Autonomous and Assisted Control for Synthetic Microbiology
Abstract
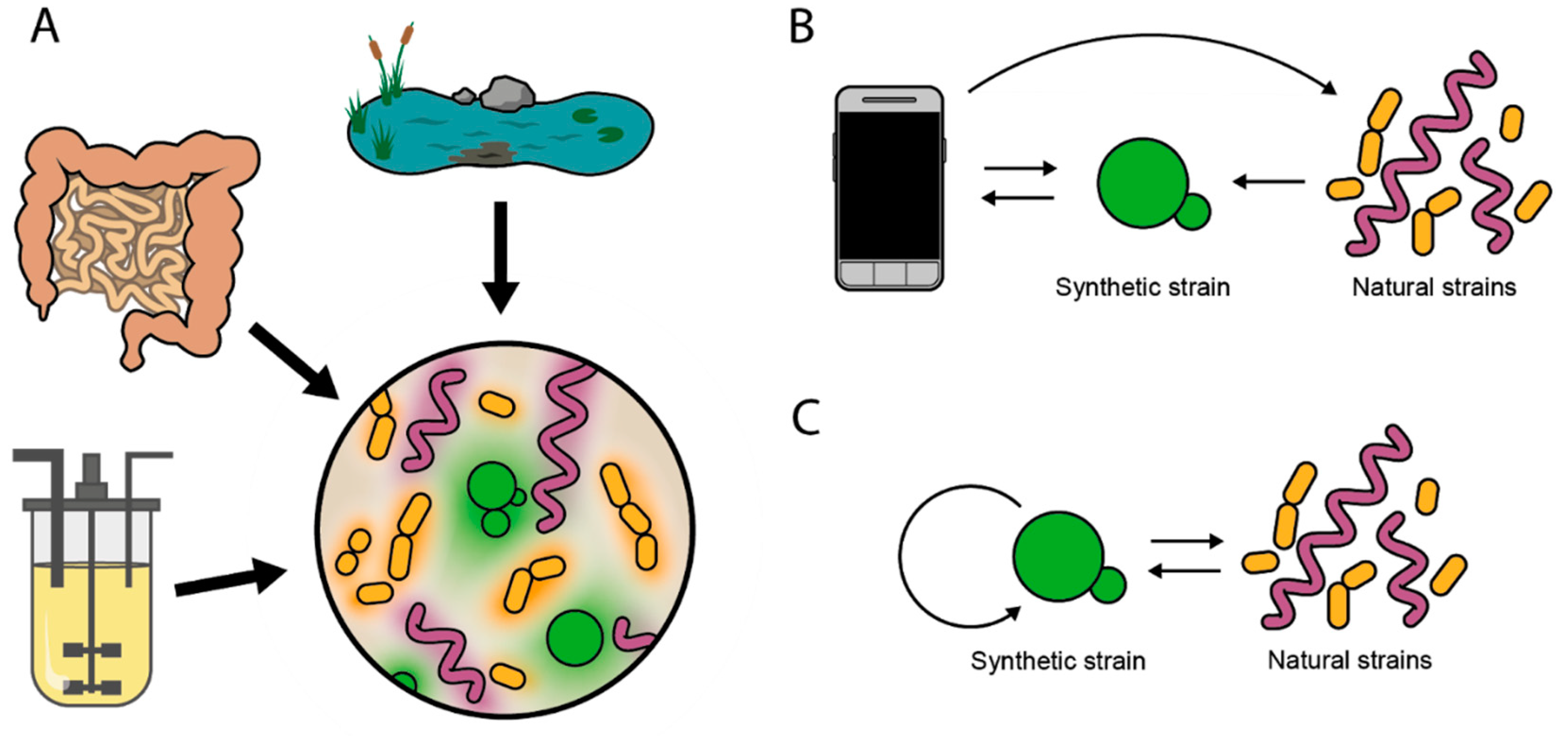
1. Introduction
2. Natural Robust Control
2.1. Perfect Adaptation and Relative Sensing of Stimuli
2.2. Sensing Relative Population Composition
3. Synthetic Population Control
4. Cybergenetic Control
4.1. External (Computer-Aided) Control
4.2. Internal Cybergenetic Control
5. Interactions between Controllers and Natural Populations
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wood, J.M. Bacterial osmoregulation: A paradigm for the study of cellular homeostasis. Annu. Rev. Microbiol. 2011, 65, 215–238. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, J.; Boersma, A.J.; Poolman, B. Microorganisms maintain crowding homeostasis. Nat. Rev. Microbiol. 2017, 15, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Kitano, H. Towards a theory of biological robustness. Mol. Syst. Biol. 2007, 3, 137. [Google Scholar] [CrossRef]
- Masel, J.; Siegal, M.L. Robustness: Mechanisms and consequences. Trends Genet. 2009, 25, 395–403. [Google Scholar] [CrossRef]
- Khammash, M. An engineering viewpoint on biological robustness. BMC Biol. 2016, 14, 22. [Google Scholar] [CrossRef]
- Arkin, A.P. A wise consistency: Engineering biology for conformity, reliability, predictability. Curr. Opin. Chem. Biol. 2013, 17, 893–901. [Google Scholar] [CrossRef]
- Xie, M.; Fussenegger, M. Designing cell function: Assembly of synthetic gene circuits for cell biology applications. Nat. Rev. Mol. Cell Biol. 2018, 19, 507–525. [Google Scholar] [CrossRef]
- Yi, T.M.; Huang, Y.; Simon, M.I.; Doyle, J. Robust perfect adaptation in bacterial chemotaxis through integral feedback control. Proc. Natl. Acad. Sci. USA 2000, 97, 4649–4653. [Google Scholar] [CrossRef]
- Barkai, N.; Leibler, S. Robustness in simple biochemical networks. Nature 1997, 387, 913–917. [Google Scholar] [CrossRef] [PubMed]
- Alon, U.; Surette, M.G.; Barkai, N.; Leibler, S. Robustness in bacterial chemotaxis. Nature 1999, 397, 168–171. [Google Scholar] [CrossRef]
- Adler, M.; Mayo, A.; Alon, U. Logarithmic and power law input-output relations in sensory systems with fold-change detection. PLoS Comput. Biol. 2014, 10, e1003781. [Google Scholar] [CrossRef] [PubMed]
- Ferrell, J.E. Signaling Motifs and Weber’s Law. Mol. Cell 2009, 36, 724–727. [Google Scholar] [CrossRef] [PubMed]
- Daniel, R.; Rubens, J.R.; Sarpeshkar, R.; Lu, T.K. Synthetic analog computation in living cells. Nature 2013, 497, 619–623. [Google Scholar] [CrossRef] [PubMed]
- Colin, R.; Sourjik, V. Emergent properties of bacterial chemotaxis pathway. Curr. Opin. Microbiol. 2017, 39, 24–33. [Google Scholar] [CrossRef]
- Kalinin, Y.V.; Jiang, L.; Tu, Y.; Wu, M. Logarithmic sensing in Escherichia coli bacterial chemotaxis. Biophys. J. 2009, 96, 2439–2448. [Google Scholar] [CrossRef]
- Sourjik, V.; Wingreen, N.S. Responding to chemical gradients: Bacterial chemotaxis. Curr. Opin. Cell Biol. 2012, 24, 262–268. [Google Scholar] [CrossRef]
- Shoval, O.; Goentoro, L.; Hart, Y.; Mayo, A.; Sontag, E.; Alon, U. Fold-change detection and scalar symmetry of sensory input fields. Proc. Natl. Acad. Sci. USA 2010, 107, 15995–16000. [Google Scholar] [CrossRef]
- Goentoro, L.; Shoval, O.; Kirschner, M.W.; Alon, U. The incoherent feedforward loop can provide fold-change detection in gene regulation. Mol. Cell 2009, 36, 894–899. [Google Scholar] [CrossRef]
- Adler, M.; Alon, U. Fold-change detection in biological systems. Curr. Opin. Syst. Biol. 2018, 8, 81–89. [Google Scholar] [CrossRef]
- Kim, J.; Khetarpal, I.; Sen, S.; Murray, R.M. Synthetic circuit for exact adaptation and fold-change detection. Nucleic Acids Res. 2014, 42, 6078–6089. [Google Scholar] [CrossRef]
- Banderas, A.; Koltai, M.; Anders, A.; Sourjik, V. Sensory input attenuation allows predictive sexual response in yeast. Nat. Commun. 2016, 7, 12590. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Babel, H.; Naranjo-Meneses, P.; Trauth, S.; Schulmeister, S.; Malengo, G.; Sourjik, V.; Bischofs, I.B. Ratiometric population sensing by a pump-probe signaling system in Bacillus subtilis. Nat. Commun. 2020, 11, 1176. [Google Scholar] [CrossRef] [PubMed]
- Banderas, A.; Carcano, A.; Sia, E.; Li, S.; Lindner, A.B. Ratiometric quorum sensing governs the trade-off between bacterial vertical and horizontal antibiotic resistance propagation. PLoS Biol. 2020, 18, e3000814. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Cook, L.C.C.; Shu, C.-C.; Chen, Y.; Manias, D.A.; Ramkrishna, D.; Dunny, G.M.; Hu, W.-S. Antagonistic self-sensing and mate-sensing signaling controls antibiotic-resistance transfer. Proc. Natl. Acad. Sci. USA 2013, 110, 7086–7090. [Google Scholar] [CrossRef] [PubMed]
- Antebi, Y.E.; Linton, J.M.; Klumpe, H.; Bintu, B.; Gong, M.; Su, C.; McCardell, R.; Elowitz, M.B. Combinatorial Signal Perception in the BMP Pathway. Cell 2017, 170, 1184–1196. [Google Scholar] [CrossRef] [PubMed]
- Alnahhas, R.N.; Sadeghpour, M.; Chen, Y.; Frey, A.A.; Ott, W.; Josić, K.; Bennett, M.R. Majority sensing in synthetic microbial consortia. Nat. Commun. 2020, 11, 3659. [Google Scholar] [CrossRef]
- Giri, S.; Shitut, S.; Kost, C. Harnessing ecological and evolutionary principles to guide the design of microbial production consortia. Curr. Opin. Biotechnol. 2020, 62, 228–238. [Google Scholar] [CrossRef]
- McCarty, N.S.; Ledesma-Amaro, R. Synthetic Biology Tools to Engineer Microbial Communities for Biotechnology. Trends Biotechnol. 2019, 37, 181–197. [Google Scholar] [CrossRef]
- Mee, M.T.; Collins, J.J.; Church, G.M.; Wang, H.H. Syntrophic exchange in synthetic microbial communities. Proc. Natl. Acad. Sci. USA 2014, 111, E2149–E2156. [Google Scholar] [CrossRef]
- Scott, S.R.; Din, M.O.; Bittihn, P.; Xiong, L.; Tsimring, L.S.; Hasty, J. A stabilized microbial ecosystem of self-limiting bacteria using synthetic quorum-regulated lysis. Nat. Microbiol. 2017, 2, 17083. [Google Scholar] [CrossRef]
- Du, P.; Zhao, H.; Zhang, H.; Wang, R.; Huang, J.; Tian, Y.; Luo, X.; Luo, X.; Wang, M.; Xiang, Y.; et al. De novo design of an intercellular signaling toolbox for multi-channel cell-cell communication and biological computation. Nat. Commun. 2020, 11, 4226. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; He, X.; Luo, Y.; Mu, Y.; Gu, F.; Liang, Q.; Qi, Q. Two Completely Orthogonal Quorum Sensing Systems with Self-Produced Autoinducers Enable Automatic Delayed Cascade Control. ACS Synth. Biol. 2020, 9, 2588–2599. [Google Scholar] [CrossRef] [PubMed]
- Miano, A.; Liao, M.J.; Hasty, J. Inducible cell-to-cell signaling for tunable dynamics in microbial communities. Nat. Commun. 2020, 11, 1193. [Google Scholar] [CrossRef] [PubMed]
- Kylilis, N.; Tuza, Z.A.; Stan, G.-B.; Polizzi, K.M. Tools for engineering coordinated system behaviour in synthetic microbial consortia. Nat. Commun. 2018, 9, 2677. [Google Scholar] [CrossRef]
- Khammash, M.; Di Bernardo, M.; Di Bernardo, D. Cybergenetics: Theory and Methods for Genetic Control System. In Proceedings of the 2019 IEEE 58th Conference on Decision and Control (CDC), Nice, France, 11–13 December 2019; pp. 916–926. [Google Scholar]
- Lugagne, J.-B.; Dunlop, M.J. Cell-machine interfaces for characterizing gene regulatory network dynamics. Curr. Opin. Syst. Biol. 2019, 14, 1–8. [Google Scholar] [CrossRef]
- Baetica, A.-A.; Westbrook, A.; El-Samad, H. Control theoretical concepts for synthetic and systems biology. Curr. Opin. Syst. Biol. 2019, 14, 50–57. [Google Scholar] [CrossRef]
- Carrasco-López, C.; García-Echauri, S.A.; Kichuk, T.; Avalos, J.L. Optogenetics and biosensors set the stage for metabolic cybergenetics. Curr. Opin. Biotechnol. 2020, 65, 296–309. [Google Scholar] [CrossRef]
- Lalwani, M.A.; Zhao, E.M.; Avalos, J.L. Current and future modalities of dynamic control in metabolic engineering. Curr. Opin. Biotechnol. 2018, 52, 56–65. [Google Scholar] [CrossRef]
- Harrigan, P.; Madhani, H.D.; El-Samad, H. Real-Time Genetic Compensation Defines the Dynamic Demands of Feedback Control. Cell 2018, 175, 877–886. [Google Scholar] [CrossRef]
- Rullan, M.; Benzinger, D.; Schmidt, G.W.; Milias-Argeitis, A.; Khammash, M. An Optogenetic Platform for Real-Time, Single-Cell Interrogation of Stochastic Transcriptional Regulation. Mol. Cell 2018, 70, 745–756. [Google Scholar] [CrossRef]
- Chait, R.; Ruess, J.; Bergmiller, T.; Tkačik, G.; Guet, C.C. Shaping bacterial population behavior through computer-interfaced control of individual cells. Nat. Commun. 2017, 8, 1535. [Google Scholar] [CrossRef] [PubMed]
- Perkins, M.L.; Benzinger, D.; Arcak, M.; Khammash, M. Cell-in-the-loop pattern formation with optogenetically emulated cell-to-cell signaling. Nat. Commun. 2020, 11, 1355. [Google Scholar] [CrossRef]
- Lugagne, J.-B.; Sosa Carrillo, S.; Kirch, M.; Köhler, A.; Batt, G.; Hersen, P. Balancing a genetic toggle switch by real-time feedback control and periodic forcing. Nat. Commun. 2017, 8, 1671. [Google Scholar] [CrossRef] [PubMed]
- Guarino, A.; Fiore, D.; Salzano, D.; di Bernardo, M. Balancing Cell Populations Endowed with a Synthetic Toggle Switch via Adaptive Pulsatile Feedback Control. ACS Synth. Biol. 2020, 9, 793–803. [Google Scholar] [CrossRef] [PubMed]
- Salzano, D.; Fiore, D.; di Bernardo, M. Ratiometric control for differentiation of cell populations endowed with synthetic toggle switches. In Proceedings of the 2019 IEEE 58th Conference on Decision and Control (CDC), Nice, France, 11–13 December 2019. [Google Scholar]
- Mangan, S.; Alon, U. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. USA 2003, 100, 11980–11985. [Google Scholar] [CrossRef]
- Segall-Shapiro, T.H.; Sontag, E.D.; Voigt, C.A. Engineered promoters enable constant gene expression at any copy number in bacteria. Nat. Biotechnol. 2018, 36, 352–358. [Google Scholar] [CrossRef]
- Lillacci, G.; Benenson, Y.; Khammash, M. Synthetic control systems for high performance gene expression in mammalian cells. Nucleic Acids Res. 2018, 46, 9855–9863. [Google Scholar] [CrossRef]
- Briat, C.; Gupta, A.; Khammash, M. Antithetic Integral Feedback Ensures Robust Perfect Adaptation in Noisy Biomolecular Networks. Cell Syst. 2016, 2, 15–26. [Google Scholar] [CrossRef]
- Aoki, S.K.; Lillacci, G.; Gupta, A.; Baumschlager, A.; Schweingruber, D.; Khammash, M. A universal biomolecular integral feedback controller for robust perfect adaptation. Nature 2019, 570, 533–537. [Google Scholar] [CrossRef]
- Hsiao, V.; de los Santos, E.L.C.; Whitaker, W.R.; Dueber, J.E.; Murray, R.M. Design and implementation of a biomolecular concentration tracker. ACS Synth. Biol. 2015, 4, 150–161. [Google Scholar] [CrossRef]
- Chen, D.; Arkin, A.P. Sequestration-based bistability enables tuning of the switching boundaries and design of a latch. Mol. Syst. Biol. 2012, 8, 620. [Google Scholar] [CrossRef] [PubMed]
- Devkota, S.R.; Kwon, E.; Ha, S.C.; Chang, H.W.; Kim, D.Y. Structural insights into the regulation of Bacillus subtilis SigW activity by anti-sigma RsiW. PLoS ONE 2017, 12, e0174284. [Google Scholar] [CrossRef] [PubMed]
- Schöbel, S.; Zellmeier, S.; Schumann, W.; Wiegert, T. The Bacillus subtilis sigmaW anti-sigma factor RsiW is degraded by intramembrane proteolysis through YluC. Mol. Microbiol. 2004, 52, 1091–1105. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, M.; Gómez-Schiavon, M.; Ng, A.H.; El-Samad, H. Design and Analysis of a Proportional-Integral-Derivative Controller with Biological Molecules. Cell Syst. 2019, 9, 338–353. [Google Scholar] [CrossRef]
- Fiore, D.; Salzano, D.; Cristòbal-Cóppulo, E.; Olm, J.M.; di Bernardo, M. Multicellular Feedback Control of a Genetic Toggle-Switch in Microbial Consortia. IEEE Control Syst. Lett. 2020, 5, 151–156. [Google Scholar] [CrossRef]
- Agrawal, D.K.; Marshall, R.; Noireaux, V.; Sontag, E.D. In vitro implementation of robust gene regulation in a synthetic biomolecular integral controller. Nat. Commun. 2019, 10, 5760. [Google Scholar] [CrossRef]
- Briat, C.; Khammash, M. Perfect Adaptation and Optimal Equilibrium Productivity in a Simple Microbial Biofuel Metabolic Pathway Using Dynamic Integral Control. ACS Synth. Biol. 2018, 7, 419–431. [Google Scholar] [CrossRef]
- Danino, T.; Prindle, A.; Kwong, G.A.; Skalak, M.; Li, H.; Allen, K.; Hasty, J.; Bhatia, S.N. Programmable probiotics for detection of cancer in urine. Sci. Transl. Med. 2015, 7, 289ra84. [Google Scholar] [CrossRef]
- Chowdhury, S.; Castro, S.; Coker, C.; Hinchliffe, T.E.; Arpaia, N.; Danino, T. Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat. Med. 2019, 25, 1057–1063. [Google Scholar] [CrossRef]
- Din, M.O.; Danino, T.; Prindle, A.; Skalak, M.; Selimkhanov, J.; Allen, K.; Julio, E.; Atolia, E.; Tsimring, L.S.; Bhatia, S.N.; et al. Synchronized cycles of bacterial lysis for in vivo delivery. Nature 2016, 536, 81–85. [Google Scholar] [CrossRef]
- Saeidi, N.; Wong, C.K.; Lo, T.-M.; Nguyen, H.X.; Ling, H.; Leong, S.S.J.; Poh, C.L.; Chang, M.W. Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Mol. Syst. Biol. 2011, 7, 521. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.Y.; Tan, M.H.; Koh, E.; Ho, C.L.; Poh, C.L.; Chang, M.W. Reprogramming microbes to be pathogen-seeking killers. ACS Synth. Biol. 2014, 3, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Pandi, A.; Koch, M.; Voyvodic, P.L.; Soudier, P.; Bonnet, J.; Kushwaha, M.; Faulon, J.-L. Metabolic perceptrons for neural computing in biological systems. Nat. Commun. 2019, 10, 3880. [Google Scholar] [CrossRef] [PubMed]
- Mimee, M.; Tucker, A.C.; Voigt, C.A.; Lu, T.K. Programming a Human Commensal Bacterium, to Sense and Respond to Stimuli in the Murine Gut Microbiota. Cell Syst. 2015, 1, 62–71. [Google Scholar] [CrossRef]
- Platt, T.G.; Fuqua, C. What’s in a name? The semantics of quorum sensing. Trends Microbiol. 2010, 18, 383–387. [Google Scholar] [CrossRef]
- Youk, H.; Lim, W.A. Secreting and sensing the same molecule allows cells to achieve versatile social behaviors. Science 2014, 343, 1242782. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Banderas, A.; Le Bec, M.; Cordier, C.; Hersen, P. Autonomous and Assisted Control for Synthetic Microbiology. Int. J. Mol. Sci. 2020, 21, 9223. https://doi.org/10.3390/ijms21239223
Banderas A, Le Bec M, Cordier C, Hersen P. Autonomous and Assisted Control for Synthetic Microbiology. International Journal of Molecular Sciences. 2020; 21(23):9223. https://doi.org/10.3390/ijms21239223
Chicago/Turabian StyleBanderas, Alvaro, Matthias Le Bec, Céline Cordier, and Pascal Hersen. 2020. "Autonomous and Assisted Control for Synthetic Microbiology" International Journal of Molecular Sciences 21, no. 23: 9223. https://doi.org/10.3390/ijms21239223
APA StyleBanderas, A., Le Bec, M., Cordier, C., & Hersen, P. (2020). Autonomous and Assisted Control for Synthetic Microbiology. International Journal of Molecular Sciences, 21(23), 9223. https://doi.org/10.3390/ijms21239223




