Comprehensive Analysis of SRO Gene Family in Sesamum indicum (L.) Reveals Its Association with Abiotic Stress Responses
Abstract
:1. Introduction
2. Results
2.1. Identification and Analysis of SRO Family Genes in Sesame
2.2. Phylogenetic Analysis of SiSROs
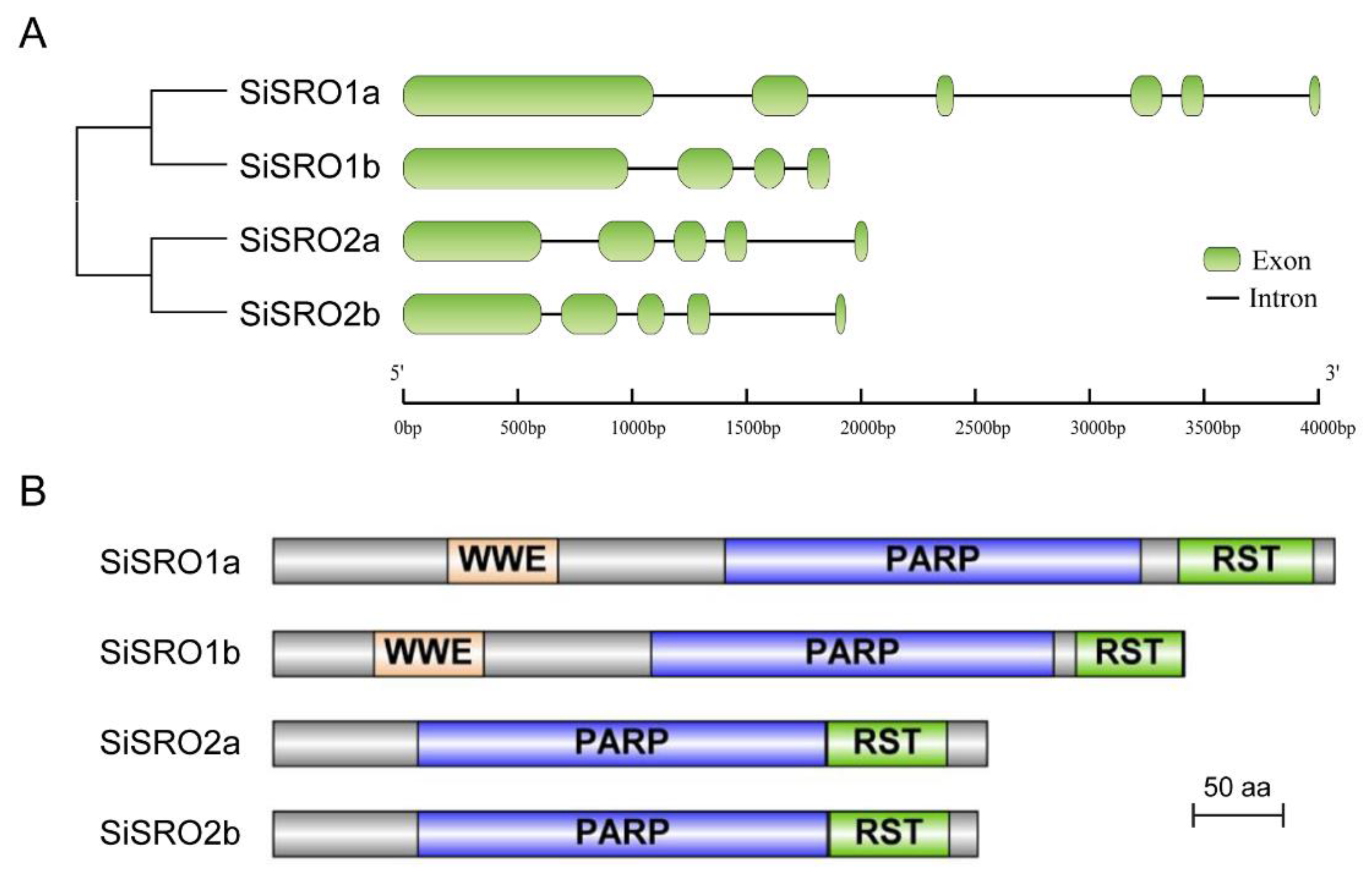
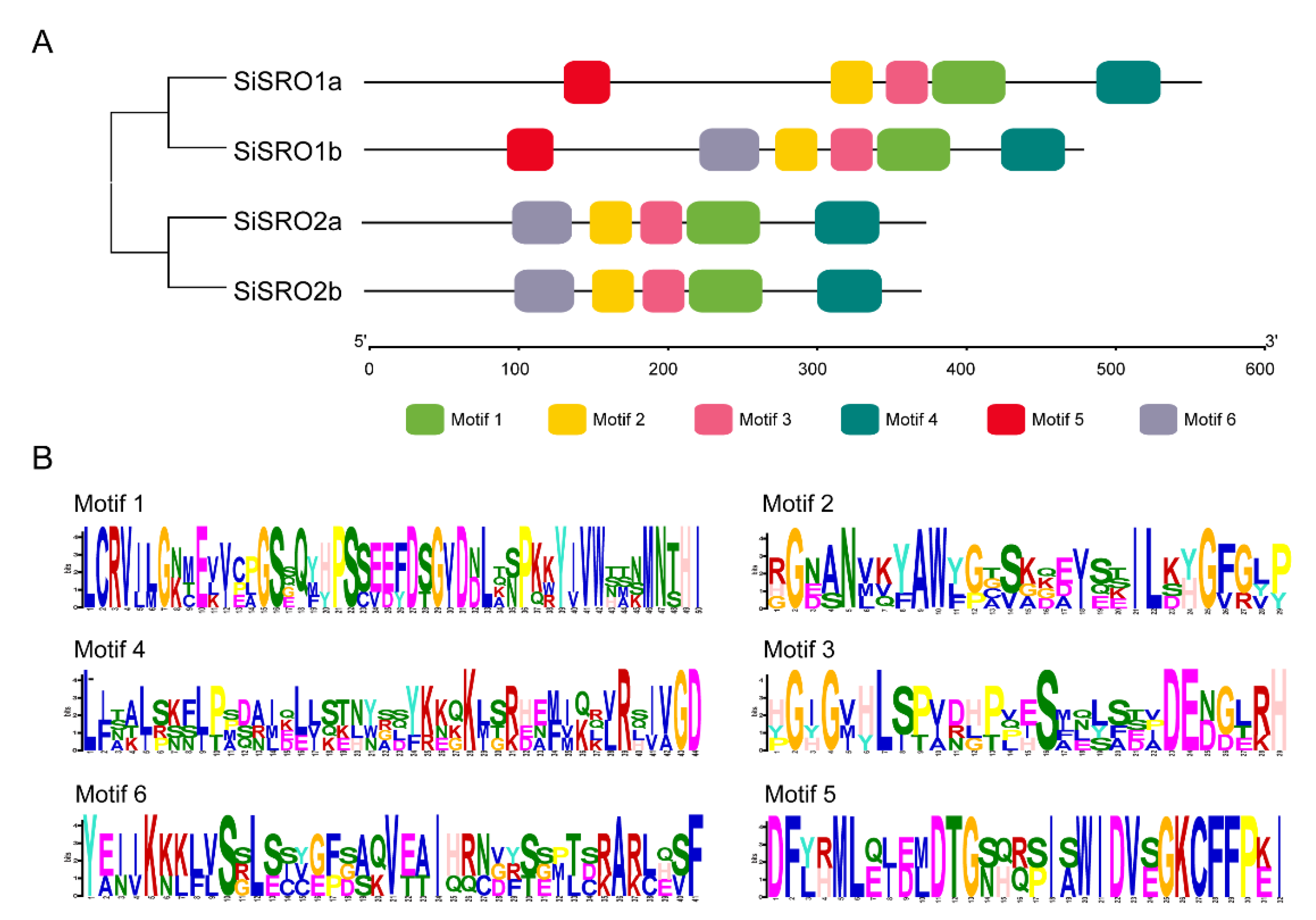
2.3. Intron-Exon Distribution and Motif Composition Analyses of SiSROs
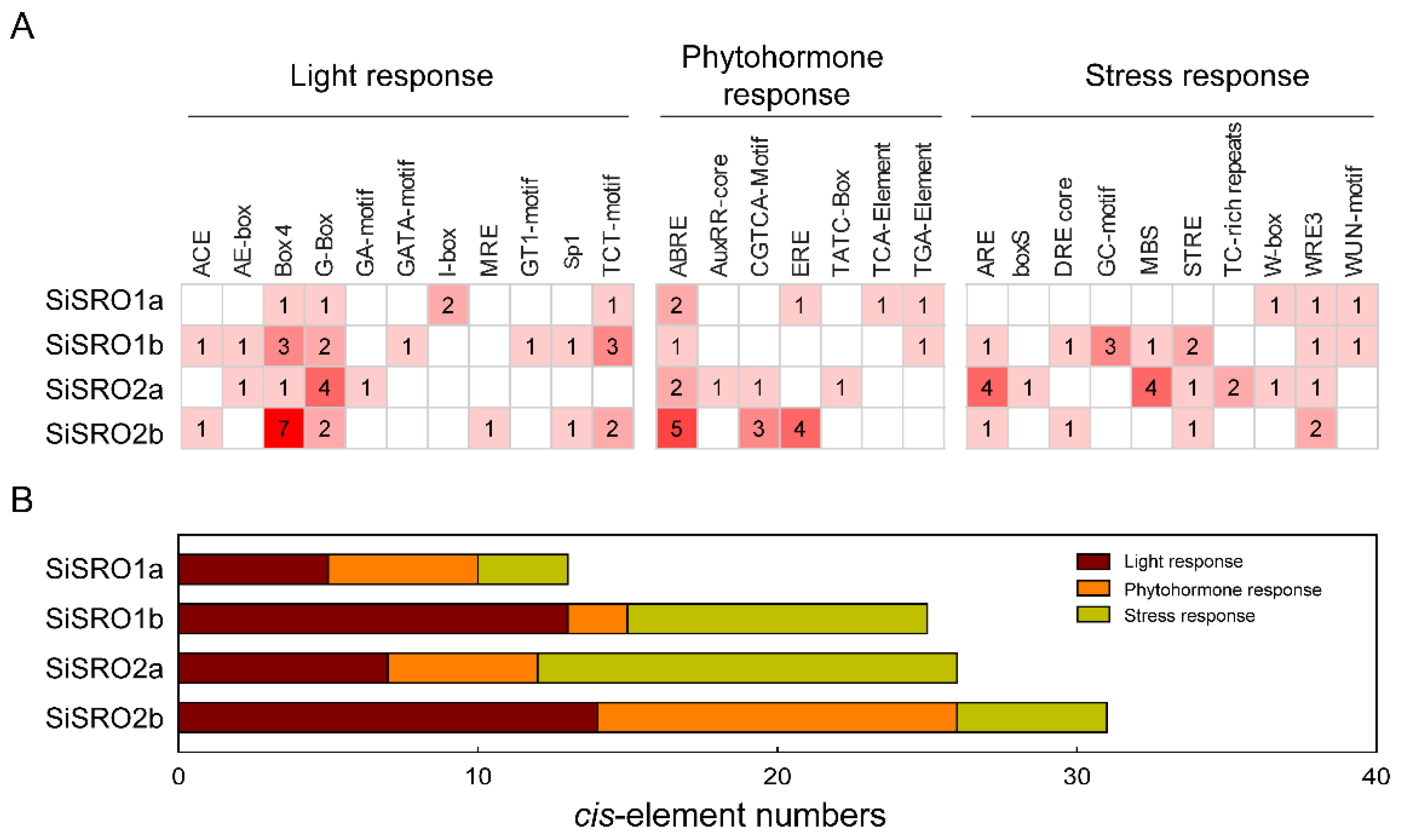
2.4. Cis-Acting Elements in the Promoters of SiSRO Genes
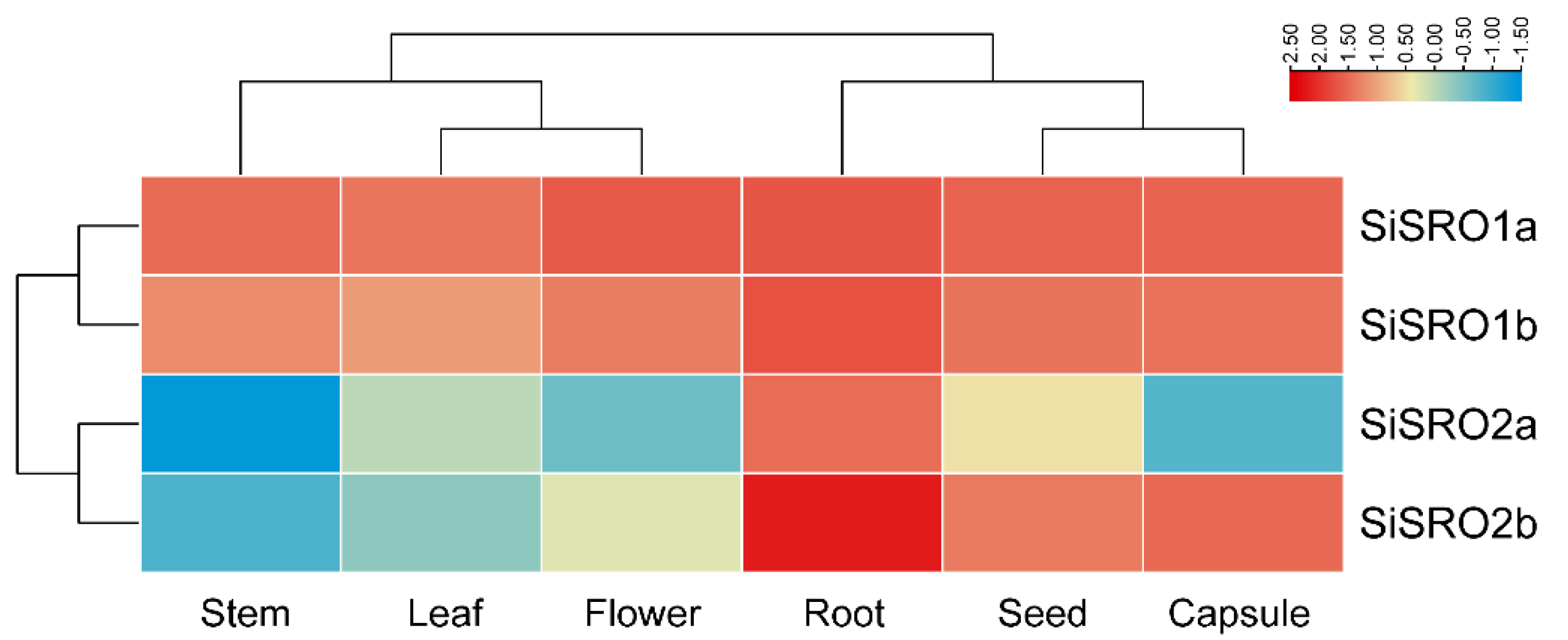
2.5. Expression Patterns of SiSROs in Various Tissues
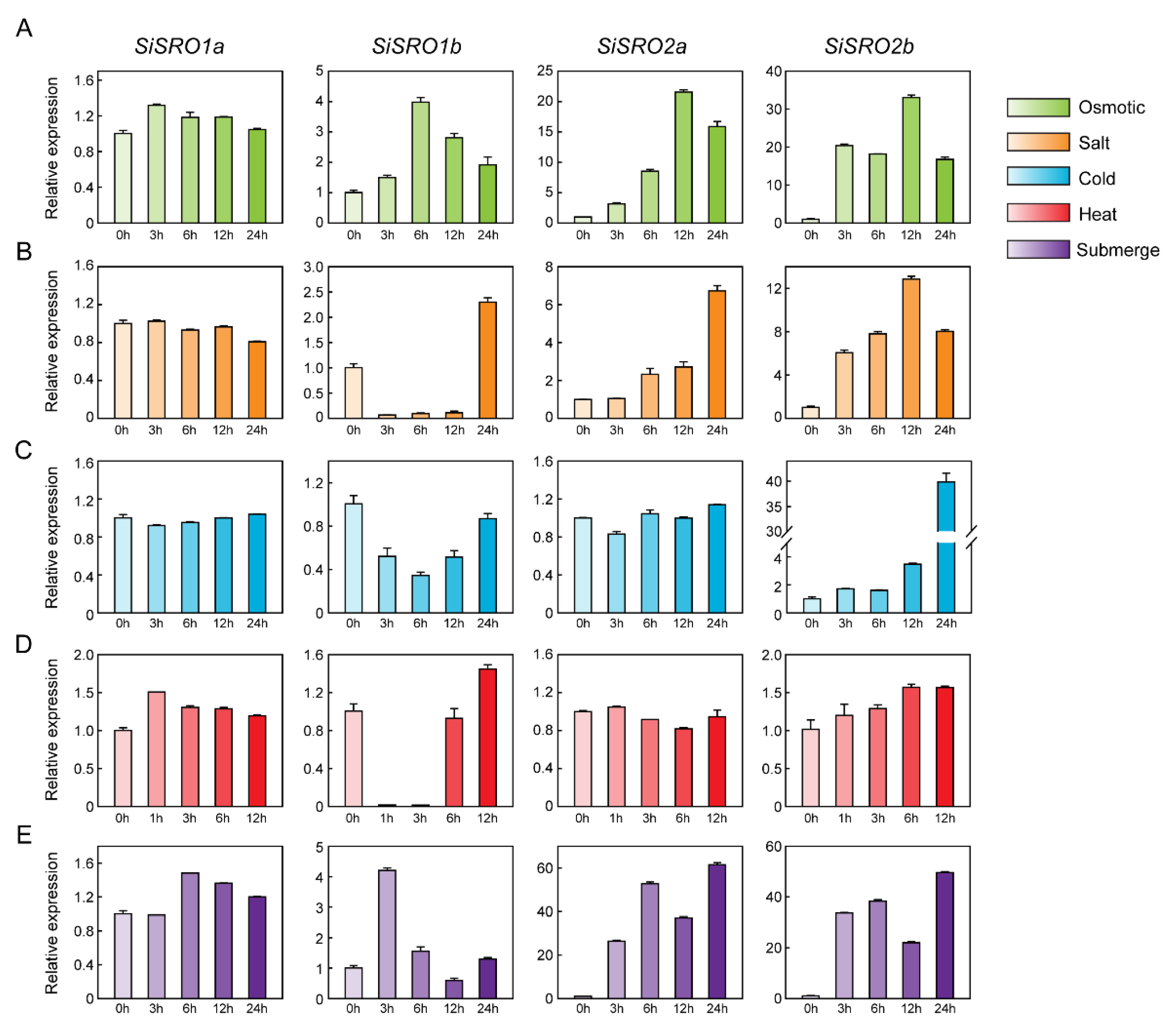
2.6. Expression Profiles of SiSROs under Abiotic Stress Treatments
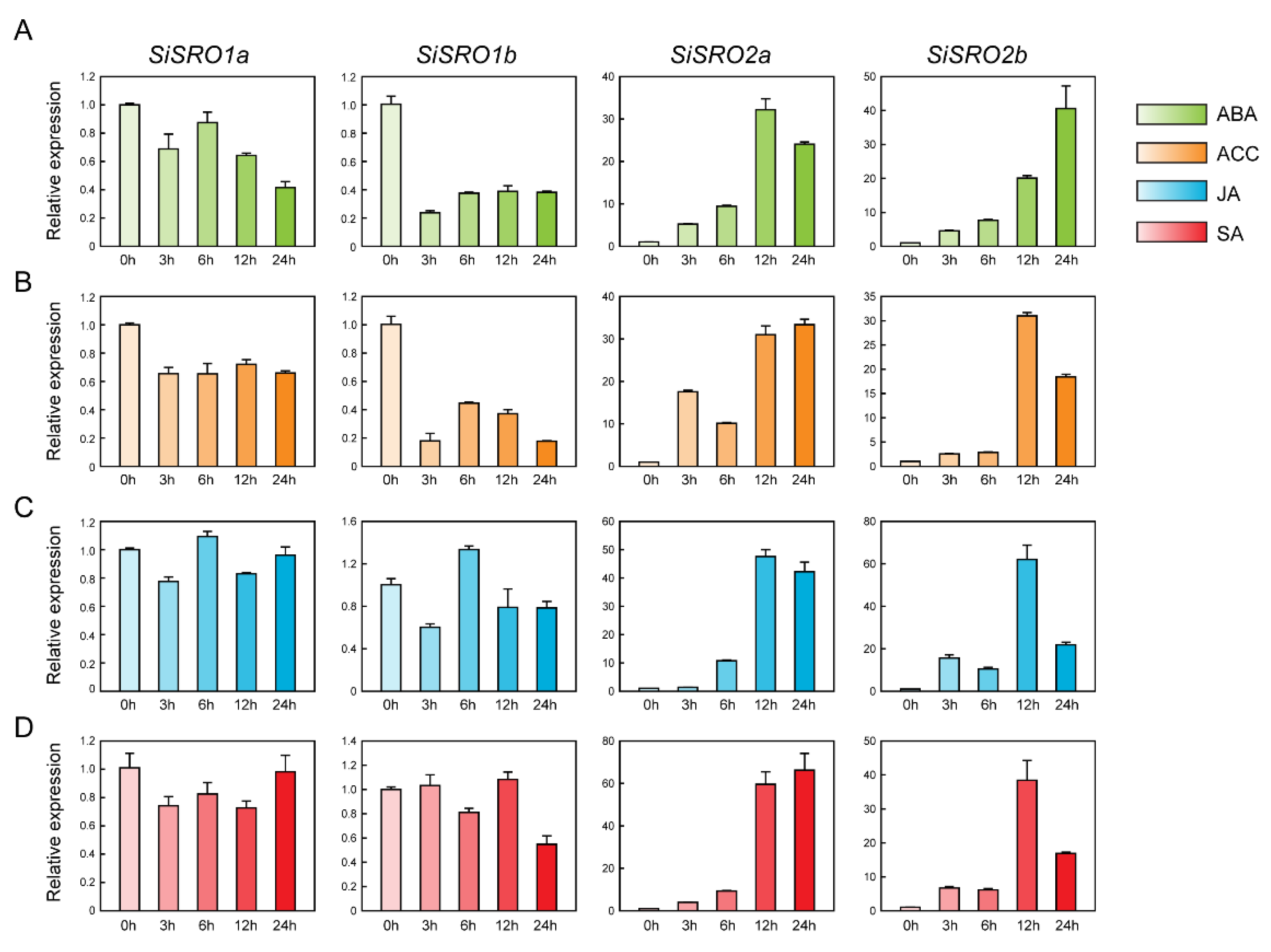
2.7. Expression Profiles of SiSROs under Phytohormone Treatments
2.8. Protein-Protein Interaction Network of SiSROs
2.9. Co-Expression Network of SiSROs under Abiotic Stress Conditions
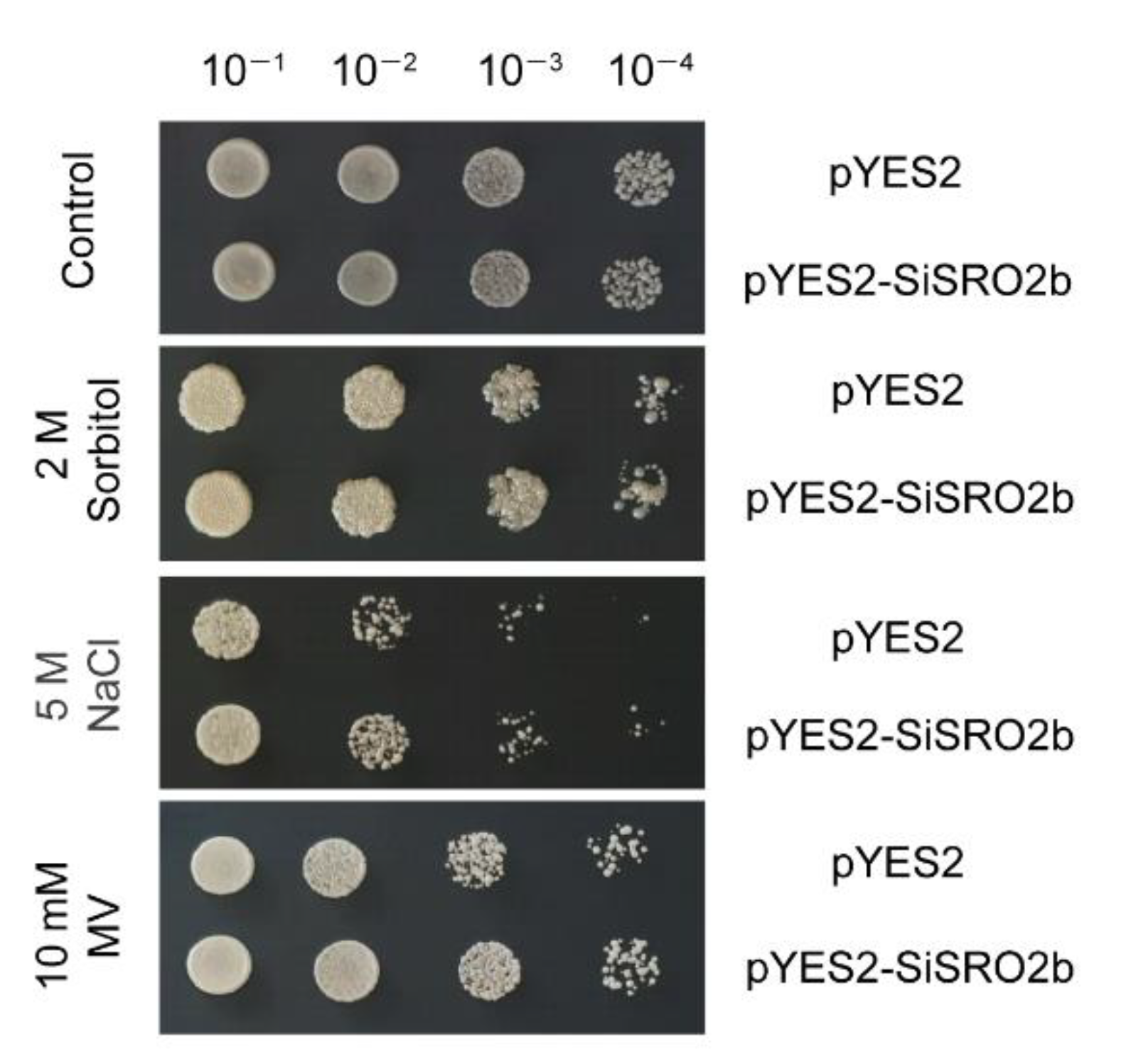
2.10. Expression of SiSRO2b in Yeast Enhances Tolerance to Multiple Abiotic Stresses
3. Discussion
4. Materials and Methods
4.1. Identification of SRO Family Genes and Phylogenetic Analysis
4.2. Gene Structure, Protein Conserved Motifs and cis-Acting Element Analysis
4.3. Protein Interaction and Co-Expression Analysis
4.4. Plant Materials and Treatments
4.5. RNA Isolation and Quantitative Real-Time RT-PCR
4.6. Cloning and Functional Characterization of SiSRO2b in Yeast
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chang, Y.; Zhu, C.; Jiang, J.; Zhang, H.; Zhu, J.; Duan, C. Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 2020, 62, 563–580. [Google Scholar] [CrossRef]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.-Y.; Li, J.; Wang, P.-Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef]
- Jaspers, P.; Overmyer, K.; Wrzaczek, M.; Vainonen, J.P.; Blomster, T.; Salojärvi, J.; A Reddy, R.; Kangasjärvi, J. The RST and PARP-like domain containing SRO protein family: Analysis of protein structure, function and conservation in land plants. BMC Genom. 2010, 11, 170. [Google Scholar] [CrossRef] [Green Version]
- Yuan, B.; Chen, M.; Li, S. Isolation and Identification of Ipomoea cairica (L.) Sweet Gene IcSRO1 Encoding a SIMILAR TO RCD-ONE Protein, Which Improves Salt and Drought Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2020, 21, 1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vainonen, J.P.; Jaspers, P.; Wrzaczek, M.; Lamminmäki, A.; Reddy, R.A.; Vaahtera, L.; Brosche, M.; Kangasjärvi, J. RCD1–DREB2A interaction in leaf senescence and stress responses in Arabidopsis thaliana. Biochem. J. 2012, 442, 573–581. [Google Scholar] [CrossRef] [Green Version]
- You, J.; Zong, W.; Du, H.; Hu, H.; Xiong, L. A special member of the rice SRO family, OsSRO1c, mediates responses to multiple abiotic stresses through interaction with various transcription factors. Plant Mol. Biol. 2014, 84, 693–705. [Google Scholar] [CrossRef]
- Jaspers, P.; Blomster, T.; Brosche, M.; Salojärvi, J.; Ahlfors, R.; Vainonen, J.P.; Reddy, R.A.; Immink, R.; Angenent, G.; Turck, F.; et al. Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant J. 2009, 60, 268–279. [Google Scholar] [CrossRef]
- You, J.; Zong, W.; Li, X.; Ning, J.; Hu, H.; Li, X.; Xiao, J.; Xiong, L. The SNAC1-targeted gene OsSRO1c modulates stomatal closure and oxidative stress tolerance by regulating hydrogen peroxide in rice. J. Exp. Bot. 2012, 64, 569–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Liu, S.; Wang, M.; Wei, T.; Meng, C.; Wang, M.; Xia, G. A Wheat SIMILAR TO RCD-ONE Gene Enhances Seedling Growth and Abiotic Stress Resistance by Modulating Redox Homeostasis and Maintaining Genomic Integrity. Plant Cell 2014, 26, 164–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Overmyer, K.; Tuominen, H.; Kettunen, R.; Betz, C.; Langebartels, C., Jr.; Kangasjärvi, J. Ozone-Sensitive Arabidopsis rcd1 Mutant Reveals Opposite Roles for Ethylene and Jasmonate Signaling Pathways in Regulating Superoxide-Dependent Cell Death. Plant Cell 2000, 12, 1849–1862. [Google Scholar] [CrossRef] [Green Version]
- Belles-Boix, E.; Babiychuk, E.; Van Montagu, M.; Inzé, D.; Kushnir, S. CEO1, a new protein fromArabidopsis thaliana, protects yeast against oxidative damage1. FEBS Lett. 2000, 482, 19–24. [Google Scholar] [CrossRef]
- Fujibe, T.; Saji, H.; Arakawa, K.; Yabe, N.; Takeuchi, Y.; Yamamoto, K.T. A Methyl Viologen-Resistant Mutant of Arabidopsis, Which Is Allelic to Ozone-Sensitive rcd1, Is Tolerant to Supplemental Ultraviolet-B Irradiation. Plant Physiol. 2004, 134, 275–285. [Google Scholar] [CrossRef] [Green Version]
- Ahlfors, R.; Lång, S.; Overmyer, K.; Jaspers, P.; Brosche, M.; Tauriainen, A.; Kollist, H.; Tuominen, H.; Belles-Boix, E.; Piippo, M.; et al. Arabidopsis RADICAL-INDUCED CELL DEATH1 Belongs to the WWE Protein–Protein Interaction Domain Protein Family and Modulates Abscisic Acid, Ethylene, and Methyl Jasmonate Responses. Plant Cell 2004, 16, 1925–1937. [Google Scholar] [CrossRef] [Green Version]
- Katiyar-Agarwal, S.; Zhu, J.; Kim, K.; Agarwal, M.; Fu, X.; Huang, A. The plasma membrane Na+/H+ antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 18816–18821. [Google Scholar] [CrossRef] [Green Version]
- Teotia, S.; Lamb, R.S. The Paralogous GenesRADICAL-INDUCED CELL DEATH1andSIMILAR TO RCD ONE1Have Partially Redundant Functions during Arabidopsis Development. Plant Physiol. 2009, 151, 180–198. [Google Scholar] [CrossRef] [Green Version]
- Teotia, S.; Lamb, R.S. RCD1 and SRO1 are necessary to maintain meristematic fate in Arabidopsis thaliana. J. Exp. Bot. 2010, 62, 1271–1284. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Su, J.; Xu, M.; Zhou, Z.; Zhu, X.; Ma, X.; Hou, J.; Tan, L.; Zhu, Z.; Cai, H.; et al. A common wild rice-derived BOC1 allele reduces callus browning in indica rice transformation. Nat. Commun. 2020, 11, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashihara, K.; Onohata, T.; Yariuchi, R.; Tanaka, S.; Akimitsu, K.; Gomi, K. The overexpression of OsSRO1a, which encodes an OsNINJA1- and OsMYC2-interacting protein, negatively affects OsMYC2-mediated jasmonate signaling in rice. Plant Cell Rep. 2020, 39, 489–500. [Google Scholar] [CrossRef]
- Li, H.; Li, R.; Qu, F.; Yao, J.; Hao, Y.; Wang, X.; You, C. Identification of the SRO gene family in apples (Malus×domestica) with a functional characterization of MdRCD1. Tree Genet. Genomes 2017, 13, 94. [Google Scholar] [CrossRef]
- Li, X.; Xu, Y.; Liu, F.; Zhao, M.; Sun, Y.; Ma, Q. Maize similar to RCD1 gene induced by salt enhances Arabidopsis thaliana abiotic stress resistance. Biochem. Biophys. Res. Commun. 2018, 503, 2625–2632. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Kaur, C.; Singla-Pareek, S.L.; Sopory, S.K. OsSRO1a Interacts with RNA Binding Domain-Containing Protein (OsRBD1) and Functions in Abiotic Stress Tolerance in Yeast. Front. Plant Sci. 2016, 7, 62. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Xiao, Y.; Zhu, S. Genome-wide identification, systematic analysis and characterization of SRO family genes in maize (Zea mays L.). Acta Physiol. Plant. 2018, 40, 176. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, D.; Hu, H.; Li, W.; Hu, Y.; Xie, J.; Huang, S.; Wang, W. Genome-wide characterization of a SRO gene family involved in response to biotic and abiotic stresses in banana (Musa spp.). BMC Plant Biol. 2019, 19, 211. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Gao, X.; Liu, Z.; Wu, Y.; Hu, L.; Yu, J. Genome-Wide Identification and Analysis of SRO Gene Family in Chinese Cabbage (Brassica rapa L.). Plants 2020, 9, 1235. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Geng, Y.; Liu, Y.; Chen, S.; Cao, S.; Li, W.; Chen, H.; Ma, D.; Yin, J. Genome-wide identification and characterization of SRO gene family in wheat: Molecular evolution and expression profiles during different stresses. Plant Physiol. Biochem. 2020, 154, 590–611. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xu, R.; Wang, B.; Wang, J.; Huang, S.; Yu, Q.; Gao, J. Genome-Wide Identification and Evolutionary Analysis of the SRO Gene Family in Tomato. Front. Genet. 2021, 12, 12. [Google Scholar] [CrossRef] [PubMed]
- Dossa, K.; Li, D.; Zhou, R.; Yu, J.; Wang, L.; Zhang, Y.; You, J.; Liu, A.; Mmadi, M.A.; Fonceka, D.; et al. The genetic basis of drought tolerance in the high oil crop Sesamum indicum. Plant Biotechnol. J. 2019, 17, 1788–1803. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Dossa, K.; You, J.; Zhang, Y.; Li, D.; Zhou, R.; Yu, J.; Wei, X.; Zhu, X.; Jiang, S.; et al. High-resolution temporal transcriptome sequencing unravels ERF and WRKY as the master players in the regulatory networks underlying sesame responses to waterlogging and recovery. Genomics 2021, 113, 276–290. [Google Scholar] [CrossRef]
- Qin, L.; Sun, L.; Wei, L.; Yuan, J.; Kong, F.; Zhang, Y.; Miao, X.; Xia, G.; Liu, S. Maize SRO1e represses anthocyanin synthesis through regulating the MBW complex in response to abiotic stress. Plant J. 2021, 105, 1010–1025. [Google Scholar] [CrossRef]
- Lee, H.K.; Cho, S.K.; Son, O.; Xu, Z.; Hwang, I.; Kim, W.T. Drought Stress-Induced Rma1H1, a RING Membrane-Anchor E3 Ubiquitin Ligase Homolog, Regulates Aquaporin Levels via Ubiquitination in TransgenicArabidopsisPlants. Plant Cell 2009, 21, 622–641. [Google Scholar] [CrossRef] [Green Version]
- Rausell, A.; Kanhonou, R.; Yenush, L.; Serrano, R.; Ros, R. The translation initiation factor eIF1A is an important determinant in the tolerance to NaCl stress in yeast and plants. Plant J. 2003, 34, 257–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alavilli, H.; Awasthi, J.P.; Rout, G.R.; Sahoo, L.; Lee, B.-H.; Panda, S.K. Overexpression of a Barley Aquaporin Gene, HvPIP2;5 Confers Salt and Osmotic Stress Tolerance in Yeast and Plants. Front. Plant Sci. 2016, 7, 7. [Google Scholar] [CrossRef] [Green Version]
- Wisecaver, J.H.; Borowsky, A.T.; Tzin, V.; Jander, G.; Kliebenstein, D.J.; Rokas, A. A Global Coexpression Network Approach for Connecting Genes to Specialized Metabolic Pathways in Plants. Plant Cell 2017, 29, 944–959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eserin, E.A.R.; Nijveen, H.; Hilhorst, H.; Eligterink, W. Learning from Co-expression Networks: Possibilities and Challenges. Front. Plant Sci. 2016, 7, 444. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Hou, C.; Zheng, K.; Li, Q.; Chen, S.; Wang, S. Overexpression of ERF96, a small ethylene response factor gene, enhances salt tolerance in Arabidopsis. Biol. Plant. 2017, 61, 693–701. [Google Scholar] [CrossRef]
- Muthusamy, M.; Kim, J.Y.; Yoon, E.K.; Kim, J.A.; Lee, S.I. BrEXLB1, a Brassica rapa Expansin-Like B1 Gene Is Associated with Root Development, Drought Stress Response, and Seed Germination. Genes 2020, 11, 404. [Google Scholar] [CrossRef] [Green Version]
- Park, H.-Y.; Seok, H.-Y.; Woo, D.-H.; Lee, S.-Y.; Tarte, V.N.; Lee, E.-H.; Lee, C.-H.; Moon, Y.-H. AtERF71/HRE2 transcription factor mediates osmotic stress response as well as hypoxia response in Arabidopsis. Biochem. Biophys. Res. Commun. 2011, 414, 135–141. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2020, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, T.J.; Eddy, S. nhmmer: DNA homology search with profile HMMs. Bioinformatics 2013, 29, 2487–2489. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools—An inte-grative toolkit developed for interactive analyses of big biological data. Molecules 2020. [Google Scholar]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, w202–w208. [Google Scholar] [CrossRef]
- You, J.; Zhang, Y.; Liu, A.; Li, D.; Wang, X.; Dossa, K.; Zhou, R.; Yu, J.; Zhang, Y.; Wang, L.; et al. Transcriptomic and metabolomic profiling of drought-tolerant and susceptible sesame genotypes in response to drought stress. BMC Plant Biol. 2019, 19, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Dossa, K.; Li, D.; Wang, L.; Zheng, X.; Liu, A.; Yu, J.; Wei, X.; Zhou, R.; Foncéka, D.; Diouf, D.; et al. Transcriptomic, biochemical and physio-anatomical investigations shed more light on responses to drought stress in two contrasting sesame genotypes. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Li, D.; Zhou, R.; Wang, X.; Dossa, K.; Wang, L.; Zhang, Y.; Yu, J.; Gong, H.; Zhang, X.; et al. Transcriptome and metabolome analyses of two contrasting sesame genotypes reveal the crucial biological pathways involved in rapid adaptive response to salt stress. BMC Plant Biol. 2019, 19, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Dossa, K.; You, J.; Wang, L.; Zhang, Y.; Li, D.; Zhou, R.; Yu, J.; Wei, X.; Zhu, X.; Jiang, S.; et al. Transcriptomic profiling of sesame during waterlogging and recovery. Sci. Data 2019, 6, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Poretsky, E.; Huffaker, A. MutRank: An R shiny web-application for exploratory targeted mutual rank-based coexpression analyses integrated with user-provided supporting information. PeerJ 2020, 8, e10264. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [Green Version]
- You, J.; Zhang, L.; Song, B.; Qi, X.; Chan, Z. Systematic Analysis and Identification of Stress-Responsive Genes of the NAC Gene Family in Brachypodium distachyon. PLoS ONE 2015, 10, e0122027. [Google Scholar] [CrossRef]
- You, J.; Wang, Y.; Zhang, Y.; Dossa, K.; Li, D.; Zhou, R.; Wang, L.; Zhang, X. Genome-wide identification and expression analyses of genes involved in raffinose accumulation in sesame. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Gietz, R.D.; Robbins, A.; Graham, K.C.; Triggs-Raine, B.; Woods, R.A. Identification of proteins that interact with a protein of interest: Applications of the yeast two-hybrid system. Mol. Cell. Biochem. 1997, 172, 67–79. [Google Scholar] [CrossRef]



| Gene Name | Gene ID | Linkage Group | Start (bp) | End (bp) | No. of Exon | Protein Length (aa) | MW (kD) | pI | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| SiSRO1a | LOC105155511 | LG2 | 6894030 | 6901957 | 6 | 560 | 62.73 | 6.36 | Chloroplast |
| SiSRO1b | LOC105159067 | LG3 | 21219139 | 21221593 | 4 | 481 | 54.39 | 8.29 | Nucleus |
| SiSRO2a | LOC105157214 | LG3 | 3993893 | 3996925 | 5 | 377 | 42.35 | 6.43 | Nucleus |
| SiSRO2b | LOC105179407 | Unplaced Scaffold NW_011628063.1 | 287017 | 290048 | 5 | 372 | 41.13 | 6.86 | Nucleus |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, A.; Wei, M.; Zhou, Y.; Li, D.; Zhou, R.; Zhang, Y.; Zhang, X.; Wang, L.; You, J. Comprehensive Analysis of SRO Gene Family in Sesamum indicum (L.) Reveals Its Association with Abiotic Stress Responses. Int. J. Mol. Sci. 2021, 22, 13048. https://doi.org/10.3390/ijms222313048
Liu A, Wei M, Zhou Y, Li D, Zhou R, Zhang Y, Zhang X, Wang L, You J. Comprehensive Analysis of SRO Gene Family in Sesamum indicum (L.) Reveals Its Association with Abiotic Stress Responses. International Journal of Molecular Sciences. 2021; 22(23):13048. https://doi.org/10.3390/ijms222313048
Chicago/Turabian StyleLiu, Aili, Mengyuan Wei, Yong Zhou, Donghua Li, Rong Zhou, Yanxin Zhang, Xiurong Zhang, Linhai Wang, and Jun You. 2021. "Comprehensive Analysis of SRO Gene Family in Sesamum indicum (L.) Reveals Its Association with Abiotic Stress Responses" International Journal of Molecular Sciences 22, no. 23: 13048. https://doi.org/10.3390/ijms222313048
APA StyleLiu, A., Wei, M., Zhou, Y., Li, D., Zhou, R., Zhang, Y., Zhang, X., Wang, L., & You, J. (2021). Comprehensive Analysis of SRO Gene Family in Sesamum indicum (L.) Reveals Its Association with Abiotic Stress Responses. International Journal of Molecular Sciences, 22(23), 13048. https://doi.org/10.3390/ijms222313048









