Microbial Upgrading of Acetate into Value-Added Products—Examining Microbial Diversity, Bioenergetic Constraints and Metabolic Engineering Approaches
Abstract
1. Introduction
2. Acetate Metabolism in Different Organisms
2.1. The Aerobic Prokaryotic Acetate Metabolism
2.2. The Aerobic Eukaryotic Acetate Metabolism
2.3. The Anaerobic Prokaryotic Acetate Metabolism
2.3.1. Clostridium kluyveri
2.3.2. Sulfate-Reducing Bacteria
3. Products from Acetate
3.1. Acetate-Based Production in Different Microorganisms
3.2. Acetate-Based Production in E. coli
| Product | Energy Balanced | Theoretical Yield | Theoretical Carbon Yield | Max. Reported Yield (Aerobic) | Reference | |
|---|---|---|---|---|---|---|
| [yes/no] | [mol/mol] | [g/g] | [%] | |||
| Acetoin | No | 0.25 | 0.37 | 50 | 0.09 g/g 1 | [115] |
| Yes | 0.29 | 0.44 | 58 | |||
| Acetone | No | 0.50 | 0.49 | 75 | 0.29 mol/mol | [113] |
| Yes | 0.39 | 0.38 | 58 | |||
| N-Acetylglutamate | No | 0.25 | 0.80 | 88 | n.a. | [116] |
| Yes | 0.22 | 0.70 | 77 | |||
| 2,3-Butanediol | No | 0.25 | 0.38 | 50 | 0.09 g/g 1 | [115] |
| Yes | 0.27 | 0.41 | 54 | |||
| β-caryophyllene | No | 0.11 | 0.38 | 83 | 0.02 g/g | [33] |
| Yes | 0.07 | 0.24 | 51 | |||
| Glycolate | No | n.a. | n.a. | n.a. | 0.58 g/g | [117] |
| Yes | 1.00 | 1.27 | 100 | |||
| 3-Hydroxybutyrate (PHB) | No | 0.50 | 0.87 | 100 | 0.25 g/g | [118] |
| Yes | 0.35 | 0.61 | 70 | |||
| 3-Hydroxypropionic acid | No | 1.00 | 1.51 | 150 | 0.30 g/g | [119] |
| Yes | 0.50 | 0.75 | 75 | |||
| Isobutanol | No | n.a. | n.a. | n.a. | 0.025 mol/mol | [120] |
| Yes | 0.25 | 0.31 | 50 | |||
| Isopropanol | No | 0.50 | 0.51 | 75 | 0.56 mol/mol | [114] |
| Yes | 0.35 | 0.36 | 53 | |||
| Itaconic acid | No | n.a. | n.a. | n.a. | 0.07 mol/mol | [60] |
| Yes | 0.33 | 0.73 | 83 | |||
| Mevalonate | No | 0.33 | 0.84 | 100 | 0.30 g/g | [121] |
| Yes | 0.23 | 0.57 | 68 | |||
| MNEI | Single chain of the sweet plant protein monellin; no stoichiometric calculation of yields possible | [122] | ||||
| Phloroglucinol | No | 0.33 | 0.71 | 100 | 0.18 g/g | [123,124] |
| Yes | 0.24 | 0.52 | 72 | |||
| Succinate | No | 0.50 | 1.00 | 100 | 0.46 mol/mol | [125,126,127] |
| Yes | 0.44 | 0.88 | 88 | |||
| Tyrosine | No | n.a. | n.a. | n.a. | 0.04 g/g | [128] |
| Yes | 0.13 | 0.38 | 56 | |||
3.3. Comparability between Products from Acetate in E. coli
4. Energetic Comparison between Acetate, Glycerol, and Glucose as Carbon Sources in E. coli under Aerobic Conditions
5. Engineering of E. coli to Optimize Productivity on Acetate
5.1. Metabolic Engineering
5.1.1. Pathway Engineering
5.1.2. Acid/Acetate Tolerance
5.1.3. Product Tolerance
5.1.4. Cofactor Engineering
5.2. Process Engineering
6. Outlook
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| TCA cycle | Tricarboxylic acid cycle |
| MTY | Maximum theoretical yield |
| MTCY | Maximum theoretical carbon yield |
References
- Clark, D.P. The fermentation pathways of Escherichia coli. FEMS Microbiol. Lett. 1989, 63, 223–234. [Google Scholar] [CrossRef]
- Landwall, P.; Holme, T. Influence of glucose and dissolved oxygen concentrations on yields of Escherichia coli B in dialysis culture. J. Gen. Microbiol. 1977, 103, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.W.; Meyer, H.-P.; Fiechter, A. Continuous production of human leukocyte interferon with Escherichia coli and continuous cell lysis in a two stage chemostat. Appl. Microbiol. Biotechnol. 1985, 23, 5–9. [Google Scholar] [CrossRef]
- Jensen, E.B.; Carlsen, S. Production of recombinant human growth hormone in Escherichia coli: Expression of different precursors and physiological effects of glucose, acetate, and salts. Biotechnol. Bioeng. 1990, 36, 1–11. [Google Scholar] [CrossRef]
- MacDonald, H.L.; Neway, J.O. Effects of medium quality on the expression of human interleukin-2 at high cell density in fermentor cultures of Escherichia coli K-12. Appl. Environ. Microbiol. 1990, 56, 640–645. [Google Scholar] [CrossRef]
- Archer, C.T.; Kim, J.F.; Jeong, H.; Park, J.H.; Vickers, C.E.; Lee, S.Y.; Nielsen, L.K. The genome sequence of E. coli W (ATCC 9637): Comparative genome analysis and an improved genome-scale reconstruction of E. coli. BMC Genom. 2011, 12, 9. [Google Scholar] [CrossRef]
- Lee, S.Y.; Chang, H.N. High cell density cultivation of Escherichia coli W using sucrose as a carbon source. Biotechnol. Lett. 1993, 15, 971–974. [Google Scholar] [CrossRef]
- Luli, G.W.; Strohl, W.R. Comparison of growth, acetate production, and acetate inhibition of Escherichia coli strains in batch and fed-batch fermentations. Appl. Environ. Microbiol. 1990, 56, 1004–1011. [Google Scholar] [CrossRef]
- Basan, M.; Hui, S.; Okano, H.; Zhang, Z.; Shen, Y.; Williamson, J.R.; Hwa, T. Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature 2015, 528, 99–104. [Google Scholar] [CrossRef]
- Valgepea, K.; Adamberg, K.; Nahku, R.; Lahtvee, P.-J.; Arike, L.; Vilu, R. Systems biology approach reveals that overflow metabolism of acetate in Escherichia coli is triggered by carbon catabolite repression of acetyl-CoA synthetase. BMC Syst. Biol. 2010, 4, 166. [Google Scholar] [CrossRef]
- Vemuri, G.N.; Altman, E.; Sangurdekar, D.P.; Khodursky, A.B.; Eiteman, M.A. Overflow metabolism in Escherichia coli during steady-state growth: Transcriptional regulation and effect of the redox ratio. Appl. Env. Microbiol. 2006, 72, 3653–3661. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Eiteman, M.A. Acetate formation during recombinant protein production in Escherichia coli K-12 with an elevated NAD(H) pool. Eng. Life Sci. 2019, 19, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Pinhal, S.; Ropers, D.; Geiselmann, J.; de Jong, H. Acetate Metabolism and the Inhibition of Bacterial Growth by Acetate. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef] [PubMed]
- Roe, A.J.; McLaggan, D.; Davidson, I.; O’Byrne, C.; Booth, I.R. Perturbation of anion balance during inhibition of growth of Escherichia coli by weak acids. J. Bacteriol. 1998, 180, 767–772. [Google Scholar] [CrossRef]
- Russell, J.B. Another explanation for the toxicity of fermentation acids at low pH: Anion accumulation versus uncoupling. J. Appl. Bacteriol. 1992, 73, 363–370. [Google Scholar] [CrossRef]
- Shiloach, J.; Kaufman, J.; Guillard, A.S.; Fass, R. Effect of glucose supply strategy on acetate accumulation, growth, and recombinant protein production by Escherichia coli BL21 (λDE3) and Escherichia coli JM109. Biotechnol. Bioeng. 1996, 49, 421–428. [Google Scholar] [CrossRef]
- Wang, H.; Wang, F.; Wang, W.; Yao, X.; Wei, D.; Cheng, H.; Deng, Z. Improving the expression of recombinant proteins in E. coli BL21 (DE3) under acetate stress: An alkaline pH shift approach. PLoS ONE 2014, 9, e112777. [Google Scholar] [CrossRef]
- de Mey, M.; Lequeux, G.J.; Beauprez, J.J.; Maertens, J.; Van Horen, E.; Soetaert, W.K.; Vanrolleghem, P.A.; Vandamme, E.J. Comparison of Different Strategies to Reduce Acetate Formation in Escherichia coli. Biotechnol. Prog. 2007, 23, 1053–1063. [Google Scholar] [CrossRef]
- Liu, L.; Duan, X.; Wu, J. L-Tryptophan Production in Escherichia coli Improved by Weakening the Pta-AckA Pathway. PLoS ONE 2016, 11, e0158200. [Google Scholar] [CrossRef]
- Liu, M.; Ding, Y.; Chen, H.; Zhao, Z.; Liu, H.; Xian, M.; Zhao, G. Improving the production of acetyl-CoA-derived chemicals in Escherichia coli BL21(DE3) through iclR and arcA deletion. BMC Microbiol. 2017, 17, 10. [Google Scholar] [CrossRef]
- Parimi, N.S.; Durie, I.A.; Wu, X.; Niyas, A.M.M.; Eiteman, M.A. Eliminating acetate formation improves citramalate production by metabolically engineered Escherichia coli. Microb. Cell Factories 2017, 16, 114. [Google Scholar] [CrossRef] [PubMed]
- Mills, T.Y.; Sandoval, N.R.; Gill, R.T. Cellulosic hydrolysate toxicity and tolerance mechanisms in Escherichia coli. Biotechnol. Biofuels 2009, 2, 26. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Z.; Zanotti, M.; Wang, X.; Ducey, C.; Liu, Y. Evaluation of lipid accumulation from lignocellulosic sugars by Mortierella isabellina for biodiesel production. Bioresour. Technol. 2012, 110, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.H.P. Reviving the carbohydrate economy via multi-product lignocellulose biorefineries. J. Ind. Microbiol. Biotechnol. 2008, 35, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Novak, K.; Pflügl, S. Towards biobased industry: Acetate as a promising feedstock to enhance the potential of microbial cell factories. Fems Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef]
- Lim, H.G.; Lee, J.H.; Noh, M.H.; Jung, G.Y. Rediscovering Acetate Metabolism: Its Potential Sources and Utilization for Biobased Transformation into Value-Added Chemicals. J. Agric. Food Chem. 2018, 66, 3998–4006. [Google Scholar] [CrossRef]
- Batlle-Vilanova, P.; Puig, S.; Gonzalez-Olmos, R.; Balaguer, M.D.; Colprim, J. Continuous acetate production through microbial electrosynthesis from CO2 with microbial mixed culture. J. Chem. Technol. Biotechnol. 2016, 91, 921–927. [Google Scholar] [CrossRef]
- Du, W.; Jongbloets, J.A.; van Boxtel, C.; Pineda Hernández, H.; Lips, D.; Oliver, B.G.; Hellingwerf, K.J.; Branco Dos Santos, F. Alignment of microbial fitness with engineered product formation: Obligatory coupling between acetate production and photoautotrophic growth. Biotechnol. Biofuels 2018, 11, 38. [Google Scholar] [CrossRef]
- Mateos, R.; Sotres, A.; Alonso, R.M.; Moran, A.; Escapa, A. Enhanced CO2 Conversion to Acetate through Microbial Electrosynthesis (MES) by Continuous Headspace Gas Recirculation. Energies 2019, 12, 3297. [Google Scholar] [CrossRef]
- Xiang, Y.; Liu, G.; Zhang, R.; Lu, Y.; Luo, H. Acetate production and electron utilization facilitated by sulfate-reducing bacteria in a microbial electrosynthesis system. Bioresour. Technol. 2017, 241, 821–829. [Google Scholar] [CrossRef]
- Bertsch, J.; Müller, V. Bioenergetic constraints for conversion of syngas to biofuels in acetogenic bacteria. Biotechnol. Biofuels 2015, 8, 210. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Luo, G.; Wang, W.; He, Y.; Zhang, R.; Liu, G. The effects of pH and temperature on the acetate production and microbial community compositions by syngas fermentation. Fuel 2018, 224, 537–544. [Google Scholar] [CrossRef]
- Yang, J.; Nie, Q. Engineering Escherichia coli to convert acetic acid to β-caryophyllene. Microb. Cell Factories 2016, 15, 74. [Google Scholar] [CrossRef]
- Hanai, T.; Atsumi, S.; Liao, J.C. Engineered Synthetic Pathway for Isopropanol Production in Escherichia coli. Appl. Environ. Microbiol. 2007, 73, 7814–7818. [Google Scholar] [CrossRef] [PubMed]
- Sandoval, N.R.; Mills, T.Y.; Zhang, M.; Gill, R.T. Elucidating acetate tolerance in E. coli using a genome-wide approach. Metab. Eng. 2011, 13, 214–224. [Google Scholar] [CrossRef]
- Yin, Y.; Zhang, Y.; Karakashev, D.B.; Wang, J.; Angelidaki, I. Biological caproate production by Clostridium kluyveri from ethanol and acetate as carbon sources. Bioresour. Technol. 2017, 241, 638–644. [Google Scholar] [CrossRef]
- Yang, S.; Li, S.; Jia, X. Production of medium chain length polyhydroxyalkanoate from acetate by engineered Pseudomonas putida KT2440. J. Ind. Microbiol. Biotechnol. 2019, 46, 793–800. [Google Scholar] [CrossRef]
- Oh, M.K.; Rohlin, L.; Kao, K.C.; Liao, J.C. Global expression profiling of acetate-grown Escherichia coli. J. Biol. Chem. 2002, 277, 13175–13183. [Google Scholar] [CrossRef]
- Kretzschmar, U.; Schobert, M.; Görisch, H. The Pseudomonas aeruginosa acsA gene, encoding an acetyl-CoA synthetase, is essential for growth on ethanol. Microbiology 2001, 147, 2671–2677. [Google Scholar] [CrossRef]
- Petushkova, E.P.; Tsygankov, A.A. Acetate metabolism in the purple non-sulfur bacterium Rhodobacter capsulatus. Biochemistry 2017, 82, 587–605. [Google Scholar] [CrossRef]
- Thauer, R.K.; Jungermann, K.; Henninger, H.; Wenning, J.; Decker, K. The energy metabolism of Clostridium kluyveri. Eur. J. Biochem. 1968, 4, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Mallick, N. Advances in cyanobacterial polyhydroxyalkanoates production. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef] [PubMed]
- Muyzer, G.; Stams, A.J.M. The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 2008, 6, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Heifetz, P.B.; Förster, B.; Osmond, C.B.; Giles, L.J.; Boynton, J.E. Effects of acetate on facultative autotrophy in Chlamydomonas reinhardtii assessed by photosynthetic measurements and stable isotope analyses. Plant Physiol. 2000, 122, 1439–1445. [Google Scholar] [CrossRef]
- Cecchin, M.; Benfatto, S.; Griggio, F.; Mori, A.; Cazzaniga, S.; Vitulo, N.; Delledonne, M.; Ballottari, M. Molecular basis of autotrophic vs mixotrophic growth in Chlorella sorokiniana. Sci. Rep. 2018, 8, 6465. [Google Scholar] [CrossRef]
- Warnecke, T.; Gill, R.T. Organic acid toxicity, tolerance, and production in Escherichia coli biorefining applications. Microb. Cell Factories 2005, 4, 25. [Google Scholar] [CrossRef]
- Martínez-Gómez, K.; Flores, N.; Castañeda, H.M.; Martínez-Batallar, G.; Hernández-Chávez, G.; Ramírez, O.T.; Gosset, G.; Encarnación, S.; Bolivar, F. New insights into Escherichia coli metabolism: Carbon scavenging, acetate metabolism and carbon recycling responses during growth on glycerol. Microb. Cell Factories 2012, 11, 46. [Google Scholar] [CrossRef]
- Hoffart, E.; Grenz, S.; Lange, J.; Nitschel, R.; Müller, F.; Schwentner, A.; Feith, A.; Lenfers-Lücker, M.; Takors, R.; Blombach, B. High substrate uptake rates empower Vibrio natriegens as production host for industrial biotechnology. Appl. Envion. Microbiol. 2017. [Google Scholar] [CrossRef]
- Dolan, S.K.; Kohlstedt, M.; Trigg, S.; Vallejo Ramirez, P.; Kaminski, C.F.; Wittmann, C.; Welch, M.; Nogales, J.; Newman, D.K. Contextual Flexibility in Pseudomonas aeruginosa Central Carbon Metabolism during Growth in Single Carbon Sources. mBio 2020, 11, e02684-19. [Google Scholar] [CrossRef]
- LaBauve, A.E.; Wargo, M.J. Growth and laboratory maintenance of Pseudomonas aeruginosa. Curr. Protoc. Microbiol. 2012. [Google Scholar] [CrossRef]
- Castaño-Cerezo, S.; Bernal, V.; Röhrig, T.; Termeer, S.; Cánovas, M. Regulation of acetate metabolism in Escherichia coli BL21 by protein Nε-lysine acetylation. Appl. Microbiol. Biotechnol. 2015, 99, 3533–3545. [Google Scholar] [CrossRef] [PubMed]
- Castaño-Cerezo, S.; Pastor, J.M.; Renilla, S.; Bernal, V.; Iborra, J.L.; Cánovas, M. An insight into the role of phosphotransacetylase (pta) and the acetate/acetyl-CoA node in Escherichia coli. Microb. Cell Factories 2009, 8, 54. [Google Scholar] [CrossRef] [PubMed]
- Gu, P.; Niu, H.; Fan, X.; Gao, J.; Li, Q. Engineering of phosphoenolpyruvate: Carbohydrate phosphotransferase system increased acetate assimilation in Escherichia coli. 3 Biotech 2019, 9, 77. [Google Scholar] [CrossRef] [PubMed]
- Enjalbert, B.; Millard, P.; Dinclaux, M.; Portais, J.C.; Létisse, F. Acetate fluxes in Escherichia coli are determined by the thermodynamic control of the Pta-AckA pathway. Sci. Rep. 2017, 7, 42135. [Google Scholar] [CrossRef] [PubMed]
- Berdalet, E.; Packard, T.; Lagace, B.; Roy, S.; St-Amand, L.; Gagne, J.P. CO2 production, O2 consumption and isocitrate dehydrogenase in the marine bacterium Vibrio natriegens. Aquat. Microb. Ecol. 1995, 9, 211–217. [Google Scholar] [CrossRef]
- Lee, H.H.; Ostrov, N.; Wong, B.G.; Gold, M.A.; Khalil, A.; Church, G.M. Vibrio natriegens, a new genomic powerhouse. bioRxiv 2016. [Google Scholar] [CrossRef]
- Görisch, H. The ethanol oxidation system and its regulation in Pseudomonas aeruginosa. Biochim. Et Biophys. Acta BBA Proteins Proteom. 2003, 1647, 98–102. [Google Scholar] [CrossRef]
- Jacob, K.; Rasmussen, A.; Tyler, P.; Servos, M.M.; Sylla, M.; Prado, C.; Daniele, E.; Sharp, J.S.; Purdy, A.E. Regulation of acetyl-CoA synthetase transcription by the CrbS/R two-component system is conserved in genetically diverse environmental pathogens. PLoS ONE 2017, 12, e0177825. [Google Scholar] [CrossRef]
- Klockgether, J.; Tümmler, B. Recent advances in understanding Pseudomonas aeruginosa as a pathogen. F1000Res 2017, 6, 1261. [Google Scholar] [CrossRef]
- Noh, M.H.; Lim, H.G.; Woo, S.H.; Song, J.; Jung, G.Y. Production of itaconic acid from acetate by engineering acid-tolerant Escherichia coli W. Biotechnol. Bioeng. 2018, 115, 729–738. [Google Scholar] [CrossRef]
- Rajaraman, E.; Agarwal, A.; Crigler, J.; Seipelt-Thiemann, R.; Altman, E.; Eiteman, M.A. Transcriptional analysis and adaptive evolution of Escherichia coli strains growing on acetate. Appl. Microbiol. Biotechnol. 2016, 100, 7777–7785. [Google Scholar] [CrossRef] [PubMed]
- Long, C.P.; Gonzalez, J.E.; Cipolla, R.M.; Antoniewicz, M.R. Metabolism of the fast-growing bacterium Vibrio natriegens elucidated by 13C metabolic flux analysis. Metab. Eng. 2017, 44, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Orr, J.S.; Christensen, D.G.; Wolfe, A.J.; Rao, C.V. Extracellular Acidic pH Inhibits Acetate Consumption by Decreasing Gene Transcription of the Tricarboxylic Acid Cycle and the Glyoxylate Shunt. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef] [PubMed]
- Ashworth, J.M.; Kornberg, H.L. The role of isocitrate lyase in Escherichia coli. Biochim. Biophys. Acta BBA Spec. Sect. Enzymol. Subj. 1964, 89, 383–384. [Google Scholar] [CrossRef]
- Chung, T.; Klumpp, D.J.; LaPorte, D.C. Glyoxylate bypass operon of Escherichia coli: Cloning and determination of the functional map. J. Bacteriol. 1988, 170, 386–392. [Google Scholar] [CrossRef]
- Kornberg, H.L. The role and control of the glyoxylate cycle in Escherichia coli. Biochem. J. 1966, 99, 1–11. [Google Scholar] [CrossRef]
- Zhao, J.; Baba, T.; Mori, H.; Shimizu, K. Effect of zwf gene knockout on the metabolism of Escherichia coli grown on glucose or acetate. Metab. Eng. 2004, 6, 164–174. [Google Scholar] [CrossRef]
- Xu, Q.; Bai, C.; Liu, Y.; Song, L.; Tian, L.; Yan, Y.; Zhou, J.; Zhou, X.; Zhang, Y.; Cai, M. Modulation of acetate utilization in Komagataella phaffii by metabolic engineering of tolerance and metabolism. Biotechnol. Biofuels 2019, 12, 61. [Google Scholar] [CrossRef]
- Sahu, U.; Rangarajan, P.N. Regulation of Acetate Metabolism and Acetyl Co-a Synthetase 1 (ACS1) Expression by Methanol Expression Regulator 1 (Mxr1p) in the Methylotrophic Yeast Pichia pastoris. J. Biol. Chem. 2016, 291, 3648–3657. [Google Scholar] [CrossRef]
- van den Berg, M.A.; de Jong-Gubbels, P.; Kortland, C.J.; van Dijken, J.P.; Pronk, J.T.; Steensma, H.Y. The Two Acetyl-coenzyme A Synthetases of Saccharomyces cerevisiae Differ with Respect to Kinetic Properties and Transcriptional Regulation. J. Biol. Chem. 1996, 271, 28953–28959. [Google Scholar] [CrossRef]
- Almeida, J.R.; Modig, T.; Petersson, A.; Hähn-Hägerdal, B.; Lidén, G.; Gorwa-Grauslund, M.F. Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J. Chem. Technol. Biotechnol. 2007, 82, 340–349. [Google Scholar] [CrossRef]
- Wei, N.; Quarterman, J.; Kim, S.R.; Cate, J.H.D.; Jin, Y.-S. Enhanced biofuel production through coupled acetic acid and xylose consumption by engineered yeast. Nat. Commun. 2013, 4, 2580. [Google Scholar] [CrossRef]
- Lee, F.-J.S.; Lin, L.-W.; Smith, J.A. Acetyl-CoA hydrolase involved in acetate utilization in Saccharomyces cerevisiae. Biochim. Biophys. Acta BBA Protein Struct. Mol. Enzymol. 1996, 1297, 105–109. [Google Scholar] [CrossRef]
- Chen, Y.; Siewers, V.; Nielsen, J. Profiling of Cytosolic and Peroxisomal Acetyl-CoA Metabolism in Saccharomyces cerevisiae. PLoS ONE 2012, 7, e42475. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, F.A.G.; Colen, G.; Takahashi, J.A. Yarrowia lipolytica and its multiple applications in the biotechnological industry. Sci. World J. 2014, 2014, 476207. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Qiao, K.; Stephanopoulos, G. 13C Metabolic Flux Analysis of acetate conversion to lipids by Yarrowia lipolytica. Metab. Eng. 2016, 38, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Kunze, M.; Pracharoenwattana, I.; Smith, S.M.; Hartig, A. A central role for the peroxisomal membrane in glyoxylate cycle function. Biochim. Biophys. Acta BBA Mol. Cell Res. 2006, 1763, 1441–1452. [Google Scholar] [CrossRef] [PubMed]
- Kabran, P.; Rossignol, T.; Gaillardin, C.; Nicaud, J.M.; Neuvéglise, C. Alternative splicing regulates targeting of malate dehydrogenase in Yarrowia lipolytica. DNA Res. 2012, 19, 231–244. [Google Scholar] [CrossRef]
- Unden, G.; Bongaerts, J. Alternative respiratory pathways of Escherichia coli: Energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta BBA Bioenerg. 1997, 1320, 217–234. [Google Scholar] [CrossRef]
- Higgins, T.E.; Johnson, M.J. Pathways of anaerobic acetate utilization in Escherichia coli and Aerobacter cloacae. J. Bacteriol. 1970, 101, 885–891. [Google Scholar] [CrossRef]
- Zelcbuch, L.; Lindner, S.N.; Zegman, Y.; Vainberg Slutskin, I.; Antonovsky, N.; Gleizer, S.; Milo, R.; Bar-Even, A. Pyruvate Formate-Lyase Enables Efficient Growth of Escherichia coli on Acetate and Formate. Biochemistry 2016, 55, 2423–2426. [Google Scholar] [CrossRef] [PubMed]
- Steinbusch, K.J.J.; Hamelers, H.V.M.; Plugge, C.M.; Buisman, C.J.N. Biological formation of caproate and caprylate from acetate: Fuel and chemical production from low grade biomass. Energy Environ. Sci. 2011, 4, 216–224. [Google Scholar] [CrossRef]
- Goevert, D.; Conrad, R. Stable carbon isotope fractionation by acetotrophic sulfur-reducing bacteria. FEMS Microbiol. Ecol. 2010, 71, 218–225. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Oude Elferink, S.J.W.H.; Akkermans-van Vliet, W.M.; Bogte, J.J.; Stams, A.J.M. Desulfobacca acetoxidans gen. nov., sp. nov., a novel acetate-degrading sulfate reducer isolated from sulfidogenic granular sludge. Int. J. Syst. Evol. Microbiol. 1999, 49, 345–350. [Google Scholar] [CrossRef] [PubMed]
- van Niftrik, L.; Jetten, M.S.M. Anaerobic Ammonium-Oxidizing Bacteria: Unique Microorganisms with Exceptional Properties. Microbiol. Mol. Biol. Rev. 2012, 76, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Gildemyn, S.; Molitor, B.; Usack, J.G.; Nguyen, M.; Rabaey, K.; Angenent, L.T. Upgrading syngas fermentation effluent using Clostridium kluyveri in a continuous fermentation. Biotechnol. Biofuels 2017, 10, 83. [Google Scholar] [CrossRef]
- Yan, S.; Dong, D. Improvement of caproic acid production in a Clostridium kluyveri H068 and Methanogen 166 co-culture fermentation system. AMB Express 2018, 8, 175. [Google Scholar] [CrossRef]
- Angenent, L.T.; Richter, H.; Buckel, W.; Spirito, C.M.; Steinbusch, K.J.J.; Plugge, C.M.; Strik, D.P.B.T.B.; Grootscholten, T.I.M.; Buisman, C.J.N.; Hamelers, H.V.M. Chain Elongation with Reactor Microbiomes: Open-Culture Biotechnology To Produce Biochemicals. Environ. Sci. Technol. 2016, 50, 2796–2810. [Google Scholar] [CrossRef]
- Seedorf, H.; Fricke, W.F.; Veith, B.; Brüggemann, H.; Liesegang, H.; Strittmatter, A.; Miethke, M.; Buckel, W.; Hinderberger, J.; Li, F.; et al. The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc. Natl. Acad. Sci. USA 2008, 105, 2128–2133. [Google Scholar] [CrossRef]
- Buckel, W.; Thauer, R.K. Flavin-Based Electron Bifurcation, Ferredoxin, Flavodoxin, and Anaerobic Respiration With Protons (Ech) or NAD(+) (Rnf) as Electron Acceptors: A Historical Review. Front Microbiol. 2018, 9, 401. [Google Scholar] [CrossRef]
- Thauer, R.K. Citric-acid cycle, 50 years on. Modifications and an alternative pathway in anaerobic bacteria. Eur. J. Biochem. 1988, 176, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Ragsdale, S.W.; Pierce, E. Acetogenesis and the Wood–Ljungdahl pathway of CO2 fixation. Biochim. Biophys. Acta BBA Proteins Proteom. 2008, 1784, 1873–1898. [Google Scholar] [CrossRef] [PubMed]
- Schauder, R.; Preuß, A.; Jetten, M.; Fuchs, G. Oxidative and reductive acetyl CoA/carbon monoxide dehydrogenase pathway in Desulfobacterium autotrophicum. Arch. Microbiol. 1988, 151, 84–89. [Google Scholar] [CrossRef]
- Bhattacharya, S.K.; Uberoi, V.; Dronamraju, M.M. Interaction between acetate fed sulfate reducers and methanogens. Water Res. 1996, 30, 2239–2246. [Google Scholar] [CrossRef]
- Oremland, R.S.; Polcin, S. Methanogenesis and Sulfate Reduction: Competitive and Noncompetitive Substrates in Estuarine Sediments. Appl. Environ. Microbiol. 1982, 44, 1270–1276. [Google Scholar] [CrossRef]
- Sela-Adler, M.; Ronen, Z.; Herut, B.; Antler, G.; Vigderovich, H.; Eckert, W.; Sivan, O. Co-existence of Methanogenesis and Sulfate Reduction with Common Substrates in Sulfate-Rich Estuarine Sediments. Front Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Chai, B.; Wang, Y.; Wang, W.; Fan, P. Effect of carbon source on lipid accumulation and biodiesel production of Yarrowia lipolytica. Environ. Sci. Pollut. Res. 2019, 26, 31234–31242. [Google Scholar] [CrossRef]
- Gong, Z.; Shen, H.; Zhou, W.; Wang, Y.; Yang, X.; Zhao, Z.K. Efficient conversion of acetate into lipids by the oleaginous yeast Cryptococcus curvatus. Biotechnol. Biofuels 2015, 8, 189. [Google Scholar] [CrossRef]
- Assawamongkholsiri, T.; Reungsang, A.; Sittijunda, S. Photo-hydrogen and lipid production from lactate, acetate, butyrate, and sugar manufacturing wastewater with an alternative nitrogen source by Rhodobacter sp. KKU-PS1. PeerJ 2019, 7, e6653. [Google Scholar] [CrossRef]
- Perez, C.M.T.; Watanabe, K.; Okamura, Y.; Nakashimada, Y.; Aki, T. Metabolite Profile Analysis of Aurantiochytrium limacinum SR21 Grown on Acetate-based Medium for Lipid Fermentation. J. Oleo Sci. 2019, 68, 541–549. [Google Scholar] [CrossRef]
- Xiao, N.; Jiao, N. Formation of polyhydroxyalkanoate in aerobic anoxygenic phototrophic bacteria and its relationship to carbon source and light availability. Appl. Environ. Microbiol. 2011, 77, 7445–7450. [Google Scholar] [CrossRef] [PubMed]
- Dutt, V.; Srivastava, S. Novel quantitative insights into carbon sources for synthesis of poly hydroxybutyrate in Synechocystis PCC 6803. Photosynth. Res. 2018, 136, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.-L.; Da, Y.-Y.; Zheng, W.-T.; Chen, G.-Q.; Li, Z.-J. Production of polyhydroxyalkanoate from acetate by metabolically engineered Aeromonas hydrophilia. J. Biosci. Bioeng. 2020. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-J.; Qiao, K.; Liu, N.; Stephanopoulos, G. Engineering Yarrowia lipolytica for poly-3-hydroxybutyrate production. J. Ind. Microbiol. Biotechnol. 2017, 44, 605–612. [Google Scholar] [CrossRef]
- Shimizu, H.; Shimizu, N.; Shioya, S. Roles of glucose and acetate as carbon sources inl-histidine production withBrevibacterium flavum FERM1564 revealed by metabolic flux analysis. Biotechnol. Bioprocess Eng. 2002, 7, 171. [Google Scholar] [CrossRef]
- Oswald, F.; Dörsam, S.; Veith, N.; Zwick, M.; Neumann, A.; Ochsenreither, K.; Syldatk, C. Sequential Mixed Cultures: From Syngas to Malic Acid. Front Microbiol. 2016, 7, 891. [Google Scholar] [CrossRef]
- Shimizu, T.; Teramoto, H.; Inui, M. Introduction of Glyoxylate Bypass Increases Hydrogen Gas Yield from Acetate and l-Glutamate in Rhodobacter sphaeroides. Appl. Environ. Microbiol. 2019, 85, e01873-18. [Google Scholar] [CrossRef]
- Friman, H.; Schechter, A.; Ioffe, Y.; Nitzan, Y.; Cahan, R. Current production in a microbial fuel cell using a pure culture of Cupriavidus basilensis growing in acetate or phenol as a carbon source. Microb. Biotechnol. 2013, 6, 425–434. [Google Scholar] [CrossRef]
- Town, J.R.; Links, M.G.; Fonstad, T.A.; Dumonceaux, T.J. Molecular characterization of anaerobic digester microbial communities identifies microorganisms that correlate to reactor performance. Bioresour. Technol. 2014, 151, 249–257. [Google Scholar] [CrossRef]
- Jain, S.; Jain, S.; Wolf, I.T.; Lee, J.; Tong, Y.W. A comprehensive review on operating parameters and different pretreatment methodologies for anaerobic digestion of municipal solid waste. Renew. Sustain. Energy Rev. 2015, 52, 142–154. [Google Scholar] [CrossRef]
- Tian, Y.; Zhang, H.; Zheng, L.; Li, S.; Hao, H.; Huang, H. Effect of Zn Addition on the Cd-Containing Anaerobic Fermentation Process: Biodegradation and Microbial Communities. Int. J. Env. Res. Public Health 2019, 16, 2998. [Google Scholar] [CrossRef] [PubMed]
- Walker, T.E.; London, R.E. Biosynthetic preparation of L-[13C]- and [15N]glutamate by Brevibacterium flavum. Appl. Environ. Microbiol. 1987, 53, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Huang, B.; Lai, N.; Gu, Y.; Li, Z.; Ye, Q.; Wu, H. Metabolic engineering of Escherichia coli carrying the hybrid acetone-biosynthesis pathway for efficient acetone biosynthesis from acetate. Microb. Cell Fact. 2019, 18, 6. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, C.; Lai, N.; Huang, B.; Fei, P.; Ding, D.; Hu, P.; Gu, Y.; Wu, H. Efficient isopropanol biosynthesis by engineered Escherichia coli using biologically produced acetate from syngas fermentation. Bioresour. Technol. 2020, 296, 122337. [Google Scholar] [CrossRef] [PubMed]
- Novak, K.; Kutscha, R.; Pflügl, S. Microbial upgrading of acetate into 2,3-butanediol and acetoin by E. coli W. Biotechnol. Biofuels 2020, 13, 177. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, W.; Chen, H.; Liu, B.; Lin, B.; Tao, Y. Metabolic engineering for efficient supply of acetyl-CoA from different carbon sources in Escherichia coli. Microb. Cell Fact. 2019, 18, 130. [Google Scholar] [CrossRef]
- Li, W.; Chen, J.; Liu, C.-X.; Yuan, Q.-P.; Li, Z.-J. Microbial production of glycolate from acetate by metabolically engineered Escherichia coli. J. Biotechnol. 2019, 291, 41–45. [Google Scholar] [CrossRef]
- Chen, J.; Li, W.; Zhang, Z.Z.; Tan, T.W.; Li, Z.J. Metabolic engineering of Escherichia coli for the synthesis of polyhydroxyalkanoates using acetate as a main carbon source. Microb. Cell Fact. 2018, 17, 102. [Google Scholar] [CrossRef]
- Lee, J.; Cha, S.; Kang, C.; Lee, G.; Lim, H.; Jung, G. Efficient Conversion of Acetate to 3-Hydroxypropionic Acid by Engineered Escherichia coli. Catalysts 2018, 8, 525. [Google Scholar] [CrossRef]
- Song, H.-S.; Seo, H.-M.; Jeon, J.-M.; Moon, Y.-M.; Hong, J.W.; Hong, Y.G.; Bhatia, S.K.; Ahn, J.; Lee, H.; Kim, W.; et al. Enhanced isobutanol production from acetate by combinatorial overexpression of acetyl-CoA synthetase and anaplerotic enzymes in engineered Escherichia coli. Biotechnol. Bioeng. 2018, 115, 1971–1978. [Google Scholar] [CrossRef]
- Xu, X.; Xie, M.; Zhao, Q.; Xian, M.; Liu, H. Microbial production of mevalonate by recombinant Escherichia coli using acetic acid as a carbon source. Bioengineered 2018, 9, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Leone, S.; Sannino, F.; Tutino, M.L.; Parrilli, E.; Picone, D. Acetate: Friend or foe? Efficient production of a sweet protein in Escherichia coli BL21 using acetate as a carbon source. Microb. Cell Factories 2015, 14, 106. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Xian, M.; Liu, H. Efficient conversion of acetate into phloroglucinol by recombinant Escherichia coli. RSC Adv. 2017, 7, 50942–50948. [Google Scholar] [CrossRef]
- Yu, S.; Guo, L.; Zhao, L.; Chen, Z.; Huo, Y. Metabolic engineering of E. coli for producing phloroglucinol from acetate. Appl. Microbiol. Biotechnol. 2020, 104, 7787–7799. [Google Scholar] [CrossRef]
- Huang, B.; Yang, H.; Fang, G.; Zhang, X.; Wu, H.; Li, Z.; Ye, Q. Central pathway engineering for enhanced succinate biosynthesis from acetate in Escherichia coli. Biotechnol. Bioeng. 2018, 115, 943–954. [Google Scholar] [CrossRef]
- Li, Y.; Huang, B.; Wu, H.; Li, Z.; Ye, Q.; Zhang, Y.H.P. Production of Succinate from Acetate by Metabolically Engineered Escherichia coli. ACS Synth. Biol. 2016, 5, 1299–1307. [Google Scholar] [CrossRef]
- Niu, H.; Li, R.; Wu, J.; Cai, Z.; Yang, D.; Gu, P.; Li, Q. Production of succinate by recombinant Escherichia coli using acetate as the sole carbon source. 3 Biotech 2018, 8, 421. [Google Scholar] [CrossRef]
- Jo, M.; Noh, M.H.; Lim, H.G.; Kang, C.W.; Im, D.K.; Oh, M.K.; Jung, G.Y. Precise tuning of the glyoxylate cycle in Escherichia coli for efficient tyrosine production from acetate. Microb. Cell Fact. 2019, 18, 57. [Google Scholar] [CrossRef]
- Taymaz-Nikerel, H.; Borujeni, A.E.; Verheijen, P.J.T.; Heijnen, J.J.; van Gulik, W.M. Genome-derived minimal metabolic models for Escherichia coli MG1655 with estimated in vivo respiratory ATP stoichiometry. Biotechnol. Bioeng. 2010, 107, 369–381. [Google Scholar] [CrossRef]
- Szenk, M.; Dill, K.A.; de Graff, A.M.R. Why Do Fast-Growing Bacteria Enter Overflow Metabolism? Testing the Membrane Real Estate Hypothesis. Cell Syst. 2017, 5, 95–104. [Google Scholar] [CrossRef]
- Steigmiller, S.; Turina, P.; Graber, P. The thermodynamic H+/ATP ratios of the H+-ATPsynthases from chloroplasts and Escherichia coli. Proc. Natl. Acad. Sci. USA 2008, 105, 3745–3750. [Google Scholar] [CrossRef] [PubMed]
- Mitsumori, F.; Rees, D.; Brindle, K.M.; Radda, G.K.; Campbell, I.D. 31P-NMR saturation transfer studies of aerobic Escherichia coli cells. Biochim. Et Biophys. Acta (Bba) Mol. Cell Res. 1988, 969, 185–193. [Google Scholar] [CrossRef]
- Nakanishi-Matsui, M.; Sekiya, M.; Futai, M. ATP synthase from Escherichia coli: Mechanism of rotational catalysis, and inhibition with the ε subunit and phytopolyphenols. Biochim. Biophys. Acta BBA Bioenerg. 2016, 1857, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Bekker, M.; de Vries, S.; Ter Beek, A.; Hellingwerf, K.J.; de Mattos, M.J. Respiration of Escherichia coli can be fully uncoupled via the nonelectrogenic terminal cytochrome bd-II oxidase. J Bacteriol. 2009, 191, 5510–5517. [Google Scholar] [CrossRef] [PubMed]
- Holms, H. Flux analysis and control of the central metabolic pathways in Escherichia coli. FEMS Microbiol. Rev. 1996, 19, 85–116. [Google Scholar] [CrossRef]
- Ingledew, W.J.; Poole, R.K. The respiratory chains of Escherichia coli. Microbiol. Rev. 1984, 48, 222–271. [Google Scholar] [CrossRef]
- Borisov, V.B.; Murali, R.; Verkhovskaya, M.L.; Bloch, D.A.; Han, H.; Gennis, R.B.; Verkhovsky, M.I. Aerobic respiratory chain of Escherichia coli is not allowed to work in fully uncoupled mode. Proc. Natl. Acad. Sci. USA 2011, 108, 17320–17324. [Google Scholar] [CrossRef]
- Spaans, S.K.; Weusthuis, R.A.; van der Oost, J.; Kengen, S.W.M. NADPH-generating systems in bacteria and archaea. Front Microbiol. 2015, 6, 742. [Google Scholar] [CrossRef]
- Jan, J.; Martinez, I.; Wang, Y.; Bennett, G.N.; San, K.-Y. Metabolic engineering and transhydrogenase effects on NADPH availability in escherichia coli. Biotechnol. Prog. 2013, 29, 1124–1130. [Google Scholar] [CrossRef]
- Wang, Y.; San, K.-Y.; Bennett, G.N. Improvement of NADPH bioavailability in Escherichia coli by replacing NAD+-dependent glyceraldehyde-3-phosphate dehydrogenase GapA with NADP+-dependent GapB from Bacillus subtilis and addition of NAD kinase. J. Ind. Microbiol. Biotechnol. 2013, 40, 1449–1460. [Google Scholar] [CrossRef]
- Wenk, S.; Schann, K.; He, H.; Rainaldi, V.; Kim, S.; Lindner, S.N.; Bar-Even, A. An “energy-auxotroph” Escherichia coli provides an in vivo platform for assessing NADH regeneration systems. Biotechnol. Bioeng. 2020. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, H.; Zhao, G.; Caiyin, Q.; Qiao, J. Redox cofactor engineering in industrial microorganisms: Strategies, recent applications and future directions. J. Ind. Microbiol. Biotechnol. 2018, 45, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Inokuma, K.; Liao, J.C.; Okamoto, M.; Hanai, T. Improvement of isopropanol production by metabolically engineered Escherichia coli using gas stripping. J. Biosci. Bioeng. 2010, 110, 696–701. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.C.; Min, B.E.; Hwang, H.G.; Seo, S.W.; Jung, G.Y. Pathway optimization by re-design of untranslated regions for L-tyrosine production in Escherichia coli. Sci. Rep. 2015, 5, 13853. [Google Scholar] [CrossRef]
- Seo, H.-M.; Jeon, J.-M.; Lee, J.H.; Song, H.-S.; Joo, H.-B.; Park, S.-H.; Choi, K.-Y.; Kim, Y.H.; Park, K.; Ahn, J.; et al. Combinatorial application of two aldehyde oxidoreductases on isobutanol production in the presence of furfural. J. Ind. Microbiol. Biotechnol. 2016, 43, 37–44. [Google Scholar] [CrossRef]
- Kuhn, M.L.; Zemaitaitis, B.; Hu, L.I.; Sahu, A.; Sorensen, D.; Minasov, G.; Lima, B.P.; Scholle, M.; Mrksich, M.; Anderson, W.F.; et al. Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation. PLoS ONE 2014, 9, e94816. [Google Scholar] [CrossRef]
- Lima, B.P.; Thanh Huyen, T.T.; Bäsell, K.; Becher, D.; Antelmann, H.; Wolfe, A.J. Inhibition of acetyl phosphate-dependent transcription by an acetylatable lysine on RNA polymerase. J. Biol. Chem. 2012, 287, 32147–32160. [Google Scholar] [CrossRef]
- Batchelor, E.; Walthers, D.; Kenney, L.J.; Goulian, M. The Escherichia coli CpxA-CpxR envelope stress response system regulates expression of the porins ompF and ompC. J. Bacteriol. 2005, 187, 5723–5731. [Google Scholar] [CrossRef]
- Seo, S.W.; Gao, Y.; Kim, D.; Szubin, R.; Yang, J.; Cho, B.-K.; Palsson, B.O. Revealing genome-scale transcriptional regulatory landscape of OmpR highlights its expanded regulatory roles under osmotic stress in Escherichia coli K-12 MG1655. Sci. Rep. 2017, 7, 2181. [Google Scholar] [CrossRef]
- Shinagawa, H.; Makino, K.; Nakata, A.; Brenner, S. Regulation of the pho regulon in Escherichia coli K-12: Genetic and physiological regulation of the positive regulatory gene phoB. J. Mol. Biol. 1983, 168, 477–488. [Google Scholar] [CrossRef]
- Sourjik, V.; Berg, H.C. Binding of the Escherichia coli response regulator CheY to its target measured in vivo by fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. USA 2002, 99, 12669–12674. [Google Scholar] [CrossRef] [PubMed]
- Tovilla-Coutiño, D.B.; Momany, C.; Eiteman, M.A. Engineered citrate synthase alters Acetate Accumulation in Escherichia coli. Metab. Eng. 2020, 61, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Soma, Y.; Tsuruno, K.; Wada, M.; Yokota, A.; Hanai, T. Metabolic flux redirection from a central metabolic pathway toward a synthetic pathway using a metabolic toggle switch. Metab. Eng. 2014, 23, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Chong, H.; Yeow, J.; Wang, I.; Song, H.; Jiang, R. Improving Acetate Tolerance of Escherichia coli by Rewiring Its Global Regulator cAMP Receptor Protein (CRP). PLoS ONE 2013, 8, e77422. [Google Scholar] [CrossRef] [PubMed]
- Kanjee, U.; Houry, W.A. Mechanisms of Acid Resistance in Escherichia coli. Annu. Rev. Microbiol. 2013, 67, 65–81. [Google Scholar] [CrossRef] [PubMed]
- Richard, H.; Foster, J.W. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J. Bacteriol. 2004, 186, 6032–6041. [Google Scholar] [CrossRef]
- De Biase, D.; Pennacchietti, E. Glutamate decarboxylase-dependent acid resistance in orally acquired bacteria: Function, distribution and biomedical implications of the gadBC operon. Mol. Microbiol. 2012, 86, 770–786. [Google Scholar] [CrossRef]
- Woo, J.-M.; Kim, J.-W.; Song, J.-W.; Blank, L.M.; Park, J.-B. Activation of the Glutamic Acid-Dependent Acid Resistance System in Escherichia coli BL21 (DE3) Leads to Increase of the Fatty Acid Biotransformation Activity. PLoS ONE 2016, 11. [Google Scholar] [CrossRef]
- Royce, L.A.; Boggess, E.; Fu, Y.; Liu, P.; Shanks, J.V.; Dickerson, J.; Jarboe, L.R. Transcriptomic Analysis of Carboxylic Acid Challenge in Escherichia coli: Beyond Membrane Damage. PLoS ONE 2014, 9, e89580. [Google Scholar] [CrossRef]
- Roe, A.J.; O’Byrne, C.; McLaggan, D.; Booth, I.R. Inhibition of Escherichia coli growth by acetic acid: A problem with methionine biosynthesis and homocysteine toxicity. Microbiology 2002, 148, 2215–2222. [Google Scholar] [CrossRef]
- Mordukhova, E.A.; Pan, J.-G. Evolved cobalamin-independent methionine synthase (MetE) improves the acetate and thermal tolerance of Escherichia coli. Appl. Environ. Microbiol. 2013, 79, 7905–7915. [Google Scholar] [CrossRef]
- Klein, A.H.; Shulla, A.; Reimann, S.A.; Keating, D.H.; Wolfe, A.J. The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. J. Bacteriol. 2007, 189, 5574–5581. [Google Scholar] [CrossRef] [PubMed]
- Castaño-Cerezo, S.; Bernal, V.; Post, H.; Fuhrer, T.; Cappadona, S.; Sánchez-Díaz, N.C.; Sauer, U.; Heck, A.J.R.; Altelaar, A.F.M.; Cánovas, M. Protein acetylation affects acetate metabolism, motility and acid stress response in Escherichia coli. Mol. Syst. Biol. 2014, 10, 762. [Google Scholar] [CrossRef] [PubMed]
- Weinert Brian, T.; Iesmantavicius, V.; Wagner Sebastian, A.; Schölz, C.; Gummesson, B.; Beli, P.; Nyström, T.; Choudhary, C. Acetyl-Phosphate Is a Critical Determinant of Lysine Acetylation in E. coli. Mol. Cell 2013, 51, 265–272. [Google Scholar] [CrossRef]
- Fernández-Sandoval, M.T.; Huerta-Beristain, G.; Trujillo-Martinez, B.; Bustos, P.; González, V.; Bolivar, F.; Gosset, G.; Martinez, A. Laboratory metabolic evolution improves acetate tolerance and growth on acetate of ethanologenic Escherichia coli under non-aerated conditions in glucose-mineral medium. Appl. Microbiol. Biotechnol. 2012, 96, 1291–1300. [Google Scholar] [CrossRef] [PubMed]
- Seong, W.; Han, G.H.; Lim, H.S.; Baek, J.I.; Kim, S.-J.; Kim, D.; Kim, S.K.; Lee, H.; Kim, H.; Lee, S.-G.; et al. Adaptive laboratory evolution of Escherichia coli lacking cellular byproduct formation for enhanced acetate utilization through compensatory ATP consumption. Metab. Eng. 2020, 62, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Sankaranarayanan, M.; Ashok, S.; Park, S. Production of 3-hydroxypropionic acid from glycerol by acid tolerant Escherichia coli. J. Ind. Microbiol. Biotechnol. 2014, 41, 1039–1050. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Cao, Y.; Liu, W.; Xian, M.; Liu, H. Improving phloroglucinol tolerance and production in Escherichia coli by GroESL overexpression. Microb. Cell Factories 2017, 16, 227. [Google Scholar] [CrossRef]
- Cao, Y.; Jiang, X.; Zhang, R.; Xian, M. Improved phloroglucinol production by metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 2011, 91, 1545–1552. [Google Scholar] [CrossRef]
- Horinouchi, T.; Sakai, A.; Kotani, H.; Tanabe, K.; Furusawa, C. Improvement of isopropanol tolerance of Escherichia coli using adaptive laboratory evolution and omics technologies. J. Biotechnol. 2017, 255, 47–56. [Google Scholar] [CrossRef]
- Satowa, D.; Fujiwara, R.; Uchio, S.; Nakano, M.; Otomo, C.; Hirata, Y.; Matsumoto, T.; Noda, S.; Tanaka, T.; Kondo, A. Metabolic engineering of E. coli for improving mevalonate production to promote NADPH regeneration and enhance acetyl-CoA supply. Biotechnol. Bioeng. 2020, 117, 2153–2164. [Google Scholar] [CrossRef] [PubMed]
- de Arroyo Garcia, L.; Jones, P.R. In silico co-factor balance estimation using constraint-based modelling informs metabolic engineering in Escherichia coli. PLoS Comput. Biol. 2020, 16, e1008125. [Google Scholar] [CrossRef] [PubMed]
- Han, G.H.; Kim, S.K.; Yoon, P.K.-S.; Kang, Y.; Kim, B.S.; Fu, Y.; Sung, B.H.; Jung, H.C.; Lee, D.-H.; Kim, S.-W.; et al. Fermentative production and direct extraction of (-)-α-bisabolol in metabolically engineered Escherichia coli. Microb. Cell Factories 2016, 15, 185. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, K.H. Effects of minimal media vs. complex media on the metabolite profiles of Escherichia coli and Saccharomyces cerevisiae. Process Biochem. 2017, 57, 64–71. [Google Scholar] [CrossRef]
- Ederer, M.; Steinsiek, S.; Stagge, S.; Rolfe, M.; Ter Beek, A.; Knies, D.; Teixeira de Mattos, J.; Sauter, T.; Green, J.; Poole, R.; et al. A mathematical model of metabolism and regulation provides a systems-level view of how Escherichia coli responds to oxygen. Front Microbiol. 2014, 5. [Google Scholar] [CrossRef]
- Enjalbert, B.; Letisse, F.; Portais, J.-C. Physiological and Molecular Timing of the Glucose to Acetate Transition in Escherichia coli. Metabolites 2013, 3, 820–837. [Google Scholar] [CrossRef]
- Zeng, H.; Yang, A. Modelling overflow metabolism in Escherichia coli with flux balance analysis incorporating differential proteomic efficiencies of energy pathways. BMC Syst. Biol. 2019, 13, 3. [Google Scholar] [CrossRef]
- Gutmann, B.; Cantillo, D.; Kappe, C.O. Continuous-Flow Technology—A Tool for the Safe Manufacturing of Active Pharmaceutical Ingredients. Angew. Chem. Int. Ed. 2015, 54, 6688–6728. [Google Scholar] [CrossRef]
- Guan, N.; Liu, L. Microbial response to acid stress: Mechanisms and applications. Appl. Microbiol. Biotechnol. 2020, 104, 51–65. [Google Scholar] [CrossRef]
- David, A.; Tripathi, A.K.; Sani, R.K. Acetate Production from Cafeteria Wastes and Corn Stover Using a Thermophilic Anaerobic Consortium: A Prelude Study for the Use of Acetate for the Production of Value-Added Products. Microorganisms 2020, 8, 353. [Google Scholar] [CrossRef]
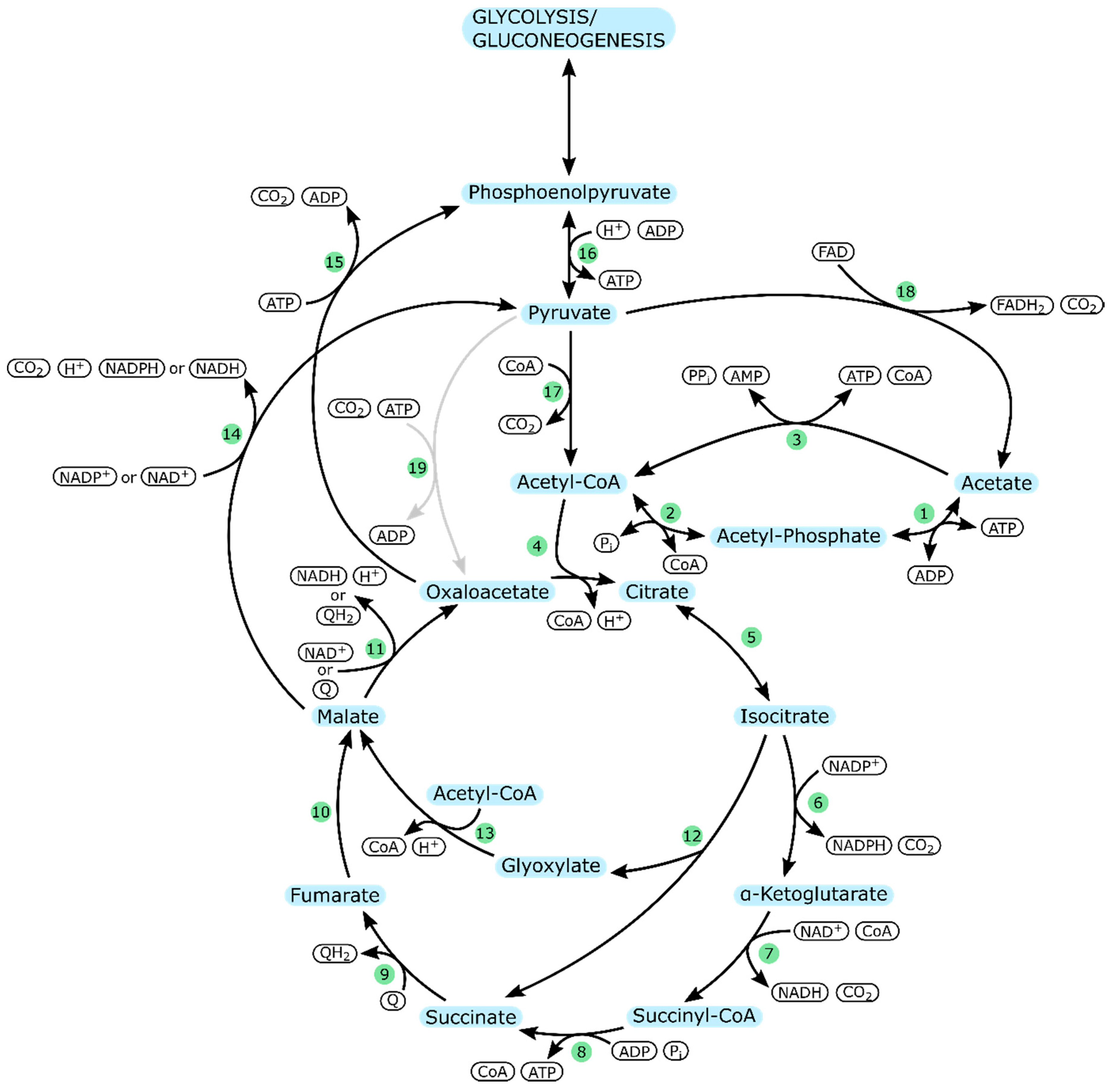

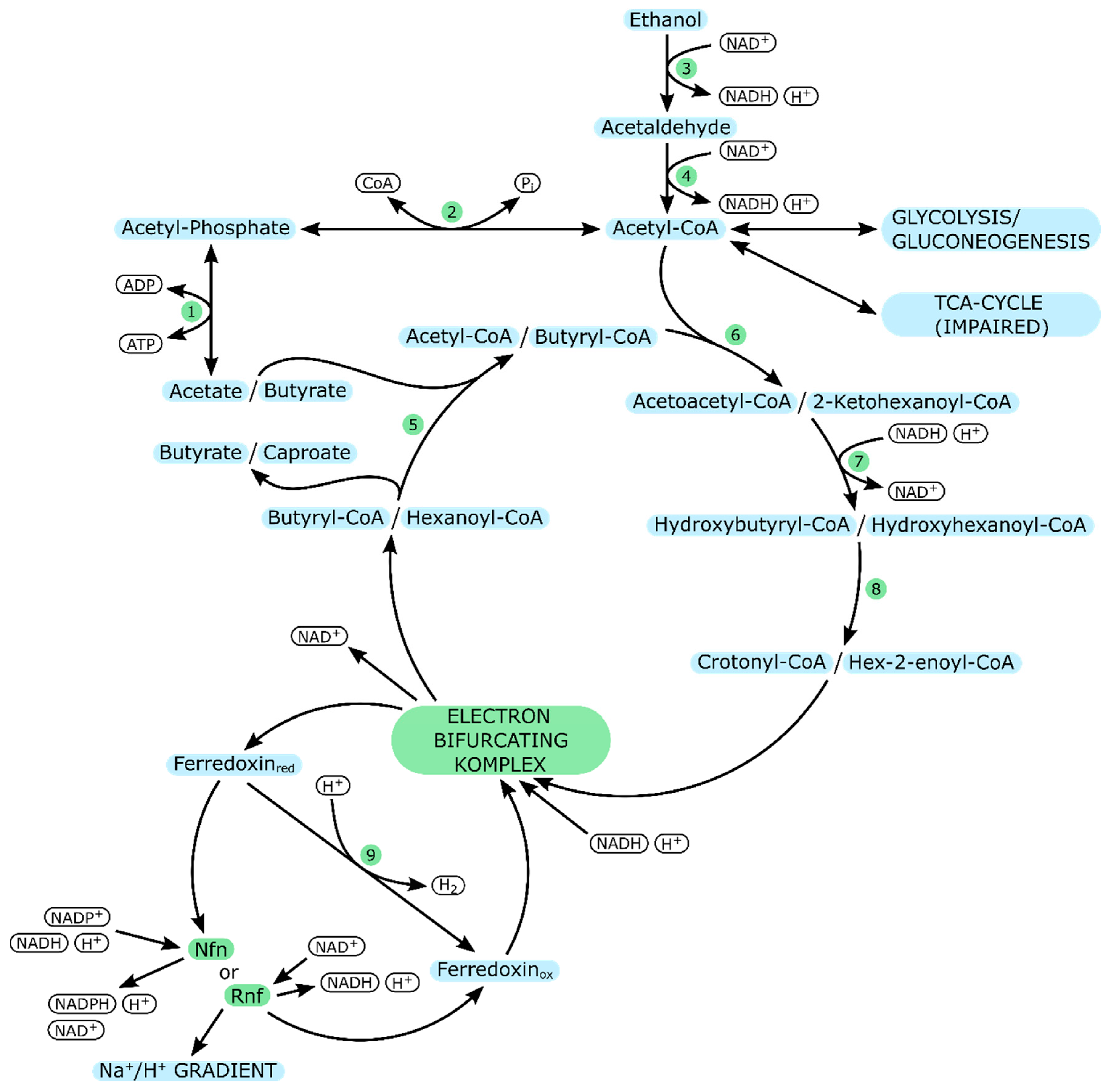

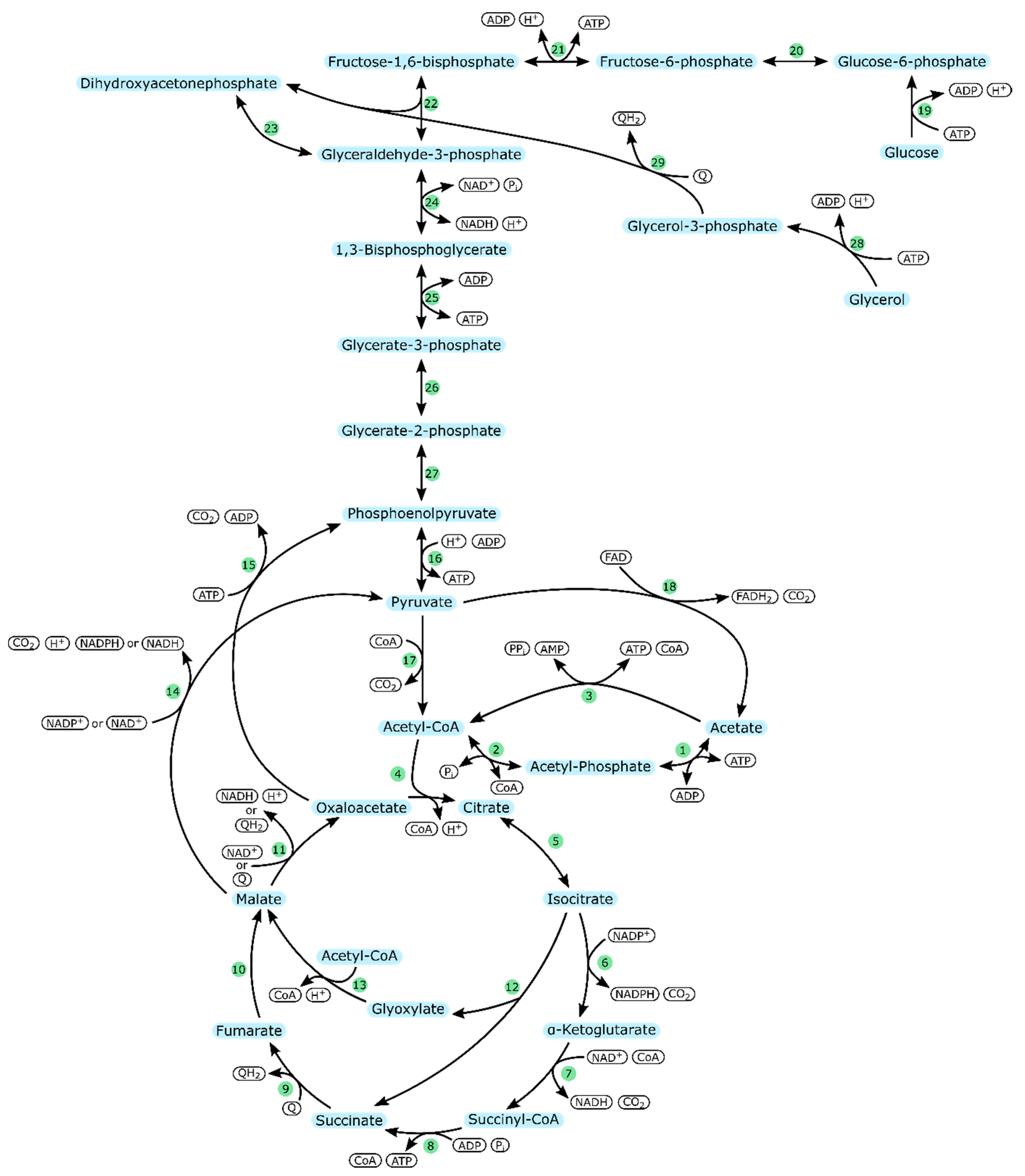
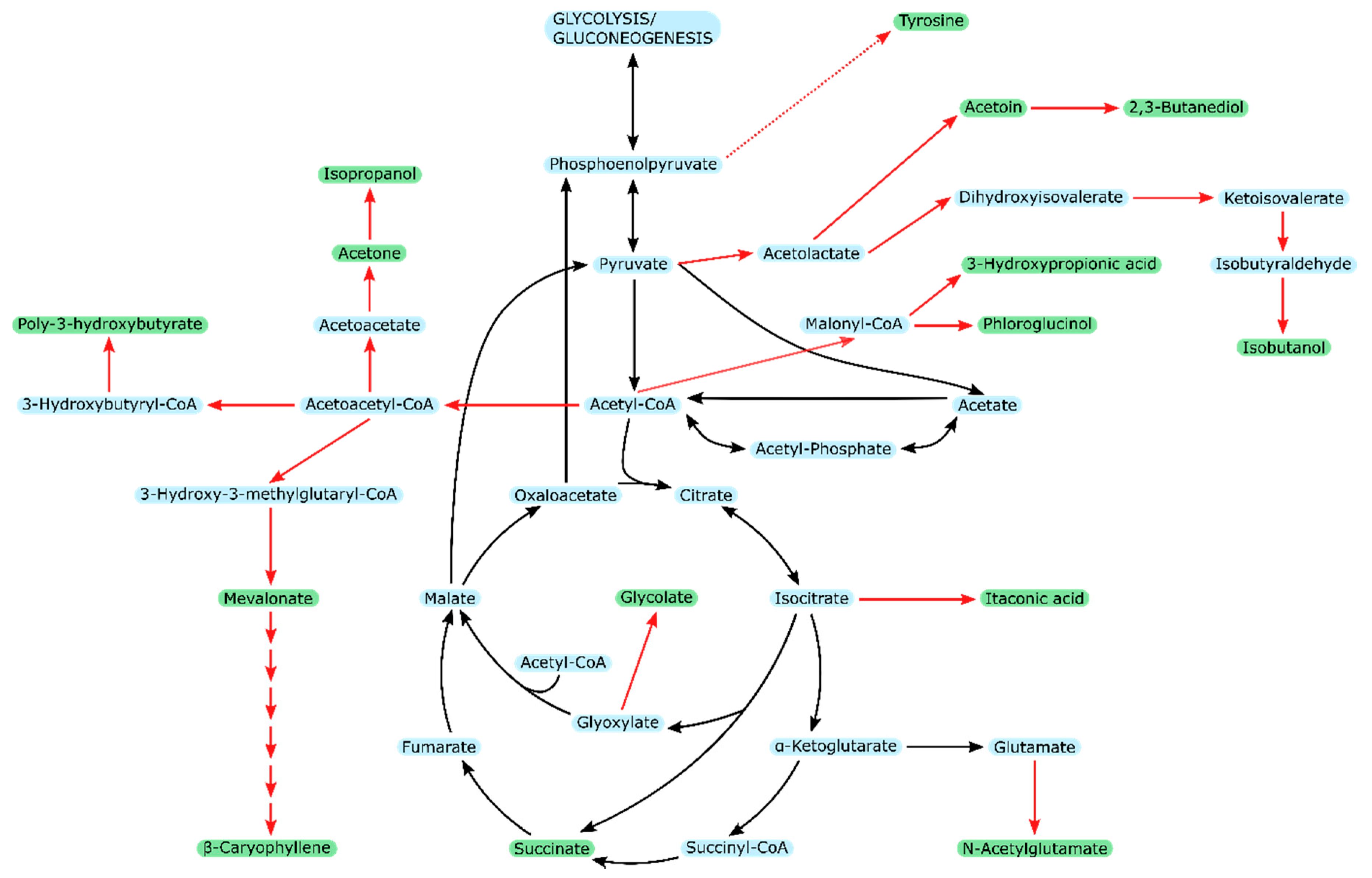
| Organism & Strain | Acetate Concentration [mM] | Specific Growth Rate | Reference |
|---|---|---|---|
| E. coli MC4100 | 42 | 0.33 ± 0.05 h−1 | [38] |
| E. coli BW25113 | 60 | 0.28 ± 0.03 h−1 | [52] |
| E. coli W | 169 | 0.46 h−1 | [60] |
| E. coli W | 85 | 0.37 ± 0.01 h−1 | [61] |
| E. coli C | 85 | 0.41 ± 0.01 h−1 | [61] |
| V. natriegens DSM 759 | 42 | 0.45 ± 0.03 h−1 | [48] |
| P. aeruginosa PAO1 | 20 | 0.80 ± 0.01 h−1 | [49,50] |
| E. coli, V. natriegens, and P. aeruginosa | ||
|---|---|---|
| Enzyme Number | Enzyme | Part in Metabolism |
| 1 | Acetate kinase (ackA) | Acetate uptake |
| 2 | Phosphotransacetylase (pta) | |
| 3 | Acetyl-CoA synthetase (acs) | |
| 4 | Citrate synthase (gltA) | TCA cycle |
| 5 | Aconitate hydratase (acnA/acnB) | |
| 6 | Isocitrate dehydrogenase (icd) | |
| 7 | α-ketoglutarate dehydrogenase (sucA/sucB) | |
| 8 | Succinyl-CoA synthetase (sucC/sucD) | |
| 9 | Succinate dehydrogenase (aerobic: sdhCDAB)/Fumarate reductase (anaerobic: frdABCD) | |
| 10 | Fumarate hydratase (aerobic: fumA/ anaerobic: fumB) | |
| 11 | Malate dehydrogenase (mdh/mqo) | |
| 12 | Isocitrate lyase (aceA) | Glyoxylate cycle |
| 13 | Malate synthase (glcB) | |
| 14 | Malate dehydrogenase (maeA/maeB) | Pyruvate metabolism |
| 15 | Phosphoenolpyruvate carboxykinase (ATP-dependent) (pckA) | |
| 16 | Pyruvate kinase (pykA/pykF) | |
| 17 | Pyruvate dehydrogenase (aceE/aceF) | |
| 18 | Pyruvate dehydrogenase (poxB) | |
| 19 | Pyruvate carboxylase (pycA/pycB) | P. aeruginosa specific; pyruvate metabolism |
| Y. lipolytica | ||
|---|---|---|
| Enzyme Number | Enzyme | Part in Metabolism |
| 1 | Acetyl-CoA synthetase (c) | Acetate uptake |
| 2 | Citrate synthase (m) | TCA cycle |
| 3 | Aconitate hydratase (m) | |
| 4 | Isocitrate dehydrogenase (m) | |
| 5 | α-ketoglutarate dehydrogenase (m) | |
| 6 | Succinyl-CoA synthetase (m) | |
| 7 | Succinate dehydrogenase (m) | |
| 8 | Fumarate hydratase (m) | |
| 9 | Malate dehydrogenase (m) | |
| 10 | Malate dehydrogenase (c) | Pyruvate metabolism |
| 11 | Pyruvate carboxylase (c) | |
| 12 | Pyruvate kinase (c) | |
| 13 | Pyruvate dehydrogenase (m) | |
| 14 | Aconitate hydratase (c) | Glyoxylate cycle |
| 15 | Isocitrate lyase (c) | |
| 16 | Malate synthase (p) | |
| 17 | Malate dehydrogenase (p) | |
| 18 | Malate dehydrogenase (c) | |
| C. kluyveri | |
|---|---|
| Enzyme Number | Enzyme |
| 1 | Acetate kinase |
| 2 | Phosphotransacetylase |
| 3 | Alcohol dehydrogenase |
| 4 | Aldehyde dehydrogenase |
| 5 | Butyryl-CoA: Acetate CoA transferase |
| 6 | Acetoacetyl-CoA thiolase |
| 7 | 3-Hydroxybutyryl-CoA dehydrogenase |
| 8 | 3-Hydroxybutyryl-CoA dehydratase |
| 9 | Ferredoxin-dependent hydrogenase |
| Nfn | NADH-dependent reduced ferredoxin:NADP oxidoreductase |
| Rnf | Ferredoxin-NAD reductase complex |
| Sulfate Reducers | ||
|---|---|---|
| Enzyme Number | Enzyme | Pathway |
| 1 | ATP citrate lyase | Modified TCA cycle |
| 2 | Aconitate hydratase | |
| 3 | Isocitrate dehydrogenase | |
| 4 | 2-Oxoglutarate synthase | |
| 5 | Succinyl-CoA:acetate CoA-transferase | |
| 6 | Succinate dehydrogenase | |
| 7 | Fumarate hydratase | |
| 8 | Malate dehydrogenase | |
| 9 | Acetate kinase | Reverse Wood–Ljungdahl pathway |
| 10 | Phosphotransacetylase | |
| 11 | Acetyl-CoA synthase | |
| 12 | Carbon monoxide dehydrogenase | |
| 13 | Methyltransferase | |
| Product | Organism | Comment | Reference |
|---|---|---|---|
| Lipids | Y. lipolytica | [97] | |
| Cryptococcus curvatus ATCC 20509 | Study also screened various other oleaginous yeasts for acetate-based production | [98] | |
| Rhodobacter sp. KKU-PS1 | [99] | ||
| Aurantiochytrium limacinum SR21 | [100] | ||
| Polyhydroxyalkanoate/ polyhydroxybutyrate | Dinoroseobacter sp. JL1447 | [101] | |
| Pseudomonas putida KT2440 | [37] | ||
| Synechocystis PCC 6803 | Acetate and glucose | [102] | |
| Aeromonas hydrophilia 4AK4 | Metabolically engineered | [103] | |
| Y. lipolytica Po1 g | Heterologous pathway for PHB production introduced | [104] | |
| Histidine | Brevibacterium flavum FERM1564 (Corynebacterium glutamicum) | Glucose added; uracil auxotroph mutant strain | [105] |
| Caproate | Clostridium kluyveri DSM 555 | Ethanol as additional carbon source | [36] |
| Malic acid | Aspergillus oryzae DSM1863 | Sequential culture with Clostridium ljungdahlii for acetate production from syngas | [106] |
| H2 | Rhodobacter sphaeroides ATCC 17023 | Glutamate as N-source | [107] |
| Electricity | Cupriavidus basilensis 9750 | As part of a microbial fuel cell | [108] |
| Methane/biogas | Various methanogens in communities with acetogens | Different wastes/ biowastes as substrates for acetate and H2 production | [109,110,111] |
| Isotope labeled L-glutamate | Brevibacterium flavum ATCC 14067 (Corynebacterium glutamicum) | 13C-acetate for labelled L-glutamate | [112] |
| Enzyme Number | Enzyme | Part in Metabolism |
|---|---|---|
| 1 | Acetate kinase (ackA) | Acetate uptake |
| 2 | Phosphotransacetylase (pta) | |
| 3 | Acetyl-CoA synthetase (acs) | |
| 4 | Citrate synthase (gltA) | TCA cycle |
| 5 | Aconitate hydratase (acnA/acnB) | |
| 6 | Isocitrate dehydrogenase (icd) | |
| 7 | α-ketoglutarate dehydrogenase (sucA/sucB) | |
| 8 | Succinyl-CoA synthetase (sucC/sucD) | |
| 9 | Succinate dehydrogenase (sdhCDAB) | |
| 10 | Fumarate hydratase (aerobic: fumA) | |
| 11 | Malate dehydrogenase (mdh/mqo) | |
| 12 | Isocitrate lyase (aceA) | Glyoxylate cycle |
| 13 | Malate synthase (glcB) | |
| 14 | Malate dehydrogenase (maeA/maeB) | Pyruvate metabolism |
| 15 | Phosphoenolpyruvate carboxykinase (ATP-dependent) (pckA) | |
| 16 | Pyruvate kinase (pykA/pykF) | |
| 17 | Pyruvate dehydrogenase (aceE/aceF) | |
| 18 | Pyruvate dehydrogenase (poxB) | |
| 19 | Glucokinase (glk)or PTS system | Glycolysis; glucose |
| 20 | Glucose-6-phosphate isomerase (pgi) | |
| 21 | ATP-dependent 6-phosphofructokinase (pfkB) | |
| 22 | Fructose-bisphosphate aldolase (fbaB) | |
| 23 | Triosephosphate isomerase (tpiA) | Glycolysis |
| 24 | Glyceraldehyde-3-phosphate dehydrogenase A (gapA) | |
| 25 | Phosphoglycerate kinase (pgk) | |
| 26 | 2,3-Bisphosphoglycerate-dependent/independent phosphoglycerate mutase (gpmA/gpmI) | |
| 27 | Enolase (eno) | |
| 28 | Glycerol kinase (glpK) | Glycolysis; glycerol |
| 29 | Aerobic glycerol-3-phosphate dehydrogenase (glpD) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kutscha, R.; Pflügl, S. Microbial Upgrading of Acetate into Value-Added Products—Examining Microbial Diversity, Bioenergetic Constraints and Metabolic Engineering Approaches. Int. J. Mol. Sci. 2020, 21, 8777. https://doi.org/10.3390/ijms21228777
Kutscha R, Pflügl S. Microbial Upgrading of Acetate into Value-Added Products—Examining Microbial Diversity, Bioenergetic Constraints and Metabolic Engineering Approaches. International Journal of Molecular Sciences. 2020; 21(22):8777. https://doi.org/10.3390/ijms21228777
Chicago/Turabian StyleKutscha, Regina, and Stefan Pflügl. 2020. "Microbial Upgrading of Acetate into Value-Added Products—Examining Microbial Diversity, Bioenergetic Constraints and Metabolic Engineering Approaches" International Journal of Molecular Sciences 21, no. 22: 8777. https://doi.org/10.3390/ijms21228777
APA StyleKutscha, R., & Pflügl, S. (2020). Microbial Upgrading of Acetate into Value-Added Products—Examining Microbial Diversity, Bioenergetic Constraints and Metabolic Engineering Approaches. International Journal of Molecular Sciences, 21(22), 8777. https://doi.org/10.3390/ijms21228777





