Altered Root Growth, Auxin Metabolism and Distribution in Arabidopsis thaliana Exposed to Salt and Osmotic Stress
Abstract
:1. Introduction
2. Results and Discussion
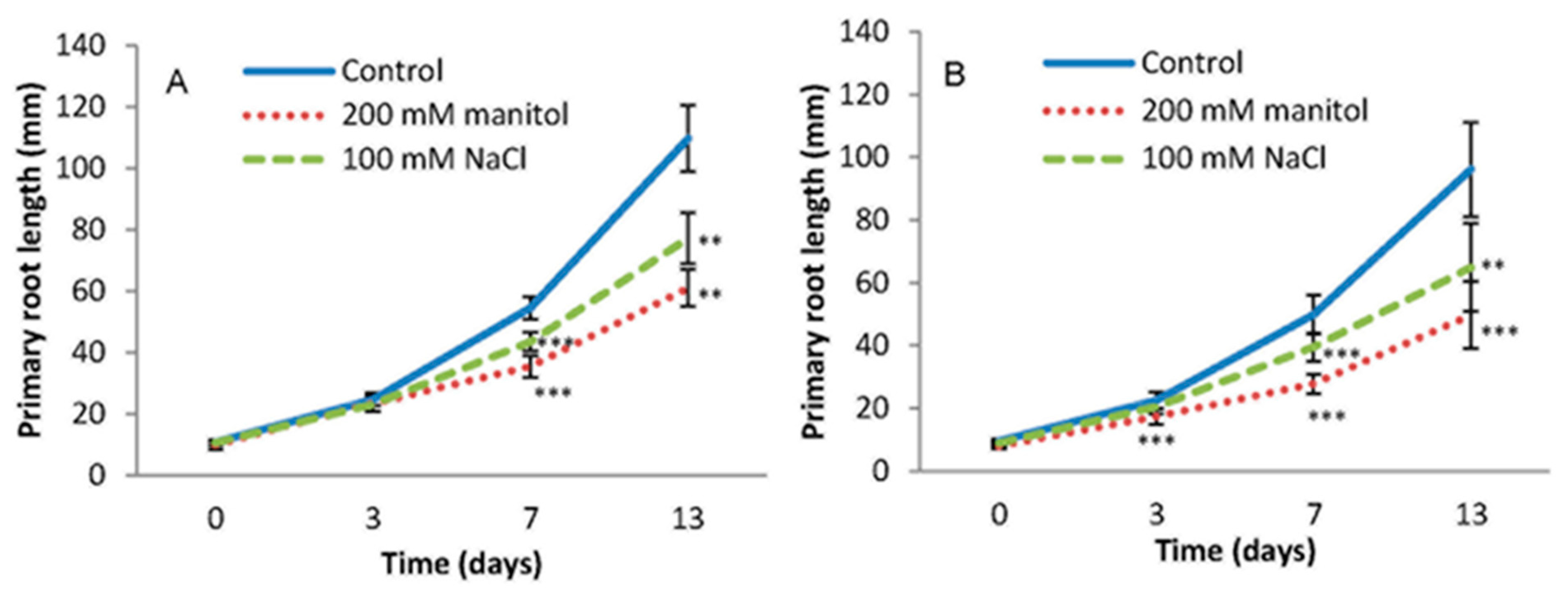
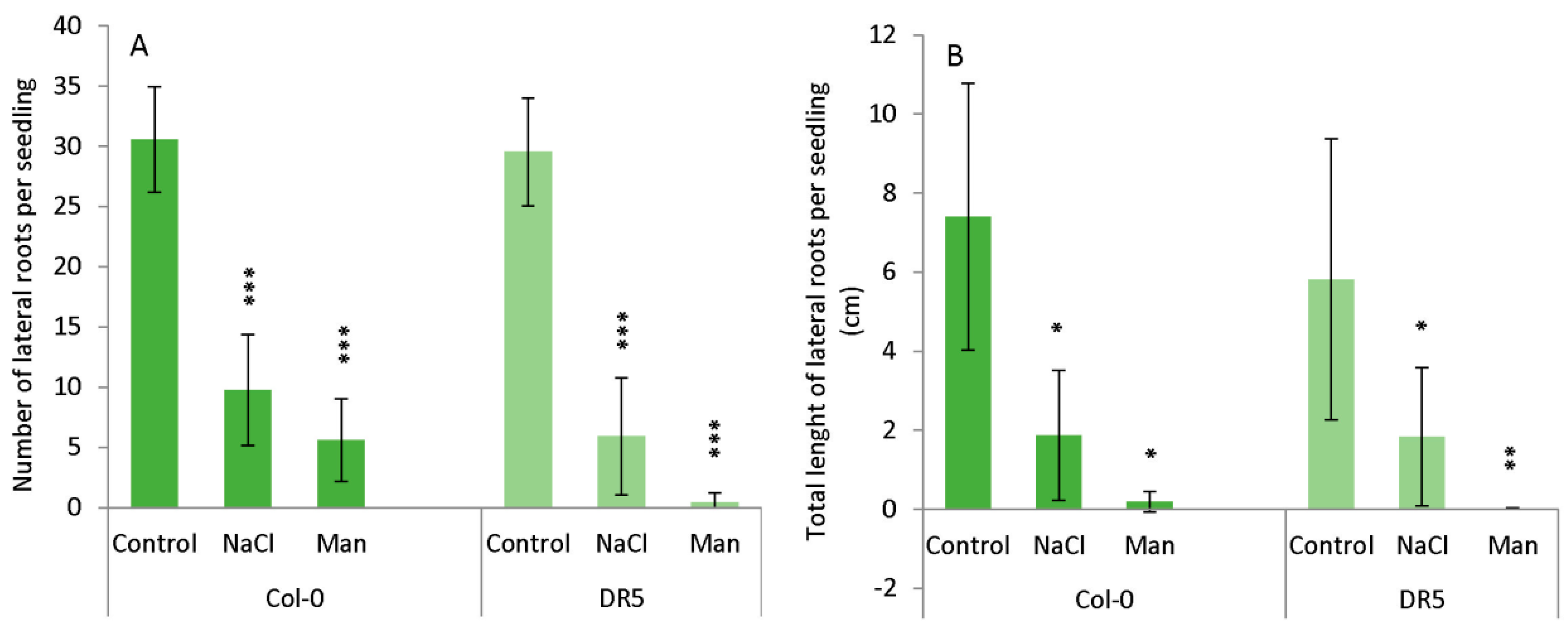
2.1. Root Growth under Abiotic Stress
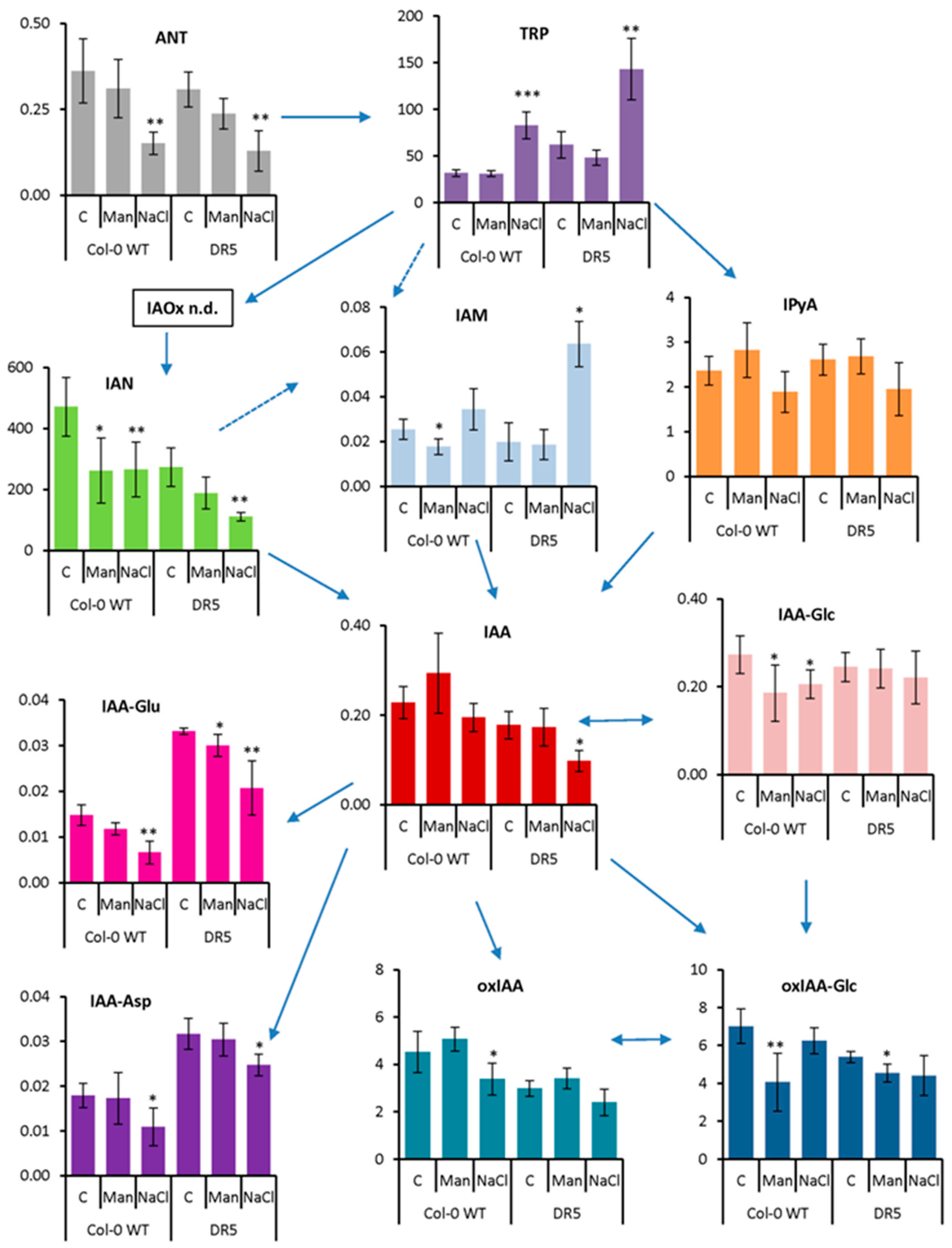
2.2. Auxin Profile in Arabidopsis after Short-Term and Prolonged Stress Conditions
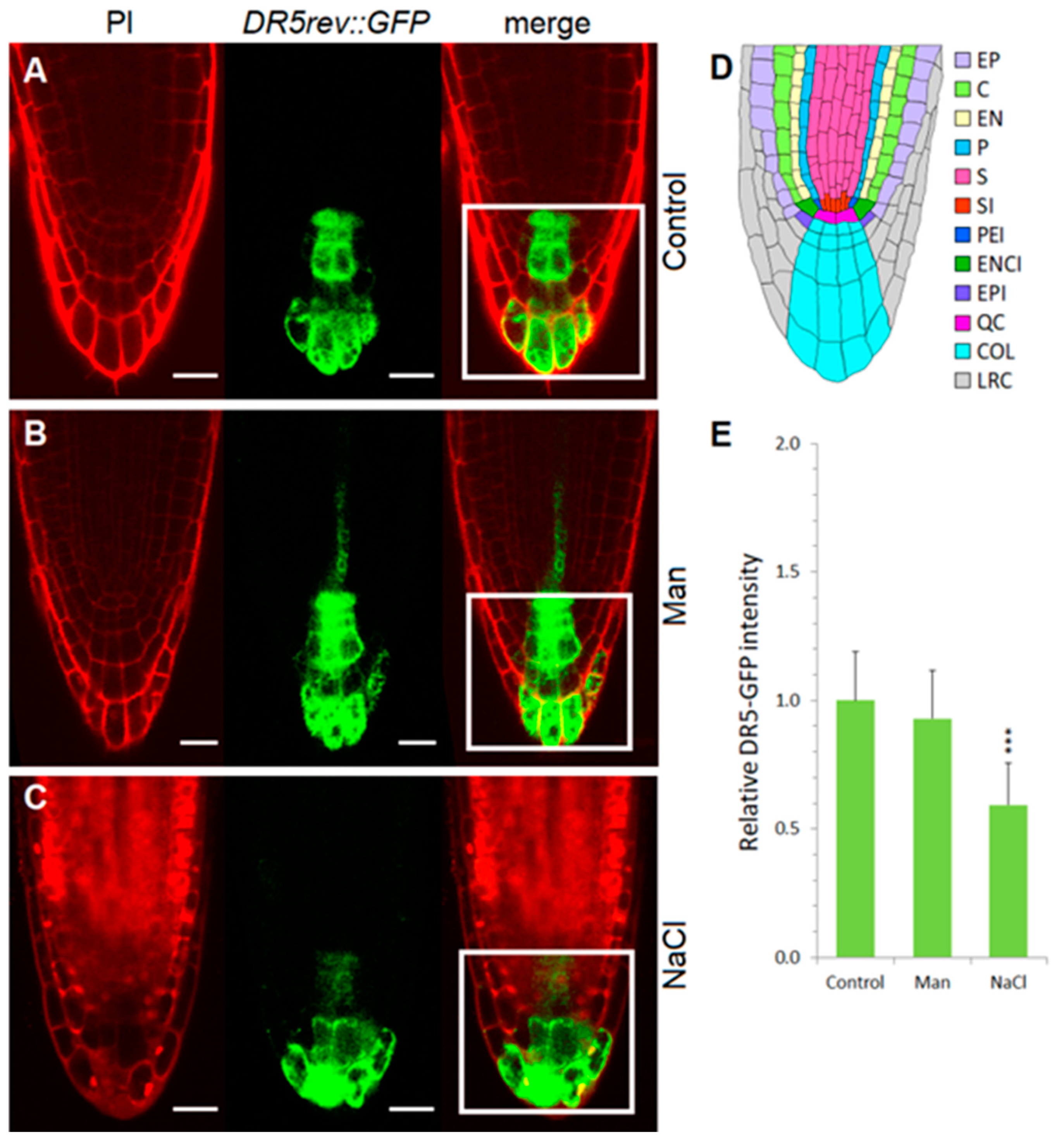
2.3. Early Auxin Redistribution in Root under Stress Conditions
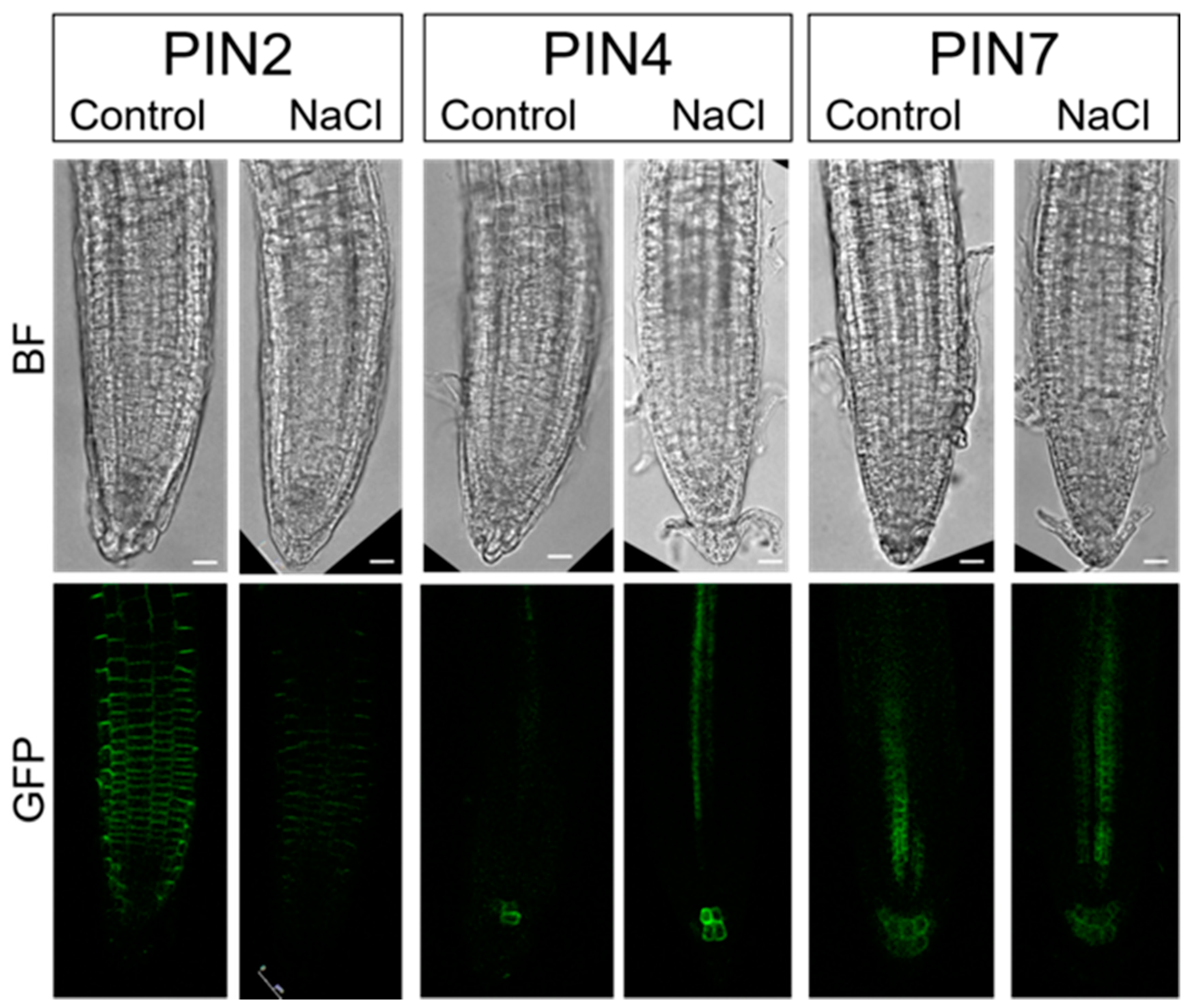
2.4. PINs Redistribution under Salinity Stress
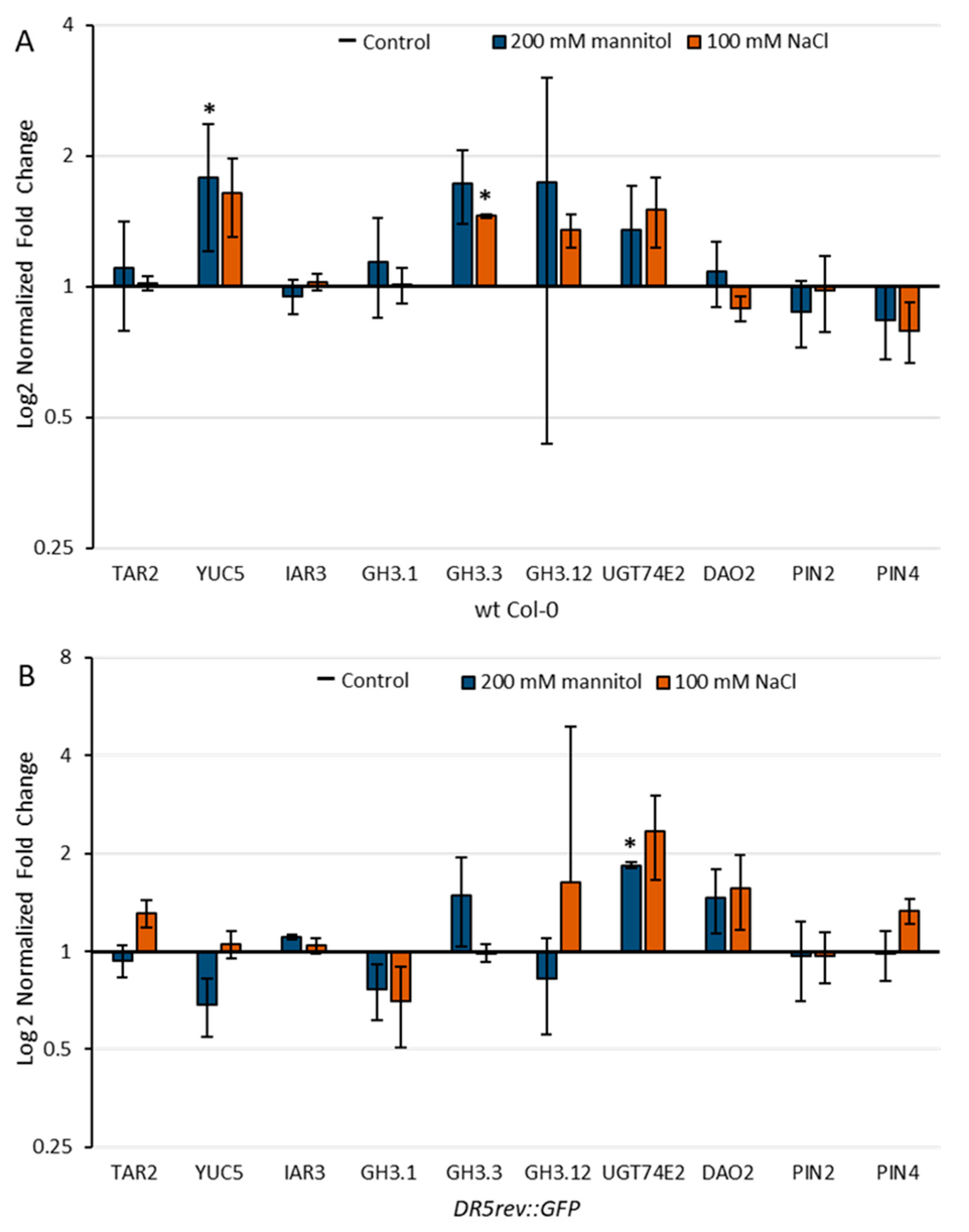
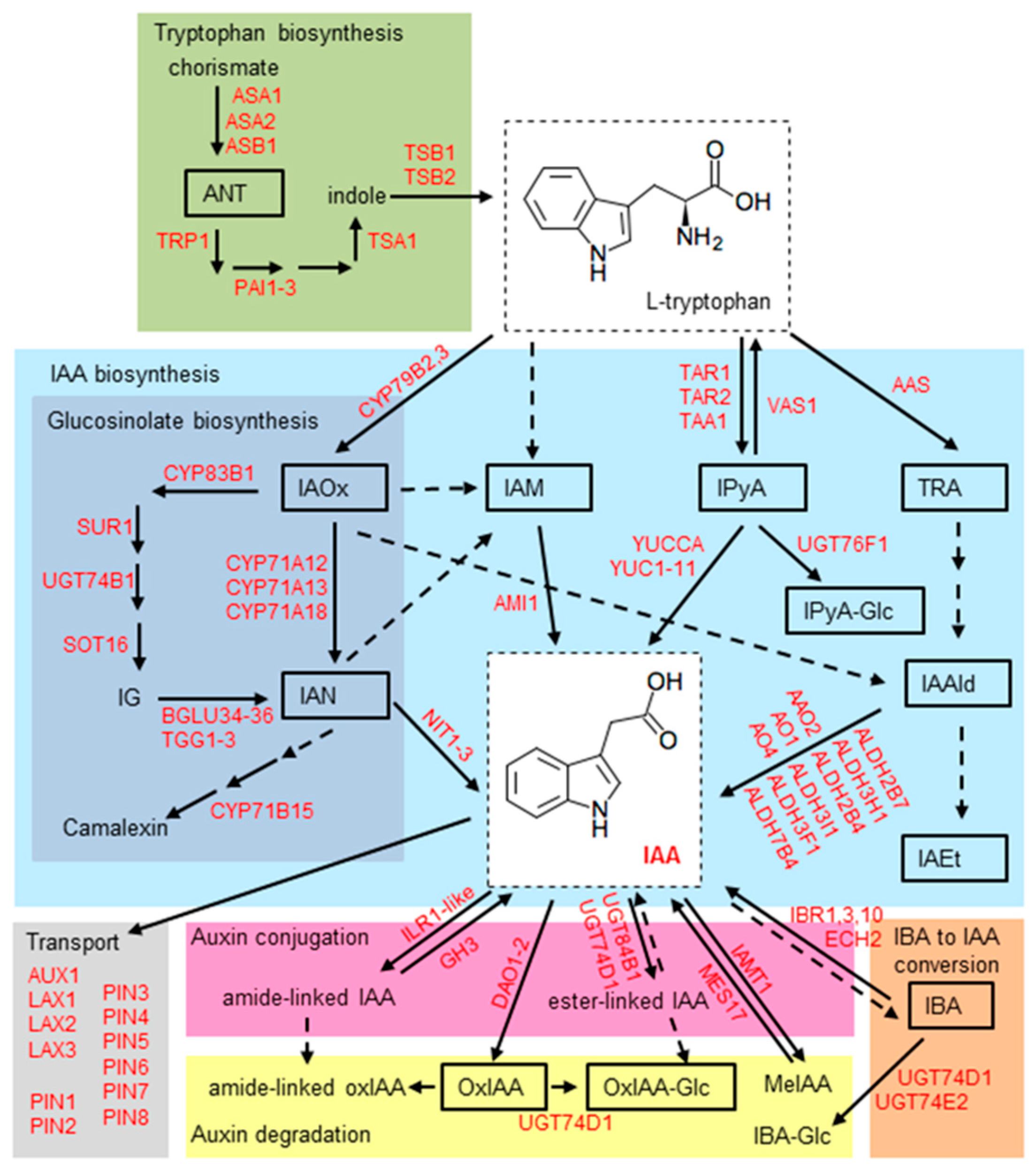
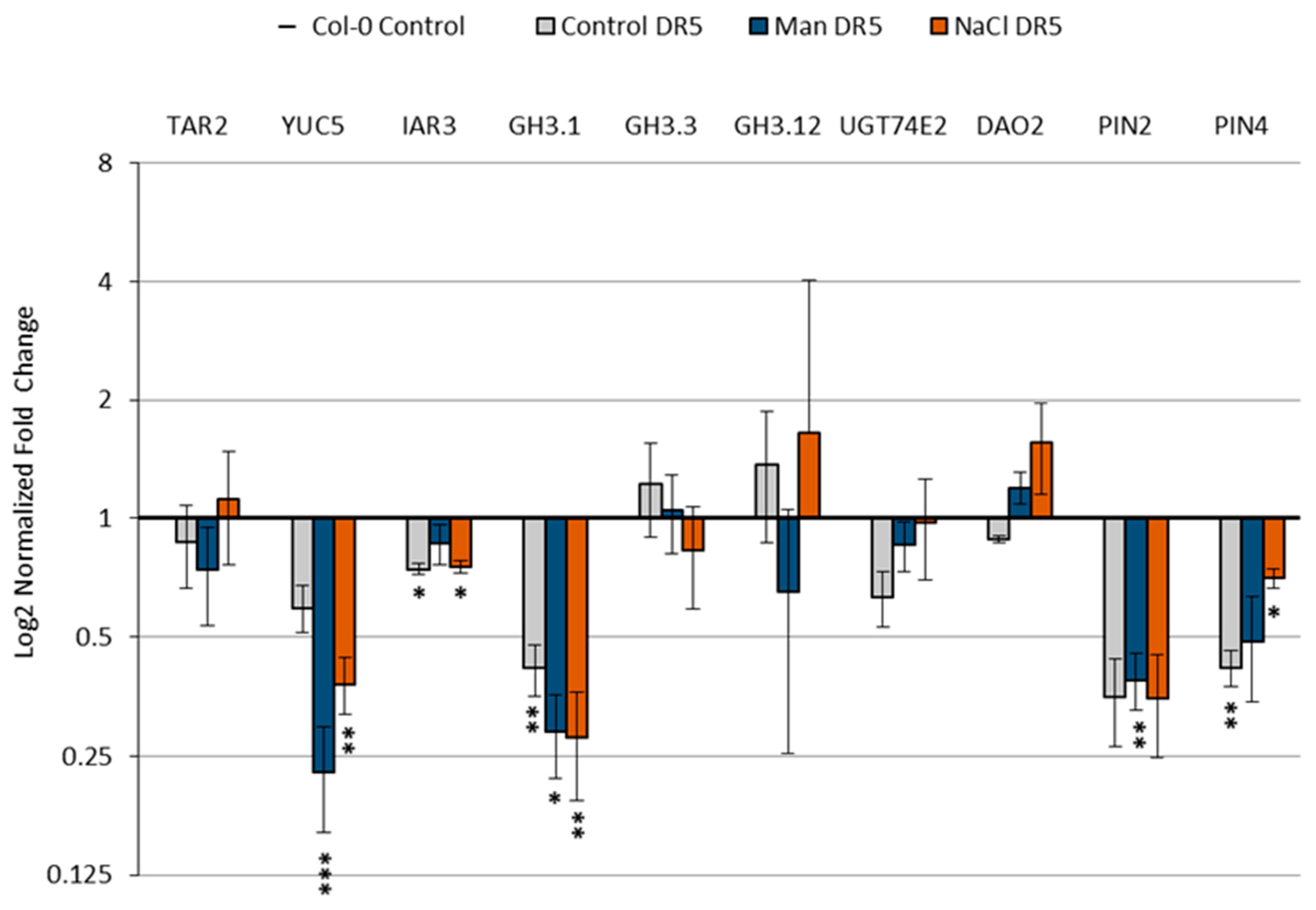
2.5. Effect of Stress Conditions on the Expression of Genes Involved in Auxin Metabolism
3. Materials and Methods
3.1. Reagents and Standards
3.2. Plant Material and Growth Conditions
3.3. Stress Treatments of Seedlings
3.4. Root Growth Bioassay
3.5. Confocal Laser Scanning Microscopy and Image Analyses
3.6. Auxin Metabolite Analysis
3.7. Real-Time PCR of Selected Genes
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Daliakopoulos, I.N.; Panagea, I.S.; Tsanis, I.K.; Grillakis, M.G.; Koutroulis, A.G.; Hessel, R.; Mayor, A.G.; Ritsema, C.J. Yield response of Mediterranean rangelands under a changing climate. Land Degrad. Dev. 2017, 28, 1962–1972. [Google Scholar] [CrossRef]
- Petersson, S.V.; Johansson, A.I.; Kowalczyk, M.; Makoveychuk, A.; Wang, J.Y.; Moritz, T.; Grebe, M.; Benfey, P.N.; Sandberg, G.; Ljung, K. An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 2009, 21, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Clark, N.M.; de Luis Balaguer, M.A.; Sozzani, R. Experimental data and computational modeling link auxin gradient and development in the Arabidopsis root. Front. Plant Sci. 2014, 5, 328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korver, R.A.; Koevoets, I.T.; Testerink, C. Out of shape during stress: A key role for auxin. Trends Plant Sci. 2018, 23, 783–793. [Google Scholar] [CrossRef]
- Ljung, K.; Hull, A.K.; Celenza, J.; Yamada, M.; Estelle, M.; Normanly, J.; Sandberg, G. Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 2005, 17, 1090–1104. [Google Scholar] [CrossRef]
- Du, Y.; Scheres, B. Lateral root formation and the multiple roles of auxin. J. Exp. Bot. 2018, 69, 155–167. [Google Scholar] [CrossRef]
- Tivendale, N.D.; Ross, J.J.; Cohen, J.D. The shifting paradigms of auxin biosynthesis. Trends Plant Sci. 2014, 19, 44–51. [Google Scholar] [CrossRef]
- Di, D.-W.; Zhang, C.; Luo, P.; An, C.-W.; Guo, G.-Q. The biosynthesis of auxin: How many paths truly lead to IAA? Plant Growth Regul. 2016, 78, 275–285. [Google Scholar] [CrossRef]
- Olatunji, D.; Geelen, D.; Verstraeten, I. Control of endogenous auxin levels in plant root development. Int. J. Mol. Sci. 2017, 18, 2587. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Peer, W.A. Auxin homeostasis: The DAO of catabolism. J. Exp. Bot. 2017, 68, 3145–3154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smolko, A.; Ludwig-Müller, J.; Salopek-Sondi, B. Auxin Amidohydrolases—From Structure to Function: Revisited. Croat. Chem. Acta 2018, 91, 233–239. [Google Scholar] [CrossRef]
- Casanova-Sáez, R.; Mateo-Bonmatí, E.; Ljung, K. Auxin metabolism in plants. Cold Spring Harb. Perspect. Biol. 2021, 13, a039867. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Normanly, J.; Slovin, J.P.; Cohen, J.D. Auxin biosynthesis and metabolism. In Plant Hormones; Davies, P.J., Ed.; Springer: Dordrecht, The Netherlands, 2010; pp. 36–62. [Google Scholar]
- Ludwig-Müller, J. Auxin conjugates: Their role for plant development and in the evolution of land plants. J. Exp. Bot. 2011, 62, 1757–1773. [Google Scholar] [CrossRef] [Green Version]
- Bartel, B.; LeClere, S.; Magidin, M.; Zolman, B.K. Inputs to the active indole-3-acetic acid pool: De novo synthesis, conjugate hydrolysis, and indole-3-butyric acid β-oxidation. J. Plant Growth Regul. 2001, 20, 198–216. [Google Scholar] [CrossRef]
- Sánchez-García, A.B.; Ibáñez, S.; Cano, A.; Acosta, M.; Pérez-Pérez, J.M. A comprehensive phylogeny of auxin homeostasis genes involved in adventitious root formation in carnation stem cuttings. PLoS ONE 2018, 13, e0196663. [Google Scholar] [CrossRef] [Green Version]
- Korasick, D.A.; Enders, T.A.; Strader, L.C. Auxin biosynthesis and storage forms. J. Exp. Bot. 2013, 64, 2541–2555. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K.; Hayashi, K.; Natsume, M.; Kamiya, Y.; Sakakibara, H.; Kawaide, H.; Kasahara, H. UGT74D1 Catalyzes the Glucosylation of 2-Oxindole-3-Acetic Acid in the Auxin Metabolic Pathway in Arabidopsis. Plant Cell Physiol. 2014, 55, 218–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugawara, S.; Hishiyama, S.; Jikumaru, Y.; Hanada, A.; Nishimura, T.; Koshiba, T.; Zhao, Y.; Kamiya, Y.; Kasahara, H. Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 5430–5435. [Google Scholar] [CrossRef] [Green Version]
- Tzin, V.; Galili, G. The Biosynthetic Pathways for Shikimate and Aromatic Amino Acids in Arabidopsis thaliana. Arabidopsis Book 2010, 8, e0132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abualia, R.; Benkova, E.; Lacombe, B. Transporters and Mechanisms of Hormone Transport in Arabidopsis. In Membrane Transport in Plants; Maurel, C.B., Ed.; Academic Press: Cambridge, MA, USA, 2018; Volume 87, pp. 115–138. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Bashir, T.; Hashem, A.; Abd_Allah, E.F.; Khan, A.L.; Al-Harrasi, A.S. Molecular players of auxin transport systems: Advances in genomic and molecular events. J. Plant Interact. 2018, 13, 483–495. [Google Scholar] [CrossRef] [Green Version]
- Koevoets, I.T.; Venema, J.H.; Elzenga, J.T.M.; Testerink, C. Roots Withstanding their Environment: Exploiting Root System Architecture Responses to Abiotic Stress to Improve Crop Tolerance. Front. Plant Sci. 2016, 7, 1335. [Google Scholar] [CrossRef] [Green Version]
- Mroue, S.; Simeunovic, A.; Robert, H.S. Auxin production as an integrator of environmental cues for developmental growth regulation. J. Exp. Bot. 2018, 69, 201–212. [Google Scholar] [CrossRef]
- Liang, W.; Ma, X.; Wan, P.; Liu, L. Plant salt-tolerance mechanism: A review. Biochem. Biophys. Res. Commun. 2018, 495, 286–291. [Google Scholar] [CrossRef]
- Salopek-Sondi, B.; Pavlović, I.; Smolko, A.; Šamec, D. Auxin as a Mediator of Abiotic Stress Responses. In Mechanism of Plant Hormone Signaling under Stress; Pandey, G., Ed.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2017; pp. 1–36. [Google Scholar]
- Wang, Y.; Li, K.; Li, X. Auxin redistribution modulates plastic development of root system architecture under salt stress in Arabidopsis thaliana. J. Plant Physiol. 2009, 166, 1637–1645. [Google Scholar] [CrossRef] [PubMed]
- Julkowska, M.M.; Hoefsloot, H.C.J.; Mol, S.; Feron, R.; de Boer, G.-J.; Haring, M.A.; Testerink, C. Capturing Arabidopsis Root Architecture Dynamics with ROOT-FIT Reveals Diversity in Responses to Salinity. Plant Physiol. 2014, 166, 1387–1402. [Google Scholar] [CrossRef] [Green Version]
- Ryu, H.; Cho, Y.-G. Plant hormones in salt stress tolerance. J. Plant Biol. 2015, 58, 147–155. [Google Scholar] [CrossRef]
- Pavlović, I.; Pěnčík, A.; Novák, O.; Vujčić, V.; Radić Brkanac, S.; Lepeduš, H.; Strnad, M.; Salopek-Sondi, B. Short-term salt stress in Brassica rapa seedlings causes alterations in auxin metabolism. Plant Physiol. Biochem. 2018, 125, 74–84. [Google Scholar] [CrossRef]
- Julkowska, M.M.; Koevoets, I.T.; Mol, S.; Hoefsloot, H.; Feron, R.; Tester, M.A.; Keurentjes, J.J.B.; Korte, A.; Haring, M.A.; de Boer, G.-J.; et al. Genetic Components of Root Architecture Remodeling in Response to Salt Stress. Plant Cell 2017, 29, 3198–3213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galvan-Ampudia, C.S.; Testerink, C. Salt stress signals shape the plant root. Curr. Opin. Plant Biol. 2011, 14, 296–302. [Google Scholar] [CrossRef]
- Liu, W.; Li, R.-J.; Han, T.-T.; Cai, W.; Fu, Z.-W.; Lu, Y.-T. Salt Stress Reduces Root Meristem Size by Nitric Oxide-Mediated Modulation of Auxin Accumulation and Signaling in Arabidopsis. Plant Physiol. 2015, 168, 343–356. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Lin, J.E.; Harris, C.; Campos Mastrotti Pereira, F.; Wu, F.; Blakeslee, J.J.; Peer, W.A. DAO1 catalyzes temporal and tissue-specific oxidative inactivation of auxin in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2016, 113, 11010–11015. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Guo, G.; Wang, Z.; Ji, H.; Mu, F.; Li, X. Auxin in Plant Growth and Stress Responses. In Phytohormones: A Window to Metabolism, Signaling and Biotechnological Applications; Phan Tran, L.-S., Pal, S., Eds.; Springer: New York, NY, USA, 2014; pp. 1–35. [Google Scholar]
- Šimura, J.; Antoniadi, I.; Široká, J.; Tarkowská, D.; Strnad, M.; Ljung, K.; Novák, O. Plant hormonomics: Multiple phytohormone profiling by targeted metabolomics. Plant Physiol. 2018, 177, 476–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Křeček, P.; Skůpa, P.; Libus, J.; Naramoto, S.; Tejos, R.; Friml, J.; Zažímalová, E. The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 2009, 10, 249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, K.; Moe-Lange, J.; Hennet, L.; Feldman, L.J. Salt Stress Affects the Redox Status of Arabidopsis Root Meristems. Front. Plant Sci. 2016, 7, 81. [Google Scholar] [CrossRef] [Green Version]
- Feraru, E.; Friml, J. PIN Polar Targeting. Plant Physiol. 2008, 147, 1553–1559. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Yang, Y.; Chen, S.; Ning, N.; Hu, H. Arabidopsis IAR4 Modulates Primary Root Growth Under Salt Stress Through ROS-Mediated Modulation of Auxin Distribution. Front. Plant Sci. 2019, 10, 522. [Google Scholar] [CrossRef]
- Kilian, J.; Whitehead, D.; Horak, J.; Wanke, D.; Weinl, S.; Batistic, O.; D’Angelo, C.; Bornberg-Bauer, E.; Kudla, J.; Harter, K. The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007, 50, 347–363. [Google Scholar] [CrossRef] [PubMed]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An “Electronic Fluorescent Pictograph” Browser for Exploring and Analyzing Large-Scale Biological Data Sets. PLoS ONE 2007, 2, e718. [Google Scholar] [CrossRef]
- Boyes, D.C.; Zayed, A.M.; Ascenzi, R.; McCaskill, A.J.; Hoffman, N.E.; Davis, K.R.; Görlach, J. Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 2001, 13, 1499–1510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Che, G.; Ding, L.; Chen, Z.; Liu, X.; Wang, H.; Zhao, W.; Ning, K.; Zhao, J.; Tesfamichael, K.; et al. Different cucumber CsYUC genes regulate response to abiotic stresses and flower development. Sci. Rep. 2016, 6, 20760. [Google Scholar] [CrossRef]
- Shi, H.; Chen, L.; Ye, T.; Liu, X.; Ding, K.; Chan, Z. Modulation of auxin content in Arabidopsis confers improved drought stress resistance. Plant Physiol. Biochem. 2014, 82, 209–217. [Google Scholar] [CrossRef]
- Kim, J.I.; Baek, D.; Park, H.C.; Chun, H.J.; Oh, D.-H.; Lee, M.K.; Cha, J.-Y.; Kim, W.-Y.; Kim, M.C.; Chung, W.S.; et al. Overexpression of Arabidopsis YUCCA6 in potato results in high-auxin developmental phenotypes and enhanced resistance to water deficit. Mol. Plant 2013, 6, 337–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, Q.; Wang, Z.; Ji, C.Y.; Jeong, J.C.; Lee, H.-S.; Li, H.; Xu, B.; Deng, X.; Kwak, S.-S. Transgenic poplar expressing Arabidopsis YUCCA6 exhibits auxin-overproduction phenotypes and increased tolerance to abiotic stress. Plant Physiol. Biochem. 2015, 94, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-E.; Park, J.-Y.; Kim, Y.-S.; Staswick, P.E.; Jeon, J.; Yun, J.; Kim, S.-Y.; Kim, J.; Lee, Y.-H.; Park, C.-M. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J. Biol. Chem. 2007, 282, 10036–10046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirungu, J.N.; Magwanga, R.O.; Lu, P.; Cai, X.; Zhou, Z.; Wang, X.; Peng, R.; Wang, K.; Liu, F. Functional characterization of Gh_A08G1120 (GH3.5) gene reveal their significant role in enhancing drought and salt stress tolerance in cotton. BMC Genet. 2019, 20, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, W.; Zhong, H.; Deng, X.; Gautam, M.; Gong, Z.; Zhang, Y.; Zhao, G.; Liu, C.; Li, Y. Evolutionary analysis of GH3 genes in six Oryza species/subspecies and their expression under salinity stress in Oryza sativa ssp. japonica. Plants 2019, 8, 30. [Google Scholar] [CrossRef] [Green Version]
- Kong, W.; Zhang, Y.; Deng, X.; Li, S.; Zhang, C.; Li, Y. Comparative genomic and transcriptomic analysis suggests the evolutionary dynamic of GH3 genes in Gramineae crops. Front. Plant Sci. 2019, 10, 1297. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita, N.; Wang, H.; Kasahara, H.; Liu, J.; MacPherson, C.; Machida, Y.; Kamiya, Y.; Hannah, M.A.; Chua, N.-H. IAA-Ala Resistant3, an evolutionarily conserved target of miR167, mediates Arabidopsis root architecture changes during high osmotic stress. Plant Cell 2012, 24, 3590–3602. [Google Scholar] [CrossRef] [Green Version]
- Rehman, H.M.; Nawaz, M.A.; Shah, Z.H.; Ludwig-Müller, J.; Chung, G.; Ahmad, M.Q.; Yang, S.H.; Lee, S.I. Comparative genomic and transcriptomic analyses of Family-1 UDP glycosyltransferase in three Brassica species and Arabidopsis indicates stress-responsive regulation. Sci. Rep. 2018, 8, 1875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benková, E.; Michniewicz, M.; Sauer, M.; Teichmann, T.; Seifertová, D.; Jürgens, G.; Friml, J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 2003, 115, 591–602. [Google Scholar] [CrossRef] [Green Version]
- Friml, J.; Vieten, A.; Sauer, M.; Weijers, D.; Schwarz, H.; Hamann, T.; Offringa, R.; Jürgens, G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 2003, 426, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Scholl, R.L.; May, S.T.; Ware, D.H. Seed and Molecular Resources for Arabidopsis. Plant Physiol. 2000, 124, 1477–1480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abas, L.; Benjamins, R.; Malenica, N.; Paciorek, T.; Wišniewska, J.; Moulinier–Anzola, J.C.; Sieberer, T.; Friml, J.; Luschnig, C. Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 2006, 8, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Vieten, A.; Vanneste, S.; Wisniewska, J.; Benkova, E.; Benjamins, R.; Beeckman, T.; Luschnig, C.; Friml, J. Functional redundancy of PIN proteins is accompanied by auxin-dependentcross-regulation of PIN expression. Development 2005, 132, 4521–4531. [Google Scholar] [CrossRef] [Green Version]
- Blilou, I.; Xu, J.; Wildwater, M.; Willemsen, V.; Paponov, I.; Friml, J.; Heidstra, R.; Aida, M.; Palme, K.; Scheres, B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 2005, 433, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, Z.; Kellermeier, F.; Armstrong, E.M.; Rogers, S.; Lobet, G.; Amtmann, A.; Hills, A. EZ-Root-VIS: A software pipeline for the rapid analysis and visual reconstruction of root system architecture. Plant Physiol. 2018, 177, 1368–1381. [Google Scholar] [CrossRef] [Green Version]
- Gu, X.; Xu, T.; He, Y. A Histone H3 Lysine-27 Methyltransferase complex represses lateral root formation in Arabidopsis thaliana. Mol. Plant 2014, 7, 977–988. [Google Scholar] [CrossRef] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pěnčík, A.; Casanova-Sáez, R.; Pilařová, V.; Žukauskaitė, A.; Pinto, R.; Micol, J.L.; Ljung, K.; Novák, O. Ultra-rapid auxin metabolite profiling for high-throughput mutant screening in Arabidopsis. J. Exp. Bot. 2018, 69, 2569–2579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arvidsson, S.; Kwasniewski, M.; Riano-Pachon, D.M.; Mueller-Roeber, B. QuantPrime—A flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinform. 2008, 9, 465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.-R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [Green Version]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.B.; Moorman, A.F.M. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waese, J.; Fan, J.; Pasha, A.; Yu, H.; Fucile, G.; Shi, R.; Cumming, M.; Kelley, L.A.; Sternberg, M.J.; Krishnakumar, V.; et al. ePlant: Visualizing and exploring multiple levels of data for hypothesis generation in plant biology. Plant Cell 2017, 29, 1806–1821. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smolko, A.; Bauer, N.; Pavlović, I.; Pěnčík, A.; Novák, O.; Salopek-Sondi, B. Altered Root Growth, Auxin Metabolism and Distribution in Arabidopsis thaliana Exposed to Salt and Osmotic Stress. Int. J. Mol. Sci. 2021, 22, 7993. https://doi.org/10.3390/ijms22157993
Smolko A, Bauer N, Pavlović I, Pěnčík A, Novák O, Salopek-Sondi B. Altered Root Growth, Auxin Metabolism and Distribution in Arabidopsis thaliana Exposed to Salt and Osmotic Stress. International Journal of Molecular Sciences. 2021; 22(15):7993. https://doi.org/10.3390/ijms22157993
Chicago/Turabian StyleSmolko, Ana, Nataša Bauer, Iva Pavlović, Aleš Pěnčík, Ondřej Novák, and Branka Salopek-Sondi. 2021. "Altered Root Growth, Auxin Metabolism and Distribution in Arabidopsis thaliana Exposed to Salt and Osmotic Stress" International Journal of Molecular Sciences 22, no. 15: 7993. https://doi.org/10.3390/ijms22157993







