Evolving Escherichia coli Host Strains for Efficient Deuterium Labeling of Recombinant Proteins Using Sodium Pyruvate-d3
Abstract
:1. Introduction
2. Results and Discussion
2.1. Adaptation of E. coli for Growth in Minimal Pyruvate Medium
2.2. Observed Mutations in the Evolved Strains
2.3. Deuteration of Pyruvate
2.4. Production of Perdeuterated Proteins in the Pyruvate Adapted Strain
3. Materials and Methods
3.1. Strains and Growth Conditions
3.2. Sodium Pyruvate-d3 Production
3.3. In Situ Deuteration of Pyruvic Acid
3.4. Growth Experiments
3.5. ALE and Strain Isolation
3.6. Whole-Genome DNA Sequencing
3.7. Lysogenization
3.8. Deletion of ompF
3.9. Perdeuteration of TIM
3.10. GFP and OmpF Expression
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AHAS II | Acetohydroxyacid synthase II |
| ALE | Adaptive laboratory evolution |
| EDTA | Ethylenediaminetetraacetic acid |
| IPTG | Isopropyl β-D-1-thiogalactopyranoside |
| NMR | Nuclear magnetic resonance |
| OmpF | Outer membrane protein F |
| SANS | Small-angle neutron scattering |
| TIM | Triose-phosphate isomerase from Leishmania mexicana |
| H-M9 | M9 minimal medium with pyruvate as carbon source. |
| D-M9 | M9 minimal medium made up with D2O and pyruvate as carbon source. |
| DD-M9 | M9 minimal medium made up with D2O and deuterated pyruvate as carbon source. |
References
- Berglund, M.; Wieser, M.E. Isotopic compositions of the elements 2009 (IUPAC Technical Report). Pure Appl. Chem. 2011, 83, 397–410. [Google Scholar] [CrossRef] [Green Version]
- Zaccai, N.R.; Sandlin, C.W.; Hoopes, J.T.; Curtis, J.E.; Fleming, P.J.; Fleming, K.G.; Krueger, S. Deuterium labeling together with contrast variation small-angle neutron scattering suggests how Skp captures and releases unfolded outer membrane proteins. Methods Enzymol. 2016, 566, 159–210. [Google Scholar]
- Yu, H. Extending the size limit of protein nuclear magnetic resonance. Proc. Natl. Acad. Sci. USA 1999, 96, 332–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oksanen, E.; Chen, J.C.; Fisher, S.Z. Neutron crystallography for the study of hydrogen bonds in macromolecules. Molecules 2017, 22, 596. [Google Scholar] [CrossRef] [Green Version]
- Helliwell, J.R. Fundamentals of neutron crystallography in structural biology. Methods Enzymol. 2020, 634, 1–19. [Google Scholar] [PubMed]
- Blakeley, M. Neutron macromolecular crystallography. Crystallogr. Rev. 2009, 15, 157–218. [Google Scholar] [CrossRef]
- Crespi, H.L.; Conard, S.M.; Uphaus, R.A.; Katz, J.J.; Conrad, S.M. Cultivation of microorganisms in heavy water. Ann. N. Y. Acad. Sci. 1960, 84, 648–666. [Google Scholar] [CrossRef] [PubMed]
- Haertlein, M.; Moulin, M.; Devos, J.M.; Laux, V.; Dunne, O.; Forsyth, V.T. Biomolecular deuteration for neutron structural biology and dynamics. Methods Enzymol. 2016, 566, 113–157. [Google Scholar]
- Aidelberg, G.; Towbin, B.D.; Rothschild, D.; Dekel, E.; Bren, A.; Alon, U. Hierarchy of non-glucose sugars in Escherichia coli. BMC Syst. Biol. 2014, 8, 133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooper, R.A.; Kornberg, H.L. The direct synthesis of phospho enol pyruvate from pyruvate by Escherichia coli. Proc. R. Soc. Lond. Ser. B Boil. Sci. 1967, 168, 263–280. [Google Scholar]
- Shchepin, R.V.; Coffey, A.M.; Waddell, K.W.; Chekmenev, E.Y. Parahydrogen Induced polarization of 1-13C-phospholactate-d 2 for Biomedical imaging with >30,000,000-fold NMR signal enhancement in water. Anal. Chem. 2014, 86, 5601–5605. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, T.E.; Salazar, M.J.; Weng, L.L.; Palsson, B.O.; Feist, A.M. The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab. Eng. 2019, 56, 1–16. [Google Scholar] [CrossRef]
- Atsumi, S.; Wu, T.; Machado, I.M.P.; Huang, W.; Chen, P.-Y.; Pellegrini, M.; Liao, J. Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol. Syst. Biol. 2010, 6, 449. [Google Scholar] [CrossRef]
- Conrad, T.M.; Frazier, M.; Joyce, A.R.; Cho, B.-K.; Knight, E.M.; Lewis, N.; Landick, R.; Palsson, B.O. RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc. Natl. Acad. Sci. USA 2010, 107, 20500–20505. [Google Scholar] [CrossRef] [Green Version]
- Minty, J.J.; A Lesnefsky, A.; Lin, F.; Chen, Y.; A Zaroff, T.; Veloso, A.B.; Xie, B.; A McConnell, C.; Ward, R.J.; Schwartz, D.R.; et al. Evolution combined with genomic study elucidates genetic bases of isobutanol tolerance in Escherichia coli. Microb. Cell Factories 2011, 10, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelpsas, V.; von Wachenfeldt, C. Strain improvement of Escherichia coli K-12 for recombinant production of deuterated proteins. Sci. Rep. 2019, 9, 17694. [Google Scholar] [CrossRef] [PubMed]
- Margolis, S.A.; Coxon, B. Identification and quantitation of the impurities in sodium pyruvate. Anal. Chem. 1986, 58, 2504–2510. [Google Scholar] [CrossRef]
- Jiang, G.R.; Nikolova, S.; Clark, D.P. Regulation of the ldhA gene, encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology 2001, 147, 2437–2446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, L.; Xu, X.; Wang, L.; Dong, H.; Yu, B. The D-lactate dehydrogenase from Sporolactobacillus inulinus also possessing reversible deamination activity. PLoS ONE 2015, 10, e0139066. [Google Scholar] [CrossRef] [Green Version]
- Landini, P.; Egli, T.; Wolf, J.; Lacour, S. sigma S, a major player in the response to environmental stresses in Escherichia coli: Role, regulation and mechanisms of promoter recognition. Environ. Microbiol. Rep. 2014, 6, 1–13. [Google Scholar] [CrossRef]
- Campbell, E.A.; Muzzin, O.; Chlenov, M.; Sun, J.L.; Olson, C.; Weinman, O.; Trester-Zedlitz, M.L.; Darst, S.A. Structure of the bacterial RNA polymerase promoter specificity σ subunit. Mol. Cell 2002, 9, 527–539. [Google Scholar] [CrossRef]
- Wösten, M. Eubacterial sigma-factors. FEMS Microbiol. Rev. 1998, 22, 127–150. [Google Scholar] [CrossRef]
- Maciąg, A.; Peano, C.; Pietrelli, A.; Egli, T.; De Bellis, G.; Landini, P. In vitro transcription profiling of the σ S subunit of bacterial RNA polymerase: Re-definition of the σS regulon and identification of σS-specific promoter sequence elements. Nucleic Acids Res. 2011, 39, 5338–5355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Straus, D.B.; Walter, W.A.; Gross, C.A. The heat shock response of E. coli is regulated by changes in the concentration of σ32. Nature 1987, 329, 348. [Google Scholar] [CrossRef] [PubMed]
- Sharp, M.M.; Chan, C.L.; Lu, C.Z.; Marr, M.T.; Nechaev, S.; Merritt, E.W.; Severinov, K.; Roberts, J.W.; Gross, C.A. The interface of sigma with core RNA polymerase is extensive, conserved, and functionally specialized. Genes Dev. 1999, 13, 3015–3026. [Google Scholar] [CrossRef] [PubMed]
- Tenaillon, O.; Rodríguez-Verdugo, A.; Gaut, R.L.; McDonald, P.; Bennett, A.F.; Long, A.D.; Gaut, B.S. The molecular diversity of adaptive convergence. Science 2012, 335, 457–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deatherage, D.E.; Kepner, J.L.; Bennett, A.F.; Lenski, R.E.; Barrick, J.E. Specificity of genome evolution in experimental populations of Escherichia coli evolved at different temperatures. Proc. Natl. Acad. Sci. USA 2017, 114, E1904–E1912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, R.H. Stable isotope studies on the biosynthesis of the thiazole moiety of thiamin in Escherichia coli. Biochemistry 1978, 17, 3833–3840. [Google Scholar] [CrossRef]
- Guzman, M.I.; Colussi, A.J.; Hoffmann, M.R. Photogeneration of distant radical pairs in aqueous pyruvic acid glasses. J. Phys. Chem. A 2006, 110, 931–935. [Google Scholar] [CrossRef] [Green Version]
- Robson, S.A.; Takeuchi, K.; Boeszoermenyi, A.; Coote, P.W.; Dubey, A.; Hyberts, S.; Wagner, G.; Arthanari, H. Mixed pyruvate labeling enables backbone resonance assignment of large proteins using a single experiment. Nat. Commun. 2018, 9, 356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Studier, F.W.; Moffatt, B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986, 189, 113–130. [Google Scholar] [CrossRef]
- Kelpsas, V.; Caldararu, O.; Blakeley, M.P.; Coquelle, N.; Wierenga, R.K.; Ryde, U.; von Wachenfeldt, C.; Oksanen, E. Neutron structures of Leishmania mexicana triosephosphate isomerase in complex with reaction-intermediate mimics shed light on the proton-shuttling steps. IUCrJ 2021, 8 Pt 4, 633–643. [Google Scholar] [CrossRef]
- Kelpšas, V.; Lafumat, B.; Blakeley, M.P.; Coquelle, N.; Oksanen, E.; von Wachenfeldt, C. Perdeuteration, large crystal growth and neutron data collection of Leishmania mexicana triose-phosphate isomerase E65Q variant. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2019, 75, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Koruza, K.; Lafumat, B.; Végvári, Á.; Knecht, W.; Fisher, S. Deuteration of human carbonic anhydrase for neutron crystallography: Cell culture media, protein thermostability, and crystallization behavior. Arch. Biochem. Biophys. 2018, 645, 26–33. [Google Scholar] [CrossRef]
- Carpenter, E.P.; Beis, K.; Cameron, A.D.; Iwata, S. Overcoming the challenges of membrane protein crystallography. Curr. Opin. Struct. Biol. 2008, 18, 581–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blattner, F.R.; Plunkett, G., III; Bloch, C.A.; Perna, N.T.; Burland, V.; Riley, M.; Collado-Vides, J.; Glasner, J.D.; Rode, C.K.; Mayhew, G.F.; et al. The complete genome sequence of Escherichia coli K-12. Science 1997, 277, 1453–1462. [Google Scholar] [CrossRef] [Green Version]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
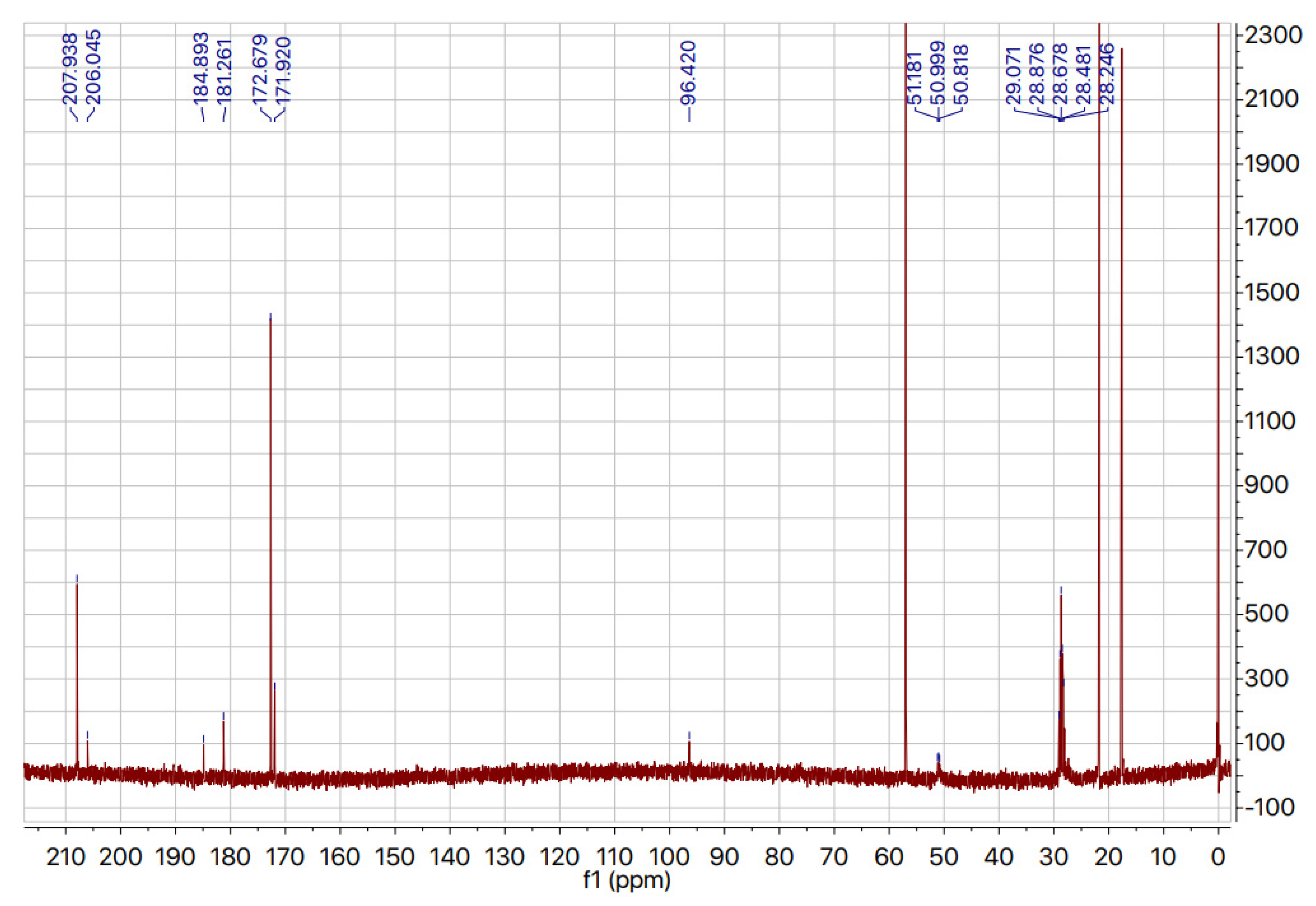
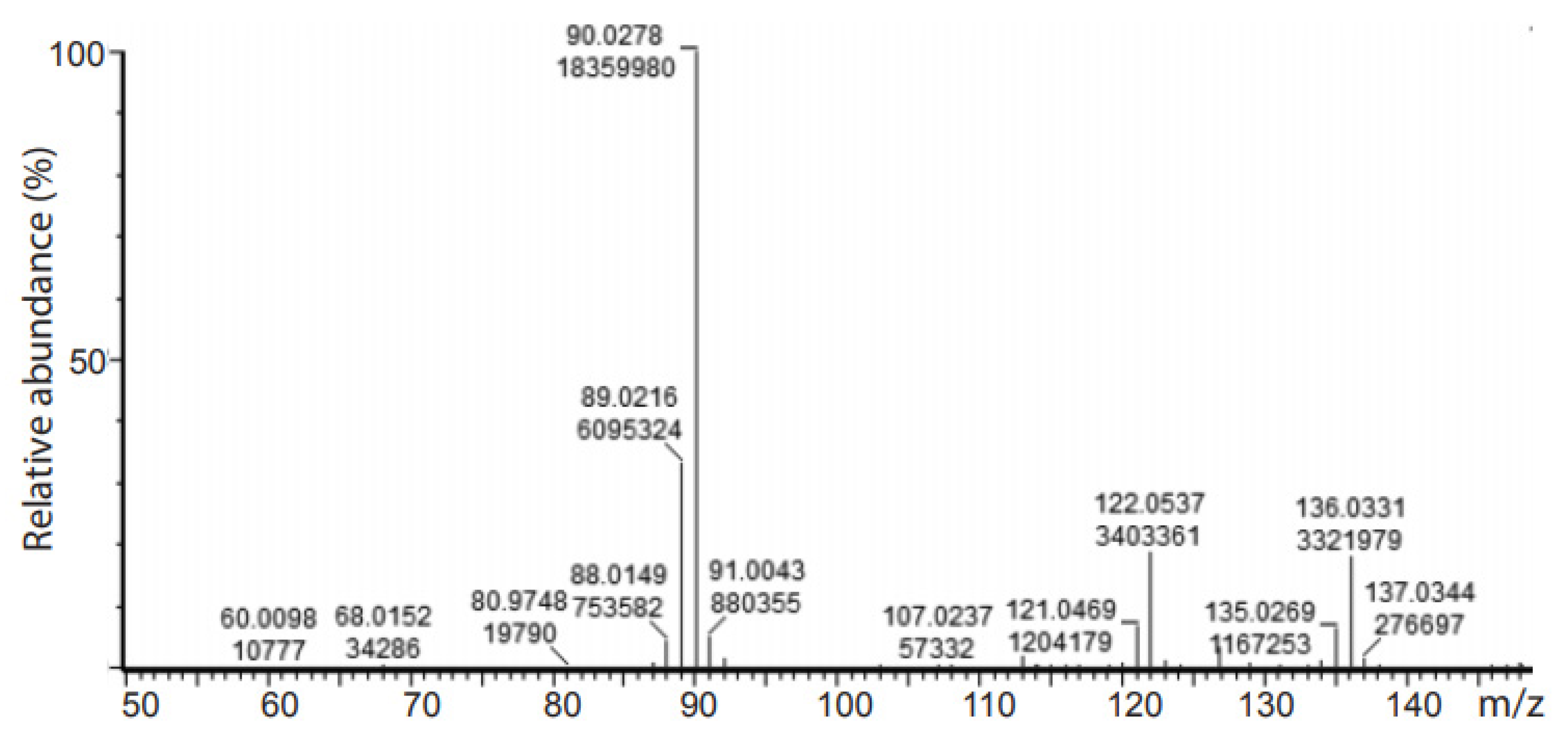

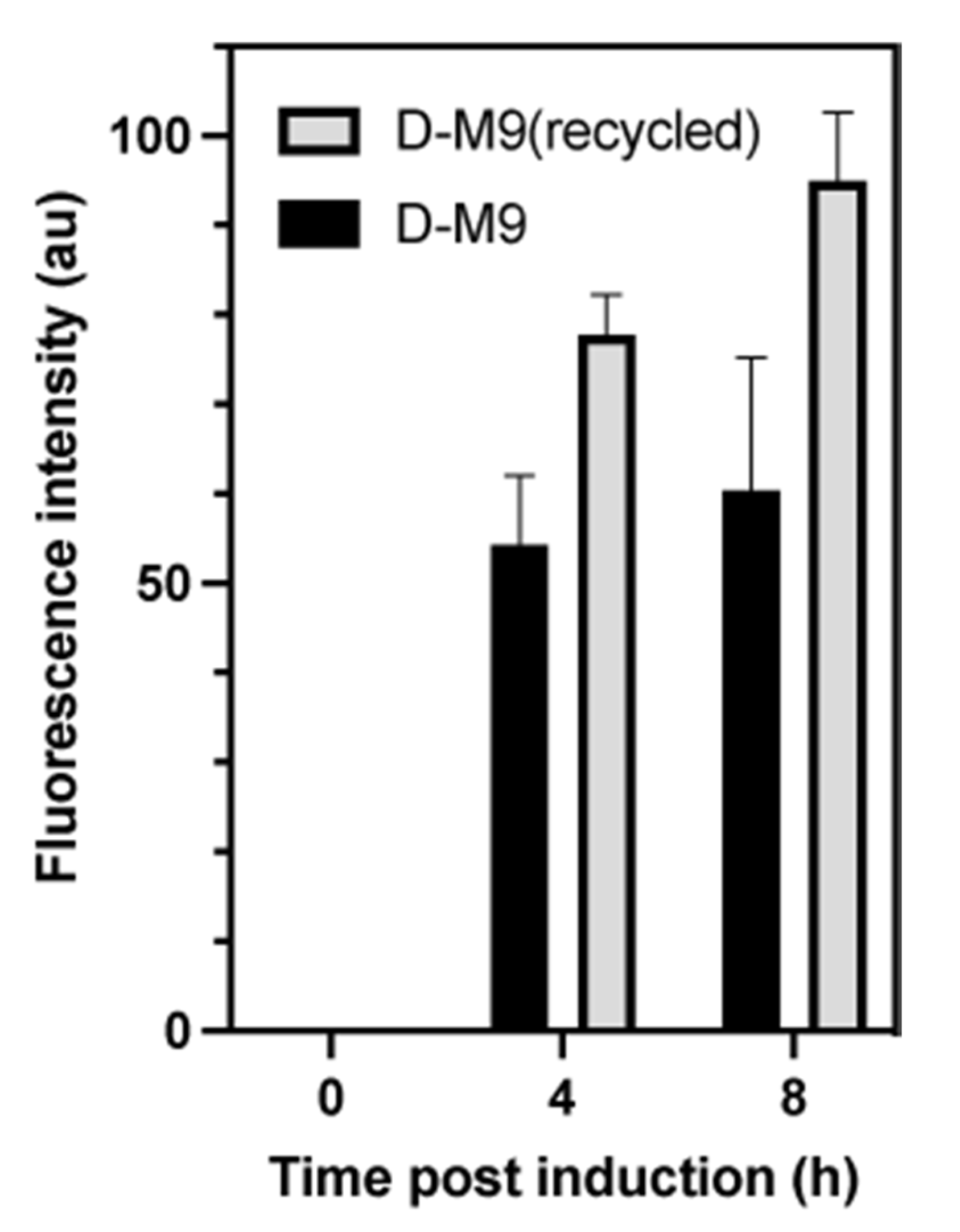
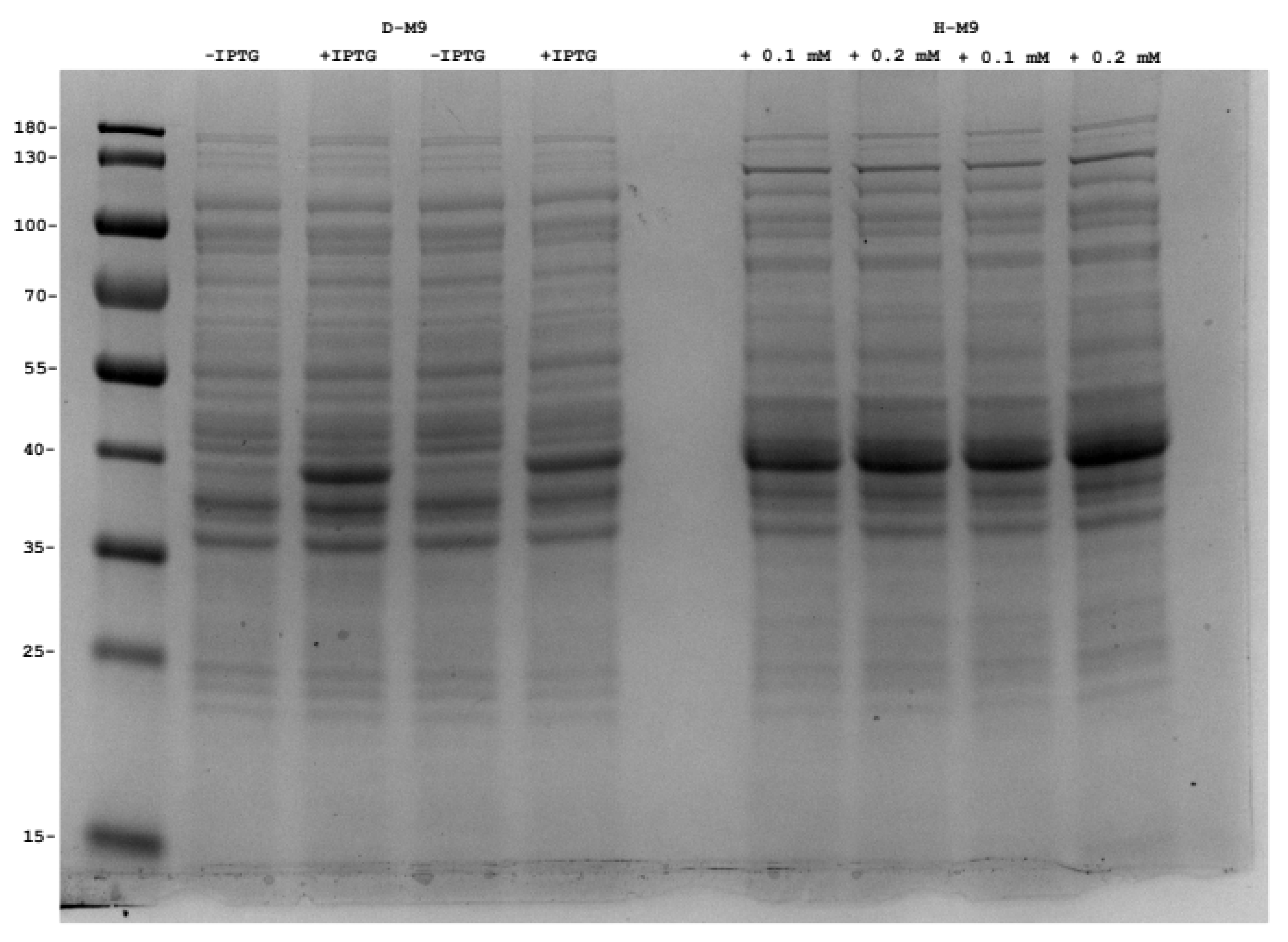
| Strain | Growth Rate (h−1) a | Generation Time (h) |
|---|---|---|
| MG1655 | 0.056 ± 0.0002 | 12.4 |
| P1.49.1 | 0.199 ± 0.0095 | 3.5 |
| P2.49.1 | 0.103 ± 0.0002 | 6.7 |
| P3.49.1 | 0.235 ± 0.004 | 2.9 |
| Position a | Gene | Strain | Coding Region Change c | Amino Acid Change d | Description | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1.49 | P2.49 | P3.49 | |||||||||||
| 1 | 2 | Frequency (%) b | 1 | 2 | Frequency (%) b | 1 | 2 | Frequency (%) b | |||||
| 1316251 | Non coding | 99.2 | A > G | NA | |||||||||
| 1330830 | Non coding | C > T | NA | ||||||||||
| 1356902 | sapA | 209T > G | Leu70Arg | Putative periplasmic binding protein | |||||||||
| 1397277 | Non coding | G > T | NA | ||||||||||
| 1442611 | ldhA | 233G > C | Gly78Ala | D-lactate dehydrogenase | |||||||||
| 1861387 | Non coding | G > A | NA | ||||||||||
| 1918327 | yebT | 818A > G | Asp273Gly | Intermembrane transport protein | |||||||||
| 2339829 | ubiG | 263T > G | Phe88Cys | Ubiquinol-8 biosynthesis protein | |||||||||
| 2423134 | yfcI | 406T > G | Trp136Gly | Recombination-promoting nuclease | |||||||||
| 2867121 | rpoS | 97.2 | 431C > T | Thr144Ile | Sigma S | ||||||||
| 2867169 | rpoS | 98.3 | 383T > A | Ile128Asn | Sigma S | ||||||||
| 2867182 | rpoS | 99.8 | 370A > T | Asn124Tyr | Sigma S | ||||||||
| 3856831 | yidl | 417C > T | NA | Putative DNA-binding transcriptional regulator | |||||||||
| 3951538 | ilvG | 66.2 | 979del | Gln327fs | Isoleucine biosynthesis | ||||||||
| 3951543 | ilvG | 44.8 | 99.7 | 984del | *328fs | Isoleucine biosynthesis | |||||||
| 3965550 | rhlB | 87.7 | 35.0 | 81dup | Gly28Fs | ATP-dependent RNA helicase RhlB | |||||||
| 4033620 | trkH | 65.7 | 476A > T | Gln159Leu | K+ transporter | ||||||||
| Sample a | Calculated Mass (Da) | Observed Mass (Da) |
|---|---|---|
| D-TIM | 28,791 | 28,772 |
| H-TIM | 27,264 | 27,285 |
| Mass difference | 1527 | 1487 |
| Sample a | Calculated Mass (Da) | Observed Mass (Da) |
|---|---|---|
| D-TIM | 28,791 | 28,793 |
| H-TIM | 27,264 | 27,285 |
| Mass difference | 1527 | 1508 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kelpšas, V.; Leung, A.; von Wachenfeldt, C. Evolving Escherichia coli Host Strains for Efficient Deuterium Labeling of Recombinant Proteins Using Sodium Pyruvate-d3. Int. J. Mol. Sci. 2021, 22, 9678. https://doi.org/10.3390/ijms22189678
Kelpšas V, Leung A, von Wachenfeldt C. Evolving Escherichia coli Host Strains for Efficient Deuterium Labeling of Recombinant Proteins Using Sodium Pyruvate-d3. International Journal of Molecular Sciences. 2021; 22(18):9678. https://doi.org/10.3390/ijms22189678
Chicago/Turabian StyleKelpšas, Vinardas, Anna Leung, and Claes von Wachenfeldt. 2021. "Evolving Escherichia coli Host Strains for Efficient Deuterium Labeling of Recombinant Proteins Using Sodium Pyruvate-d3" International Journal of Molecular Sciences 22, no. 18: 9678. https://doi.org/10.3390/ijms22189678






