Nitrate Respiration in Thermus thermophilus NAR1: from Horizontal Gene Transfer to Internal Evolution
Abstract
:1. Introduction
2. Materials and Methods
2.1. DNA Preparation and Sequencing
2.2. Genome Sequence Analysis and Assembly
2.3. Annotation of TthNAR1 Replicons
2.4. Phylogenomic Assessments
3. Results and Discussion
3.1. Sequence of the TthNAR1 Strain
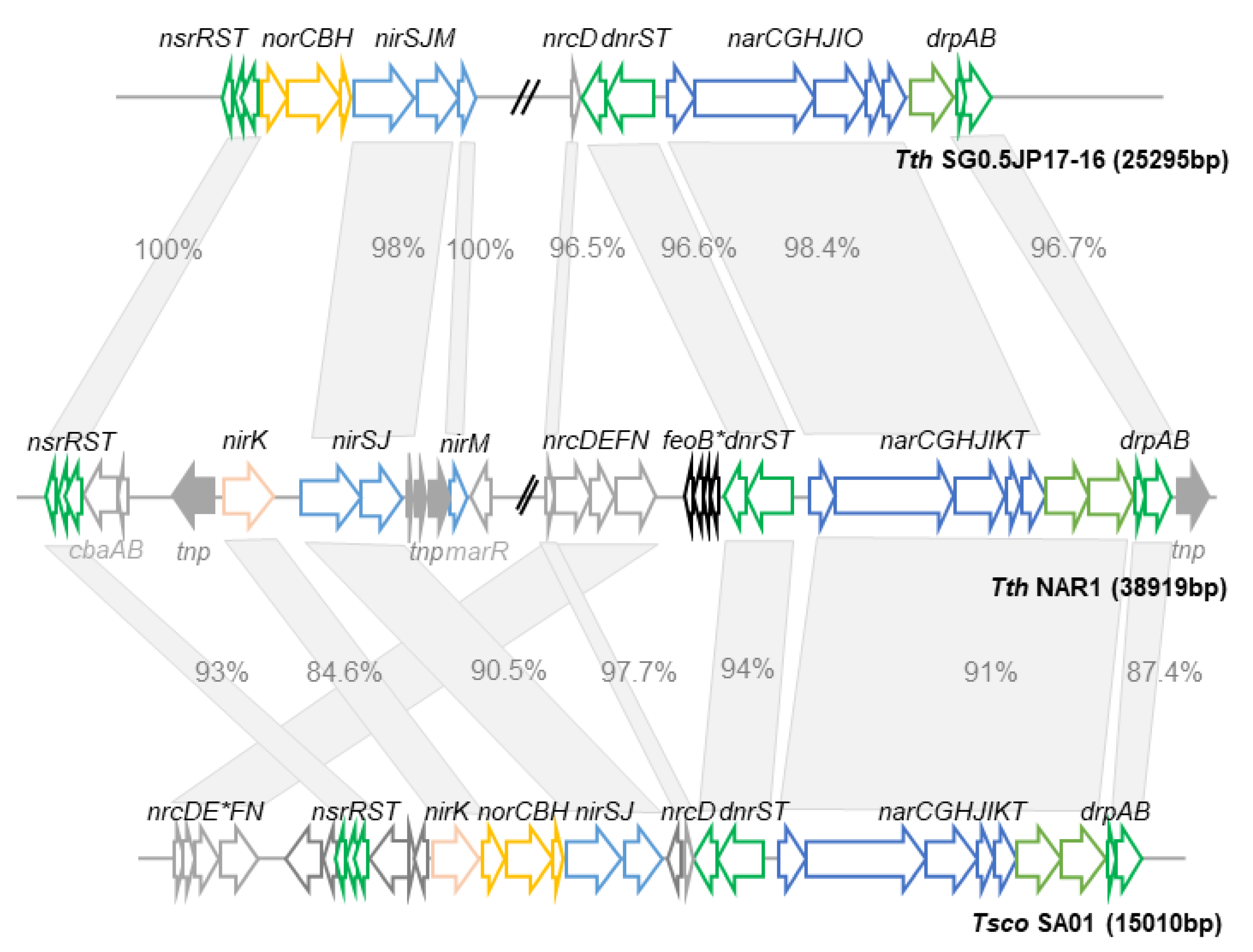
3.2. The Nitrate Reductase Gene Cluster
3.3. The Respiratory Nitrate Reductase (NR) of TthNAR1 and Other Thermus spp.
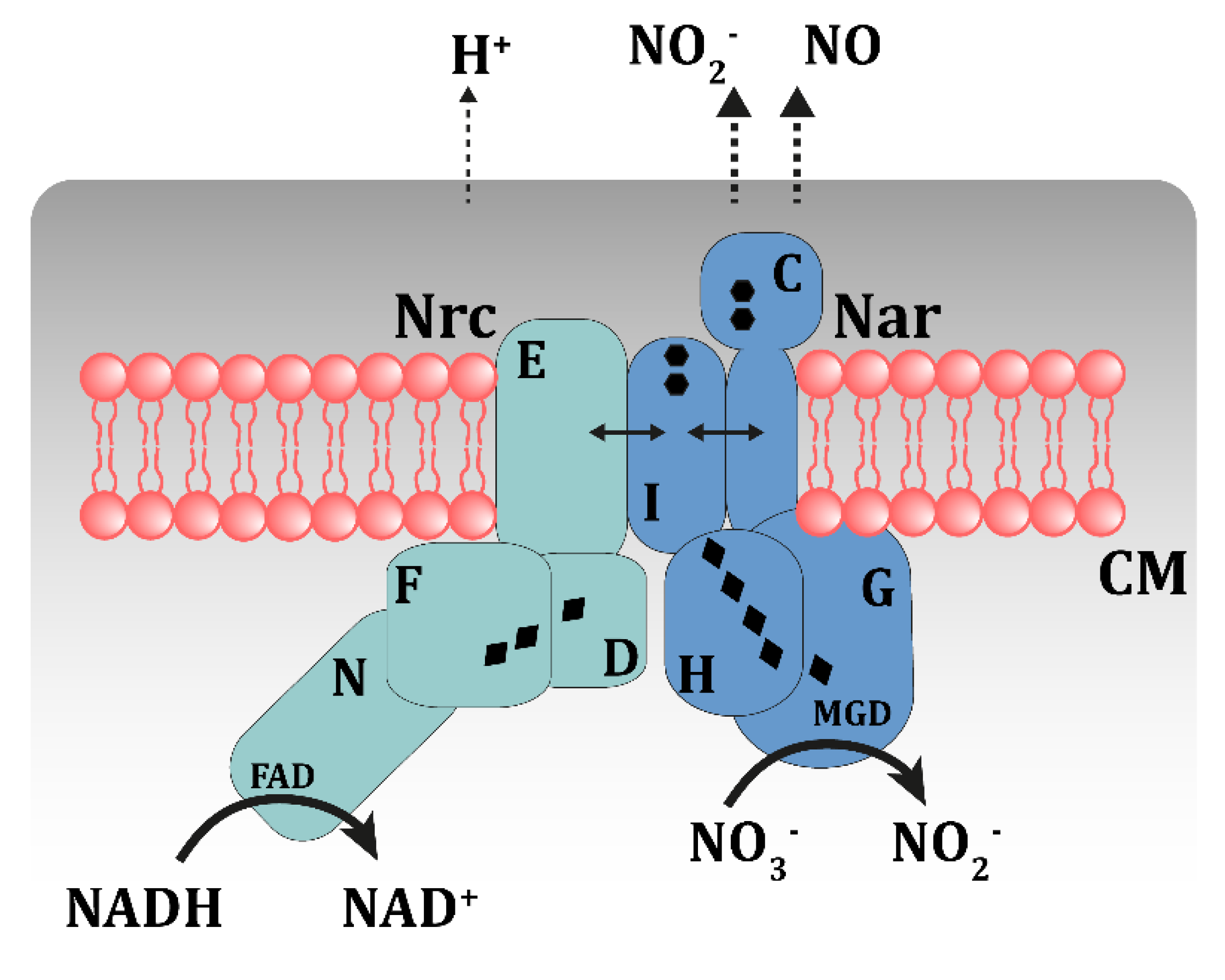
3.4. A Putative Nitrate Respirasome in the NAR1 Strain
3.5. Regulation of Nitrate Respiration in TthNAR1
3.6. Genes for Nitrite Respiration in TthNAR1
3.7. Genes for NO Respiration
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Simon, J.; Klotz, M.G. Diversity and evolution of bioenergetic systems involved in microbial nitrogen compound transformations. Biochim. Biophys. Acta 2013, 1827, 114–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiedje, J.M. Ecology of denitrification and dissimilatory nitrate reduction to ammonium. In Biology of Anaerobic Microorganisms; John Wiley & Sons: Hoboken, NY, USA, 1988; pp. 179–244. [Google Scholar]
- Zumft, W.G. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 1997, 61, 533–616. [Google Scholar] [CrossRef] [PubMed]
- Richardson, D.J. Bacterial respiration: A flexible process for a changing environment. Microbiology 2000, 146 Pt 3, 551–571. [Google Scholar] [CrossRef] [Green Version]
- Gaimster, H.; Alston, M.; Richardson, D.J.; Gates, A.J.; Rowley, G. Transcriptional and environmental control of bacterial denitrification and N2O emissions. FEMS Microbiol. Lett. 2018, 365, fnx277. [Google Scholar] [CrossRef] [PubMed]
- Betlach, M.R. Evolution of bacterial denitrification and denitrifier diversity. Antonie Van Leeuwenhoek 1982, 48, 585–607. [Google Scholar] [CrossRef] [PubMed]
- Knowles, R. Denitrification. Microbiol. Rev. 1982, 46, 43–70. [Google Scholar] [CrossRef] [PubMed]
- Richardson, D.; Felgate, H.; Watmough, N.; Thomson, A.; Baggs, E. Mitigating release of the potent greenhouse gas N2O from the nitrogen cycle—could enzymic regulation hold the key? Trends Biotechnol. 2009, 27, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Chapuis-Lardy, L.; Wrage, N.; Metay, A.; Chotte, J.L.; Bernoux, M. Soils, a sink for N2O? A review. Glob. Chang. Biol. 2007, 13, 1–17. [Google Scholar] [CrossRef]
- Philippot, L.; Andert, J.; Jones, C.M.; Bru, D.; Hallin, S. Importance of denitrifiers lacking the genes encoding the nitrous oxide reductase for N2O emissions from soil. Glob. Chang. Biol. 2011, 17, 1497–1504. [Google Scholar] [CrossRef]
- Houlton, B.Z.; Bai, E. Imprint of denitrifying bacteria on the global terrestrial biosphere. Proc. Natl. Acad. Sci. USA 2009, 106, 21713–21716. [Google Scholar] [CrossRef] [Green Version]
- Philippot, L.; Mirleau, P.; Mazurier, S.; Siblot, S.; Hartmann, A.; Lemanceau, P.; Germon, J.C. Characterization and transcriptional analysis of Pseudomonas fluorescens denitrifying clusters containing the nar, nir, nor and nos genes. Biochim. Biophys. Acta 2001, 1517, 436–440. [Google Scholar] [CrossRef]
- Vollack, K.U.; Xie, J.; Hartig, E.; Romling, U.; Zumft, W.G. Localization of denitrification genes on the chromosomal map of Pseudomonas aeruginosa. Microbiology 1998, 144 Pt 2, 441–448. [Google Scholar] [CrossRef] [Green Version]
- Zumft, W.G.; Korner, H. Enzyme diversity and mosaic gene organization in denitrification. Antonie Van Leeuwenhoek 1997, 71, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Verbaendert, I.; De Vos, P.; Boon, N.; Heylen, K. Denitrification in Gram-positive bacteria: An underexplored trait. Biochem. Soc. Trans. 2011, 39, 254–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torregrosa-Crespo, J.; Pire, C.; Martinez-Espinosa, R.M.; Bergaust, L. Denitrifying haloarchaea within the genus Haloferax display divergent respiratory phenotypes, with implications for their release of nitrogenous gases. Environ. Microbiol. 2019, 21, 427–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez, L.; Bricio, C.; Blesa, A.; Hidalgo, A.; Berenguer, J. Transferable denitrification capability of Thermus thermophilus. Appl. Environ. Microbiol. 2014, 80, 19–28. [Google Scholar] [CrossRef] [Green Version]
- Philippot, L. Denitrifying genes in bacterial and archaeal genomes. Biochim. Biophys. Acta 2002, 1577, 355–376. [Google Scholar] [CrossRef]
- Manaia, C.M.; Hoste, B.; Gutierrez, M.C.; Gillis, M.; Ventosa, A.; Kesters, K.; da Costa, M.S. Halotolerant thermus strains from marine and terrestrial hot-springs belong to Thermus thermophilus (ex Oshima, and Imahori, 1974) nom. Rev. Emend. Syst. App. Microbiol. 1995, 17, 526–532. [Google Scholar] [CrossRef]
- Cava, F.; Hidalgo, A.; Berenguer, J. Thermus thermophilus as biological model. Extremophiles 2009, 13, 213–231. [Google Scholar] [CrossRef]
- Averhoff, B. Shuffling genes around in hot environments: The unique DNA transporter of Thermus thermophilus. FEMS Microbiol. Rev. 2009, 33, 611–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verdu, C.; Sanchez, E.; Ortega, C.; Hidalgo, A.; Berenguer, J.; Mencia, M. A modular vector toolkit with a tailored set of thermosensors to regulate gene expression in Thermus thermophilus. ACS Omega 2019, 4, 14626–14632. [Google Scholar] [CrossRef] [Green Version]
- Li, H. Selection-free markerless genome manipulations in the polyploid bacterium Thermus thermophilus. 3 Biotech 2019, 9, 148. [Google Scholar] [CrossRef]
- Togawa, Y.; Shiotani, S.; Kato, Y.; Ezaki, K.; Nunoshiba, T.; Hiratsu, K. Development of a supf-based mutation-detection system in the extreme thermophile Thermus thermophilus HB27. Mol. Genet. Genom. 2019, 294, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Togawa, Y.; Nunoshiba, T.; Hiratsu, K. Cre/lox-based multiple markerless gene disruption in the genome of the extreme thermophile Thermus thermophilus. Mol. Genet. Genom. 2018, 293, 277–291. [Google Scholar] [CrossRef]
- Henne, A.; Bruggemann, H.; Raasch, C.; Wiezer, A.; Hartsch, T.; Liesegang, H.; Johann, A.; Lienard, T.; Gohl, O.; Martinez-Arias, R.; et al. The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol. 2004, 22, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Bruggemann, H.; Chen, C. Comparative genomics of Thermus thermophilus: Plasticity of the megaplasmid and its contribution to a thermophilic lifestyle. J. Biotechnol. 2006, 124, 654–661. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, N.; Tomita, M.; Itaya, M. The third plasmid pVV8 from Thermus thermophilus HB8: Isolation, characterization, and sequence determination. Extremophiles 2012, 16, 237–244. [Google Scholar] [CrossRef]
- Schwarzenlander, C.; Averhoff, B. Characterization of DNA transport in the thermophilic bacterium Thermus thermophilus HB27. FEBS J. 2006, 273, 4210–4218. [Google Scholar] [CrossRef]
- Au, K.F.; Underwood, J.G.; Lee, L.; Wong, W.H. Improving pacbio long read accuracy by short read alignment. PLoS ONE 2012, 7, e46679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramírez-Arcos, S.; Fernandez-Herrero, L.A.; Marin, I.; Berenguer, J. Anaerobic growth, a property horizontally transferred by an HFR-like mechanism among extreme thermophiles. J. Bacteriol. 1998, 180, 3137–3143. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Wingett, S.W.; Andrews, S. FASTQ screen: A tool for multi-genome mapping and quality control. F1000Res 2018, 7, 1338. [Google Scholar] [CrossRef]
- Chaisson, M.J.; Tesler, G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): Application and theory. BMC Bioinform. 2012, 13, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhoads, A.; Au, K.F. PacBio sequencing and its applications. Genom. Proteom. Bioinform. 2015, 13, 278–289. [Google Scholar] [CrossRef] [Green Version]
- Weirather, J.L.; Duggal, P.; Nascimento, E.L.; Monteiro, G.R.; Martins, D.R.; Lacerda, H.G.; Fakiola, M.; Blackwell, J.M.; Jeronimo, S.M.; Wilson, M.E. Comprehensive candidate gene analysis for symptomatic or asymptomatic outcomes of Leishmania infantum infection in Brazil. Ann. Hum. Genet. 2017, 81, 41–48. [Google Scholar] [CrossRef] [Green Version]
- De Maio, N.; Shaw, L.P.; Hubbard, A.; George, S.; Sanderson, N.D.; Swann, J.; Wick, R.; AbuOun, M.; Stubberfield, E.; Hoosdally, S.J.; et al. Comparison of long-read sequencing technologies in the hybrid assembly of complex bacterial genomes. Microb. Genom. 2019, 5. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. PROKKA: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Alam, I.; Antunes, A.; Kamau, A.A.; Ba Alawi, W.; Kalkatawi, M.; Stingl, U.; Bajic, V.B. Indigo—integrated data warehouse of microbial genomes with examples from the red sea extremophiles. PLoS ONE 2013, 8, e82210. [Google Scholar] [CrossRef] [Green Version]
- Kalkatawi, M.; Alam, I.; Bajic, V.B. BEACON: Automated tool for bacterial genome annotation comparison. BMC Genom. 2015, 16, 616. [Google Scholar] [CrossRef] [Green Version]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. MAUVE: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, M.; Blesa, A.; Sacristan-Horcajada, E.; Berenguer, J. Complete genome sequence of Mycolicibacterium hassiacum DSM 44199. Microbiol. Resour. Announc. 2019, 8, e01522-18. [Google Scholar] [CrossRef] [Green Version]
- Alvarez, L.; Sanchez-Hevia, D.; Sanchez, M.; Berenguer, J. A new family of nitrate/nitrite transporters involved in denitrification. Int. Microbiol. 2019, 22, 19–28. [Google Scholar] [CrossRef] [Green Version]
- Bertero, M.G.; Rothery, R.A.; Palak, M.; Hou, C.; Lim, D.; Blasco, F.; Weiner, J.H.; Strynadka, N.C. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase a. Nat. Struct. Biol. 2003, 10, 681–687. [Google Scholar] [CrossRef]
- Zafra, O.; Ramirez, S.; Castan, P.; Moreno, R.; Cava, F.; Valles, C.; Caro, E.; Berenguer, J. A cytochrome c encoded by the nar operon is required for the synthesis of active respiratory nitrate reductase in thermus thermophilus. FEBS Lett. 2002, 523, 99–102. [Google Scholar] [CrossRef] [Green Version]
- Zafra, O.; Cava, F.; Blasco, F.; Magalon, A.; Berenguer, J. Membrane-associated maturation of the heterotetrameric nitrate reductase of Thermus thermophilus. J. Bacteriol. 2005, 187, 3990–3996. [Google Scholar] [CrossRef] [Green Version]
- Cava, F.; Zafra, O.; Berenguer, J. A cytochrome c containing nitrate reductase plays a role in electron transport for denitrification in Thermus thermophilus without involvement of the bc respiratory complex. Mol. Microbiol. 2008, 70, 507–518. [Google Scholar] [CrossRef]
- Cava, F.; Zafra, O.; Magalon, A.; Blasco, F.; Berenguer, J. A new type of NADH dehydrogenase specific for nitrate respiration in the extreme thermophile Thermus thermophilus. J. Biol. Chem. 2004, 279, 45369–45378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venkatakrishnan, P.; Lencina, A.M.; Schurig-Briccio, L.A.; Gennis, R.B. Alternate pathways for NADH oxidation in Thermus thermophilus using type 2 NADH dehydrogenases. Biol. Chem. 2013, 394, 667–676. [Google Scholar] [CrossRef]
- Korner, H.; Sofia, H.J.; Zumft, W.G. Phylogeny of the bacterial superfamily of CRP-FNR transcription regulators: Exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 2003, 27, 559–592. [Google Scholar] [CrossRef] [Green Version]
- Schreiber, K.; Krieger, R.; Benkert, B.; Eschbach, M.; Arai, H.; Schobert, M.; Jahn, D. The anaerobic regulatory network required for Pseudomonas aeruginosa nitrate respiration. J. Bacteriol. 2007, 189, 4310–4314. [Google Scholar] [CrossRef] [Green Version]
- Alvarez, L.; Bricio, C.; Gomez, M.J.; Berenguer, J. Lateral transfer of the denitrification pathway among Thermus thermophilus strains. Appl. Environ. Microbiol. 2011, 77, 1352–1358. [Google Scholar] [CrossRef] [Green Version]
- Cava, F.; Berenguer, J. Biochemical and regulatory properties of a respiratory island encoded by a conjugative plasmid in the extreme thermophile Thermus thermophilus. Biochem. Soc. Trans. 2006, 34, 97–100. [Google Scholar] [CrossRef]
- Chahlafi, Z.; Alvarez, L.; Cava, F.; Berenguer, J. The role of conserved proteins DrpA and DrpB in nitrate respiration of Thermus thermophilus. Environ. Microbiol. 2018, 20, 3851–3861. [Google Scholar] [CrossRef]
- Nilkens, S.; Koch-Singenstreu, M.; Niemann, V.; Gotz, F.; Stehle, T.; Unden, G. Nitrate/oxygen co-sensing by an Nrea/Nreb sensor complex of Staphylococcus carnosus. Mol. Microbiol. 2014, 91, 381–393. [Google Scholar] [CrossRef]
- Alvarez, L.; Bricio, C.; Hidalgo, A.; Berenguer, J. Parallel pathways for nitrite reduction during anaerobic growth in Thermus thermophilus. J. Bacteriol. 2014, 196, 1350–1358. [Google Scholar] [CrossRef] [Green Version]
- Cava, F.; Zafra, O.; da Costa, M.S.; Berenguer, J. The role of the nitrate respiration element of Thermus thermophilus in the control and activity of the denitrification apparatus. Environ. Microbiol. 2008, 10, 522–533. [Google Scholar] [CrossRef]
- Opperman, D.J.; Murgida, D.H.; Dalosto, S.D.; Brondino, C.D.; Ferroni, F.M. A three-domain copper-nitrite reductase with a unique sensing loop. IUCrJ 2019, 6, 248–258. [Google Scholar] [CrossRef]
- Bricio, C.; Alvarez, L.; Gomez, M.J.; Berenguer, J. Partial and complete denitrification in Thermus thermophilus: Lessons from genome drafts. Biochem. Soc. Trans. 2011, 39, 249–253. [Google Scholar] [CrossRef]
- Alvarez, L.; Quintans, N.G.; Blesa, A.; Baquedano, I.; Mencia, M.; Bricio, C.; Berenguer, J. Hierarchical control of nitrite respiration by transcription factors encoded within mobile gene clusters of Thermus thermophilus. Genes 2017, 8, 361. [Google Scholar] [CrossRef] [Green Version]



| Contig | Bases | Coverage | CDS | Rep. Region (CRISPR) | rRNA | tRNA | IS |
|---|---|---|---|---|---|---|---|
| Chromosome | 2,021,843 | 339.8 | 2220 | 0 | 6 | 51 | 70 |
| pTTHNP2 | 9799 | 448.2 | 12 | 0 | 0 | 0 | 19 |
| pTTHNP3 | 77,135 | 130.5 | 106 | 2 (questionable) | 0 | 0 | 102 |
| pTTHNP4 | 370,865 | 464.9 | 429 | 4 | 0 | 0 | 100 |
| Position | ORF | IS-Type |
|---|---|---|
| 163912–162710 | TthNP4_176 + TthNP4_177 | ISTth4 |
| 167718–168215 | TthNP4_186 * | ISTth4 |
| 168356–168670 | TthNP4_187 * | ISTth4 |
| 168631–168915 | TthNP4_188 * | ISTth4 |
| 173562–174395 | TthNP4_192 | IS982-like |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Costa, M.; Blesa, A.; Berenguer, J. Nitrate Respiration in Thermus thermophilus NAR1: from Horizontal Gene Transfer to Internal Evolution. Genes 2020, 11, 1308. https://doi.org/10.3390/genes11111308
Sánchez-Costa M, Blesa A, Berenguer J. Nitrate Respiration in Thermus thermophilus NAR1: from Horizontal Gene Transfer to Internal Evolution. Genes. 2020; 11(11):1308. https://doi.org/10.3390/genes11111308
Chicago/Turabian StyleSánchez-Costa, Mercedes, Alba Blesa, and José Berenguer. 2020. "Nitrate Respiration in Thermus thermophilus NAR1: from Horizontal Gene Transfer to Internal Evolution" Genes 11, no. 11: 1308. https://doi.org/10.3390/genes11111308





