A Human Periodontal Ligament Fibroblast Cell Line as a New Model to Study Periodontal Stress
Abstract
:1. Introduction
2. Results
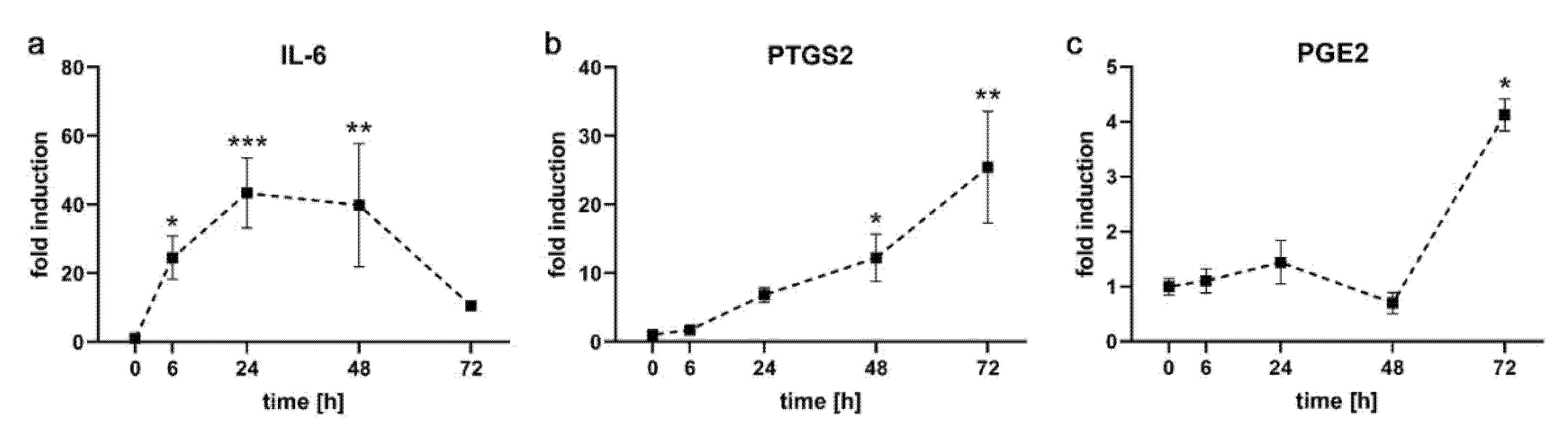
2.1. Kinetics of Pro-Inflammatory Markers in PDL-hTERT Cells
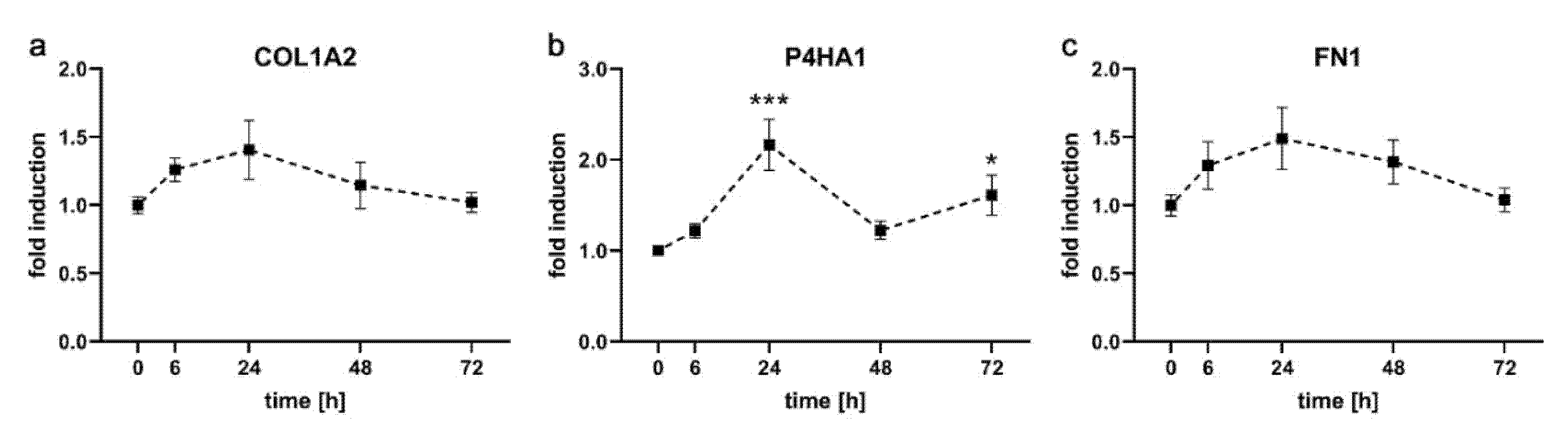
2.2. Expression Kinetics of Genes Involved in Formation of Extracellular Matrix in PDL-hTERT Cells
2.3. Expression Kinetics of Genes Involved in Angiogenesis, Osteoblastogenesis and Osteoclastogenesis
2.4. Transfection Efficiency of PDL-hTERT Cells
3. Discussion
4. Materials and Methods
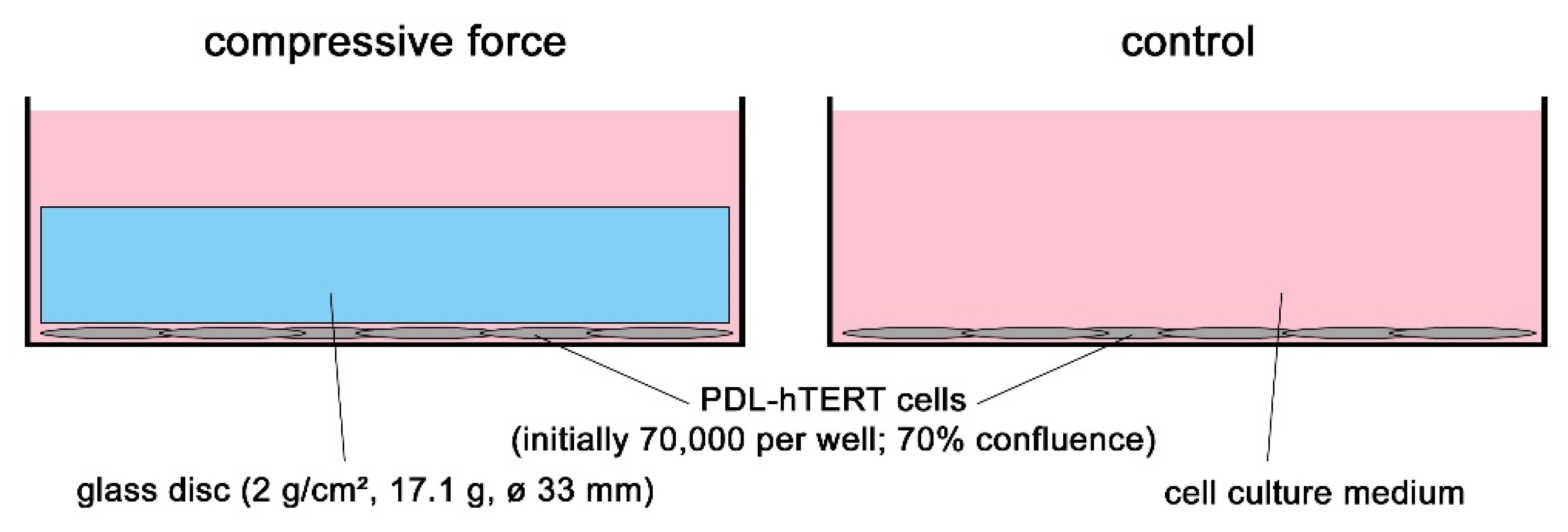
4.1. Cell Culture Experiments and In Vitro Model for Compressive Mechanical Strain
4.2. Isolation of RNA and Reverse Transcription
4.3. Quantitative Real-Time PCR
4.4. ELISA
4.5. Cell Transfection and Determination of Transfection Rate
4.6. Statistical Methods
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ELISA | enzyme-linked immunosorbent assay |
| hPDLF | human periodontal ligament fibroblasts |
| OTM | orthodontic tooth movement |
| PDL | periodontal ligament |
| PEI | Polyethylenimine |
| qRT-PCR | quantitative reverse transcriptase polymerase chain reaction |
| SEM | standard error of the mean |
References
- Lekic, P.; McCulloch, C.A.G. Periodontal ligament cell populations: The central role of fibroblasts in creating a unique tissue. Anat. Rec. 1996, 245, 327–341. [Google Scholar] [CrossRef]
- Beertsen, W.; McCulloch, C.A.; Sodek, J. The periodontal ligament: A unique, multifunctional connective tissue. Periodontol 2000 1997, 13, 20–40. [Google Scholar] [CrossRef] [PubMed]
- Kanzaki, H.; Chiba, M.; Shimizu, Y.; Mitani, H. Periodontal ligament cells under mechanical stress induce osteoclastogenesis by receptor activator of nuclear factor kappaB ligand up-regulation via prostaglandin E2 synthesis. J. Bone Miner. Res. 2002, 17, 210–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rios, H.F.; Ma, D.; Xie, Y.; Giannobile, W.V.; Bonewald, L.F.; Conway, S.J.; Feng, J.Q. Periostin is essential for the integrity and function of the periodontal ligament during occlusal loading in mice. J. Periodontol. 2008, 79, 1480–1490. [Google Scholar] [CrossRef] [PubMed]
- Gilroy, D.W.; Bishop-Bailey, D. Lipid mediators in immune regulation and resolution. Br. J. Pharmacol. 2019, 176, 1009–1023. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, J.A.; Kirkby, N.S. Eicosanoids, prostacyclin and cyclooxygenase in the cardiovascular system. Br. J. Pharmacol. 2019, 176, 1038–1050. [Google Scholar] [CrossRef]
- Kaku, M.; Motokawa, M.; Tohma, Y.; Tsuka, N.; Koseki, H.; Sunagawa, H.; Arturo Marquez Hernandes, R.; Ohtani, J.; Fujita, T.; Kawata, T.; et al. VEGF and M-CSF levels in periodontal tissue during tooth movement. Biomed. Res. 2008, 29, 181–187. [Google Scholar] [CrossRef] [Green Version]
- Meikle, M.C. The tissue, cellular, and molecular regulation of orthodontic tooth movement: 100 years after Carl Sandstedt. Eur. J. Orthod. 2006, 28, 221–240. [Google Scholar] [CrossRef]
- Brooks, P.J.; Heckler, A.F.; Wei, K.; Gong, S.G. M-CSF accelerates orthodontic tooth movement by targeting preosteoclasts in mice. Angle Orthod. 2011, 81, 277–283. [Google Scholar] [CrossRef] [Green Version]
- Theill, L.E.; Boyle, W.J.; Penninger, J.M. RANK-L and RANK: T cells, bone loss, and mammalian evolution. Annu Rev. Immunol. 2002, 20, 795–823. [Google Scholar] [CrossRef] [Green Version]
- Boyce, B.F.; Xing, L. Functions of RANKL/RANK/OPG in bone modeling and remodeling. Arch. Biochem Biophys 2008, 473, 139–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, W.E.; Huja, S.; Roberts, J.A. Bone modeling: Biomechanics, molecular mechanisms, and clinical perspectives. Semin. Orthod. 2004, 10, 123–161. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, W.; Liu, J.S.; Wang, J.; Yang, P.; Li, M.L.; Zhao, Z.H. Expression of osteoclastogenesis inducers in a tissue model of periodontal ligament under compression. J. Dent. Res. 2011, 90, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yu, M.; Jin, S.; Wang, Y.; Luo, R.; Huo, B.; Liu, D.; He, D.; Zhou, Y.; Liu, Y. Stress Distribution and Collagen Remodeling of Periodontal Ligament During Orthodontic Tooth Movement. Front. Pharm. 2019, 10, 1263. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.F.; Zhao, Y.B.; Zhang, F.M.; Han, P.Y. Comparative study of gene expression during tooth eruption and orthodontic tooth movement in mice. Oral Dis. 2009, 15, 573–579. [Google Scholar] [CrossRef]
- Schröder, A.; Bauer, K.; Spanier, G.; Proff, P.; Wolf, M.; Kirschneck, C. Expression kinetics of human periodontal ligament fibroblasts in the early phases of orthodontic tooth movement. J. Orofac. Orthop. 2018, 79, 337–351. [Google Scholar] [CrossRef]
- Janjic Rankovic, M.; Docheva, D.; Wichelhaus, A.; Baumert, U. Effect of static compressive force on in vitro cultured PDL fibroblasts: Monitoring of viability and gene expression over 6 days. Clin. Oral Investig. 2020, 24, 2497–2511. [Google Scholar] [CrossRef]
- Kanzaki, H.; Chiba, M.; Shimizu, Y.; Mitani, H. Dual regulation of osteoclast differentiation by periodontal ligament cells through RANKL stimulation and OPG inhibition. J. Dent. Res. 2001, 80, 887–891. [Google Scholar] [CrossRef]
- Kirschneck, C.; Batschkus, S.; Proff, P.; Köstler, J.; Spanier, G.; Schröder, A. Valid gene expression normalization by RT-qPCR in studies on hPDL fibroblasts with focus on orthodontic tooth movement and periodontitis. Sci. Rep. 2017, 7, 14751. [Google Scholar] [CrossRef] [Green Version]
- Memmert, S.; Damanaki, A.; Weykopf, B.; Rath-Deschner, B.; Nokhbehsaim, M.; Götz, W.; Gölz, L.; Till, A.; Deschner, J.; Jäger, A. Autophagy in periodontal ligament fibroblasts under biomechanical loading. Cell Tissue Res. 2019, 378, 499–511. [Google Scholar] [CrossRef]
- Memmert, S.; Nogueira, A.V.B.; Damanaki, A.; Nokhbehsaim, M.; Rath-Deschner, B.; Götz, W.; Gölz, L.; Cirelli, J.A.; Till, A.; Jäger, A.; et al. Regulation of the autophagy-marker Sequestosome 1 in periodontal cells and tissues by biomechanical loading. J. Orofac Orthop. 2020, 81, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Kirschneck, C.; Proff, P.; Maurer, M.; Reicheneder, C.; Römer, P. Orthodontic forces add to nicotine-induced loss of periodontal bone. J. Orofac. Orthop./Fortschr. Der. Kieferorthopädie 2015, 76, 195–212. [Google Scholar] [CrossRef] [PubMed]
- Docheva, D.; Padula, D.; Popov, C.; Weishaupt, P.; Pragert, M.; Miosge, N.; Hickel, R.; Bocker, W.; Clausen-Schaumann, H.; Schieker, M. Establishment of immortalized periodontal ligament progenitor cell line and its behavioural analysis on smooth and rough titanium surface. Eur. Cell Mater. 2010, 19, 228–241. [Google Scholar] [CrossRef] [PubMed]
- Kirschneck, C.; Meier, M.; Bauer, K.; Proff, P.; Fanghänel, J. Meloxicam medication reduces orthodontically induced dental root resorption and tooth movement velocity: A combined in vivo and in vitro study of dental-periodontal cells and tissue. Cell Tissue Res. 2017, 368, 61–78. [Google Scholar] [CrossRef]
- Schröder, A.; Nazet, U.; Neubert, P.; Jantsch, J.; Spanier, G.; Proff, P.; Kirschneck, C. Sodium-chloride-induced effects on the expression profile of human periodontal ligament fibroblasts with focus on simulated orthodontic tooth movement. Eur. J. Oral Sci. 2019, 127, 386–395. [Google Scholar] [CrossRef]
- Kirschneck, C.; Küchler, E.C.; Wolf, M.; Spanier, G.; Proff, P.; Schröder, A. Effects of the Highly COX-2-Selective Analgesic NSAID Etoricoxib on Human Periodontal Ligament Fibroblasts during Compressive Orthodontic Mechanical Strain. Mediat. Inflamm. 2019. [Google Scholar] [CrossRef]
- Sari, E.; Olmez, H.; Gürton, A.U. Comparison of some effects of acetylsalicylic acid and rofecoxib during orthodontic tooth movement. Am. J. Orthod. Dentofac. Orthop. 2004, 125, 310–315. [Google Scholar] [CrossRef]
- Shetty, N.; Patil, A.K.; Ganeshkar, S.V.; Hegde, S. Comparison of the effects of ibuprofen and acetaminophen on PGE2 levels in the GCF during orthodontic tooth movement: A human study. Prog Orthod. 2013, 14, 6. [Google Scholar] [CrossRef] [Green Version]
- Schröder, A.; Küchler, E.C.; Omori, M.; Spanier, G.; Proff, P.; Kirschneck, C. Effects of ethanol on human periodontal ligament fibroblasts subjected to static compressive force. Alcohol 2019, 77, 59–70. [Google Scholar] [CrossRef]
- Okugawa, Y.; Toiyama, Y.; Ichikawa, T.; Kawamura, M.; Yasuda, H.; Fujikawa, H.; Saigusa, S.; Ohi, M.; Araki, T.; Tanaka, K.; et al. Colony-stimulating factor-1 and colony-stimulating factor-1 receptor co-expression is associated with disease progression in gastric cancer. Int. J. Oncol. 2018, 53, 737–749. [Google Scholar] [CrossRef] [Green Version]


| Target Gene | Forward Primer (5′-…-3′) | Reverse Primer (5′-…-3′) | Annealing Temperature | Reference |
|---|---|---|---|---|
| ALPL | ACAAGCACTCCCACTTCATCTG | GGTCCGTCACGTTGTTCCTG | 60 °C | [16] |
| COL1A2 | AGAAACACGTCTGGCTAGGAG | GCATGAAGGCAAGTTGGGTAG | 60 °C | [16] |
| FN1 | GCCAGTCCTACAACCAGTATTCTC | GCTTGTTCCTCTGGATTGGAAAG | 60 °C | [16] |
| IL-6 | TGGCAGAAAACAACCTGAACC | CCTCAAACTCCAAAAGACCAGTG | 60 °C | [16] |
| M-CSF | GGAGACCTCGTGCCAAATTA | GGCATTGGGGGTGTTATCTC | 60 °C | [30] |
| P4HA1 | GCTCTCTGGCTATGAAAATCCTG | GTGCAAAGTCAAAATGGGGTTC | 60 °C | [16] |
| PPIB | TTCCATCGTGTAATCAAGGACTTC | GCTCACCGTAGATGCTCTTTC | 60 °C | [16] |
| PTGS2 | GAGCAGGCAGATGAAATACCAGTC | TGTCACCATAGAGTGCTTCCAAC | 60 °C | [16] |
| VEGF-A | TGCAGACCAAAGAAAGATAGAGC | ACGCTCCAGGACTTATACCG | 58 °C | [16] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weider, M.; Schröder, A.; Docheva, D.; Rodrian, G.; Enderle, I.; Seidel, C.L.; Andreev, D.; Wegner, M.; Bozec, A.; Deschner, J.; et al. A Human Periodontal Ligament Fibroblast Cell Line as a New Model to Study Periodontal Stress. Int. J. Mol. Sci. 2020, 21, 7961. https://doi.org/10.3390/ijms21217961
Weider M, Schröder A, Docheva D, Rodrian G, Enderle I, Seidel CL, Andreev D, Wegner M, Bozec A, Deschner J, et al. A Human Periodontal Ligament Fibroblast Cell Line as a New Model to Study Periodontal Stress. International Journal of Molecular Sciences. 2020; 21(21):7961. https://doi.org/10.3390/ijms21217961
Chicago/Turabian StyleWeider, Matthias, Agnes Schröder, Denitsa Docheva, Gabriele Rodrian, Isabel Enderle, Corinna Lesley Seidel, Darja Andreev, Michael Wegner, Aline Bozec, James Deschner, and et al. 2020. "A Human Periodontal Ligament Fibroblast Cell Line as a New Model to Study Periodontal Stress" International Journal of Molecular Sciences 21, no. 21: 7961. https://doi.org/10.3390/ijms21217961
APA StyleWeider, M., Schröder, A., Docheva, D., Rodrian, G., Enderle, I., Seidel, C. L., Andreev, D., Wegner, M., Bozec, A., Deschner, J., Kirschneck, C., Proff, P., & Gölz, L. (2020). A Human Periodontal Ligament Fibroblast Cell Line as a New Model to Study Periodontal Stress. International Journal of Molecular Sciences, 21(21), 7961. https://doi.org/10.3390/ijms21217961







