D.; Huitsing, K.; Brar, I.; Alangaden, G.J.; Ramesh, M.S.; McKinnon, J.E.; et al. Treatment with hydroxychloroquine, azithromycin, and combination in patients hospitalized with COVID. Int. J. Infect. Dis. 2020, 97, 396–403. [Google Scholar] [CrossRef]
- Catteau, L.; Dauby, N.; Montourcy, M.; Bottieau, E.; Hautekiet, J.; Goetghebeur, E.; Van Ierssel, S.; Duysburgh, E.; Van Oyen, H.; Wyndham-Thomas, C.; et al. Low-dose hydroxychloroquine therapy and mortality in hospitalised patients with COVID-19: A nationwide observational study of 8075 participants. Int. J. Antimicrob. Agents 2020, 56, 106144. [Google Scholar] [CrossRef]
- Di Castelnuovo, A.; Costanzo, S.; Antinori, A.; Berselli, N.; Blandi, L.; Bruno, R.; Cauda, R.; Guaraldi, G.; Menicanti, L.; My, I.; et al. Use of hydroxychloroquine in hospitalised COVID-19 patients is associated with reduced mortality: Findings from the observational multicentre Italian CORIST study. Eur. J. Intern. Med. 2020, in press. [Google Scholar] [CrossRef]
- VanderWeele, T.J.; Ding, P. Sensitivity Analysis in Observational Research: Introducing the E-Value. Ann. Intern. Med. 2017, 167, 268–274. [Google Scholar] [CrossRef]
- Gentry, C.A.; Humphrey, M.B.; Thind, S.K.; Hendrickson, S.C.; Kurdgelashvili, G.; Williams, R.J. Long-term hydroxychloroquine use in patients with rheumatic conditions and development of SARS-CoV-2 infection: A retrospective cohort study. Lancet Rheumatol. 2020, 2, e689–e697. [Google Scholar] [CrossRef]
- Rosenberg, E.S.; Dufort, E.M.; Udo, T.; Wilberschied, L.A.; Kumar, J.; Tesoriero, J.; Weinberg, P.; Kirkwood, J.; Muse, A.; DeHovitz, J.; et al. Association of Treatment With Hydroxychloroquine or Azithromycin With In-Hospital Mortality in Patients With COVID-19 in New York State. JAMA 2020, 323, 2493–2502. [Google Scholar] [CrossRef]
- Azoulay, E.; Fartoukh, M.; Darmon, M.; Géri, G.; Voiriot, G.; Dupont, T.; Zafrani, L.; Girodias, L.; Labbé, V.; Dres, M.; et al. Increased mortality in patients with severe SARS-CoV-2 infection admitted within seven days of disease onset. Intensiv. Care Med. 2020, 46, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Thémans, P.; Belkhir, L.; Dauby, N.; Yombi, J.-C.; De Greef, J.; Delongie, K.-A.; Vandeputte, M.; Nasreddine, R.; Wittebole, X.; Wuillaume, F.; et al. Population Pharmacokinetics of Hydroxychloroquine in COVID-19 Patients: Implications for Dose Optimization. Eur. J. Drug Metab. Pharmacokinet. 2020, 45, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Collins, F.S.; Varmus, H. A New Initiative on Precision Medicine. New Engl. J. Med. 2015, 372, 793–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letai, A. Functional precision cancer medicine—Moving beyond pure genomics. Nat. Med. 2017, 23, 1028–1035. [Google Scholar] [CrossRef]
- Voss, M.H.; Hakimi, A.A.; Pham, C.G.; Brannon, A.R.; Chen, Y.-B.; Cunha, L.F.; Akin, O.; Liu, H.; Takeda, S.; Scott, S.N.; et al. Tumor Genetic Analyses of Patients with Metastatic Renal Cell Carcinoma and Extended Benefit from mTOR Inhibitor Therapy. Clin. Cancer Res. 2014, 20, 1955–1964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagle, N.; Grabiner, B.C.; Van Allen, E.M.; Hodis, E.; Jacobus, S.; Supko, J.G.; Stewart, M.; Choueiri, T.K.; Gandhi, L.; Cleary, J.M.; et al. Activating mTOR Mutations in a Patient with an Extraordinary Response on a Phase I Trial of Everolimus and Pazopanib. Cancer Discov. 2014, 4, 546–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iyer, G.; Hanrahan, A.J.; Milowsky, M.I.; Al-Ahmadie, H.; Scott, S.N.; Janakiraman, M.; Pirun, M.; Sander, C.; Socci, N.D.; Ostrovnaya, I.; et al. Genome Sequencing Identifies a Basis for Everolimus Sensitivity. Science 2012, 338, 221. [Google Scholar] [CrossRef] [Green Version]
- De Bono, J.S.; Ashworth, A. Translating cancer research into targeted therapeutics. Nat. Cell Biol. 2010, 467, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Aronson, S.J.; Rehm, H.L. Building the foundation for genomics in precision medicine. Nat. Cell Biol. 2015, 526, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Office-Based Physician Electronic Health Record Adoption. Available online: https://dashboard.healthit.gov/quickstats/pages/physician-ehr-adoption-trends.php (accessed on 16 November 2020).
- Tunis, S.R.; Stryer, D.B.; Clancy, C.M. Practical Clinical Trials. JAMA 2003, 290, 1624–1632. [Google Scholar] [CrossRef]
- Fine, J.P.; Gray, R.J. A Proportional Hazards Model for the Subdistribution of a Competing Risk. J. Am. Stat. Assoc. 1999, 94, 496–509. [Google Scholar] [CrossRef]
- Taccone, F.S.; Gorham, J.; Vincent, J.-L. Hydroxychloroquine in the management of critically ill patients with COVID-19: The need for an evidence base. Lancet Respir. Med. 2020, 8, 539–541. [Google Scholar] [CrossRef]
- Schrezenmeier, E.; Dörner, T. Mechanisms of action of hydroxychloroquine and chloroquine: Implications for rheumatology. Nat. Rev. Rheumatol. 2020, 16, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Dauby, N.; Bottieau, E. The unfinished story of hydroxychloroquine in COVID-19: The right anti-inflammatory dose at the right moment? Int. J. Infect. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Lammers, A.; Brohet, R.; Theunissen, R.; Koster, C.; Rood, R.; Verhagen, D.; Brinkman, K.; Hassing, R.; Dofferhoff, A.; El Moussaoui, R.; et al. Early hydroxychloroquine but not chloroquine use reduces ICU admission in COVID-19 patients. Int. J. Infect. Dis. 2020, 101, 283–289. [Google Scholar] [CrossRef]
- NIH Begins Clinical Trial of Hydroxychloroquine and Azithromycin to Treat COVID-19. National Institutes of Health (NIH), 2020. Available online: https://www.nih.gov/news-events/news-releases/nih-begins-clinical-trial-hydroxychloroquine-azithromycin-treat-covid-19 (accessed on 16 November 2020).
- BULLETIN—NIH Clinical Trial Evaluating Hydroxychloroquine and Azithromycin for COVID-19 Closes Early|NIH: National Institute of Allergy and Infectious Diseases. Available online: http://www.niaid.nih.gov/news-events/bulletin-nih-clinical-trial-evaluating-hydroxychloroquine-and-azithromycin-covid-19 (accessed on 3 September 2020).
- NIH Halts Clinical Trial of Hydroxychloroquine. National Institutes of Health (NIH), 2020. Available online: https://www.nih.gov/news-events/news-releases/nih-halts-clinical-trial-hydroxychloroquine (accessed on 3 September 2020).
- Hernandez, A.V.; Roman, Y.M.; Pasupuleti, V.; Barboza-Meca, J.; White, C.M. Update Alert 2: Hydroxychloroquine or Chloroquine for the Treatment or Prophylaxis of COVID. Ann. Intern. Med. 2020, 173, W128–W129. [Google Scholar] [CrossRef] [PubMed]
- Skipper, C.P.; Pastick, K.A.; Engen, N.W.; Bangdiwala, A.S.; Abassi, M.; Lofgren, S.M.; Williams, D.A.; Okafor, E.C.; Pullen, M.F.; Nicol, M.R.; et al. Hydroxychloroquine in Nonhospitalized Adults With Early COVID. Ann. Intern. Med. 2020, 173, 623–631. [Google Scholar] [CrossRef]
- Cavalcanti, A.B.; Zampieri, F.G.; Rosa, R.G.; Azevedo, L.C.; Veiga, V.C.; Avezum, A.; Damiani, L.P.; Marcadenti, A.; Kawano-Dourado, L.; Lisboa, T.; et al. Hydroxychloroquine with or without Azithromycin in Mild-to-Moderate Covid-19. N. Engl. J. Med. 2020, 383, 2041–2052. [Google Scholar] [CrossRef]
- Global COVID-19 Prevention Trial of Hydroxychloroquine to Resume. Medscape. Available online: http://www.medscape.com/viewarticle/933174. (accessed on 1 July 2020).
- Boulware, D.R.; Pullen, M.F.; Bangdiwala, A.S.; Pastick, K.A.; Lofgren, S.M.; Okafor, E.C.; Skipper, C.P.; Nascene, A.A.; Nicol, M.R.; Abassi, M.; et al. A Randomized Trial of Hydroxychloroquine as Postexposure Prophylaxis for Covid-19. New Engl. J. Med. 2020, 383, 517–525. [Google Scholar] [CrossRef]
- WHO Solidarity Trial Consortium; Pan, H.; Peto, R.; Karim, Q.A.; Alejandria, M.; Henao-Restrepo, A.M.; García, C.H.; Kieny, M.-P.; Malekzadeh, R.; Murthy, S.; et al. Repurposed antiviral drugs for COVID-19—Interim WHO SOLIDARITY trial results. medRxiv 2020. pre-print. [Google Scholar] [CrossRef]
- Ferner, R.E.; Aronson, J.K. Chloroquine and hydroxychloroquine in Covid-19. BMJ 2020, 369, m1432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alia, E.; Grant-Kels, J.M. Does hydroxychloroquine combat COVID-19? A timeline of evidence. J. Am. Acad. Dermatol. 2020, 83, e33–e34. [Google Scholar] [CrossRef] [PubMed]
- Yazdany, J.; Kim, A.H. Use of Hydroxychloroquine and Chloroquine During the COVID-19 Pandemic: What Every Clinician Should Know. Ann. Intern. Med. 2020, 172, 754–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, D.-G.; Shin, H.-J.; Kim, M.-H.; Lee, S.; Kim, H.-S.; Myoung, J.; Kim, B.-T.; Kim, S.-J. Current Status of Epidemiology, Diagnosis, Therapeutics, and Vaccines for Novel Coronavirus Disease 2019 (COVID-19). J. Microbiol. Biotechnol. 2020, 30, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Richardson, S.; Hirsch, J.S.; Narasimhan, M.; Crawford, J.M.; McGinn, T.; Davidson, K.W. Presenting Characteristics, Comorbidities, and Outcomes Among 5700 Patients Hospitalized With COVID-19 in the New York City Area. JAMA 2020, 323, 2052–2059. [Google Scholar] [CrossRef]
- Aboughdir, M.; Kirwin, T.; Khader, A.A.; Wang, B. Prognostic Value of Cardiovascular Biomarkers in COVID-19: A Review. Viruses 2020, 12, 527. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.J.; Peltan, I.D.; Jensen, P.; Hoda, D.; Hunter, B.; Silver, A.; Starr, N.; Buckel, W.; Grisel, N.; Hummel, E.; et al. Clinical criteria for COVID-19-associated hyperinflammatory syndrome: A cohort study. Lancet Rheumatol. 2020, in press. [Google Scholar] [CrossRef]
- Mahévas, M.; Tran, V.-T.; Roumier, M.; Chabrol, A.; Paule, R.; Guillaud, C.; Fois, E.; Lepeule, R.; Szwebel, T.-A.; Lescure, F.-X.; et al. Clinical efficacy of hydroxychloroquine in patients with covid-19 pneumonia who require oxygen: Observational comparative study using routine care data. BMJ 2020, 369, m1844. [Google Scholar] [CrossRef]
- Menden, M.P.; Iorio, F.; Garnett, M.; McDermott, U.; Benes, C.H.; Ballester, P.J.; Saez-Rodriguez, J. Machine Learning Prediction of Cancer Cell Sensitivity to Drugs Based on Genomic and Chemical Properties. PLOS ONE 2013, 8, e61318. [Google Scholar] [CrossRef] [Green Version]
- Pintilie, M. Analysing and interpreting competing risk data. Stat. Med. 2007, 26, 1360–1367. [Google Scholar] [CrossRef]

| Demographics | Full Study Population | Treated with HCQ | Not Treated with HCQ | Indicated for Treatment by Algorithm | |
|---|---|---|---|---|---|
| Age | Age < 30 | 10 (3.4%) | 9 (6.3%) | 1 (0.7%) | 4 (9.3%) |
| 30–39 | 49 (16.9%) | 23 (16.2%) | 26 (17.6%) | 6 (14.0%) | |
| 50–59 | 34 (11.7%) | 21 (14.8%) | 13 (8.8%) | 3 (7.0%) | |
| 60–69 | 63 (21.7%) | 28 (19.7%) | 35 (23.6%) | 10 (23.3%) | |
| 70–79 | 70 (24.1%) | 35 (24.6%) | 35 (23.6%) | 11 (25.6%) | |
| Age > 80 | 64 (22.1%) | 26 (18.3%) | 38 (25.7%) | 9 (20.9%) | |
| Gender | Female | 129 (44.5%) | 59 (41.5%) | 70 (47.3%) | 17 (39.5%) |
| In Hospital Conditions | Average Initial O2 Sat * | 93.52 (5.52) | 92.96 (5.45) | 94.07(5.52) | 89.16(7.3) |
| Sepsis +,* | 15 (5.2%) | 10 (7.0%) | 5(3.4%) | 6(14.0%) | |
| ARDS + | 37 (12.8%) | 21 (14.8%) | 16(10.8%) | 9(20.9%) | |
| Pneumonia + | 40 (13.8%) | 30 (21.1%) | 10(6.8%) | 12(27.9%) | |
| AKI + | 26 (9.0%) | 13 (9.2%) | 13(8.8%) | 5 (11.6%) | |
| Arrhythmia + | 1 (0.3%) | 0 (0.0%) | 1 (0.7%) | 1(2.3%) | |
| Medications | Remdesivir | 16 (5.5%) | 5 (3.5%) | 11 (7.4%) | 3 (7.0%) |
| Macrolide | 130 (44.8%) | 85 (59.9%) | 45 (30.4%) | 22 (51.2%) | |
| ARB | 22 (7.6%) | 7 (4.9%) | 15 (10.1%) | 2 (4.7%) | |
| ACEI | 26 (9.0%) | 16 (11.3%) | 10 (6.8%) | 1 (2.3%) | |
| NSAID | 72 (24.8%) | 35 (24.6%) | 37 (25.0%) | 9 (20.9%) | |
| Hcq | 142 (49.0%) | 142 (100.0%) | 0 (0.0%) | 26 (60.5%) | |
| Steroids | 85 (29.3%) | 52 (36.6%) | 33 (22.3%) | 16 (37.2%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burdick, H.; Lam, C.; Mataraso, S.; Siefkas, A.; Braden, G.; Dellinger, R.P.; McCoy, A.; Vincent, J.-L.; Green-Saxena, A.; Barnes, G.; et al. Is Machine Learning a Better Way to Identify COVID-19 Patients Who Might Benefit from Hydroxychloroquine Treatment?—The IDENTIFY Trial. J. Clin. Med. 2020, 9, 3834. https://doi.org/10.3390/jcm9123834
Burdick H, Lam C, Mataraso S, Siefkas A, Braden G, Dellinger RP, McCoy A, Vincent J-L, Green-Saxena A, Barnes G, et al. Is Machine Learning a Better Way to Identify COVID-19 Patients Who Might Benefit from Hydroxychloroquine Treatment?—The IDENTIFY Trial. Journal of Clinical Medicine. 2020; 9(12):3834. https://doi.org/10.3390/jcm9123834
Chicago/Turabian StyleBurdick, Hoyt, Carson Lam, Samson Mataraso, Anna Siefkas, Gregory Braden, R. Phillip Dellinger, Andrea McCoy, Jean-Louis Vincent, Abigail Green-Saxena, Gina Barnes, and et al. 2020. "Is Machine Learning a Better Way to Identify COVID-19 Patients Who Might Benefit from Hydroxychloroquine Treatment?—The IDENTIFY Trial" Journal of Clinical Medicine 9, no. 12: 3834. https://doi.org/10.3390/jcm9123834





