Transcriptional and Metabolic Dissection of ATRA-Induced Granulocytic Differentiation in NB4 Acute Promyelocytic Leukemia Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines, Culture Conditions, and Treatments
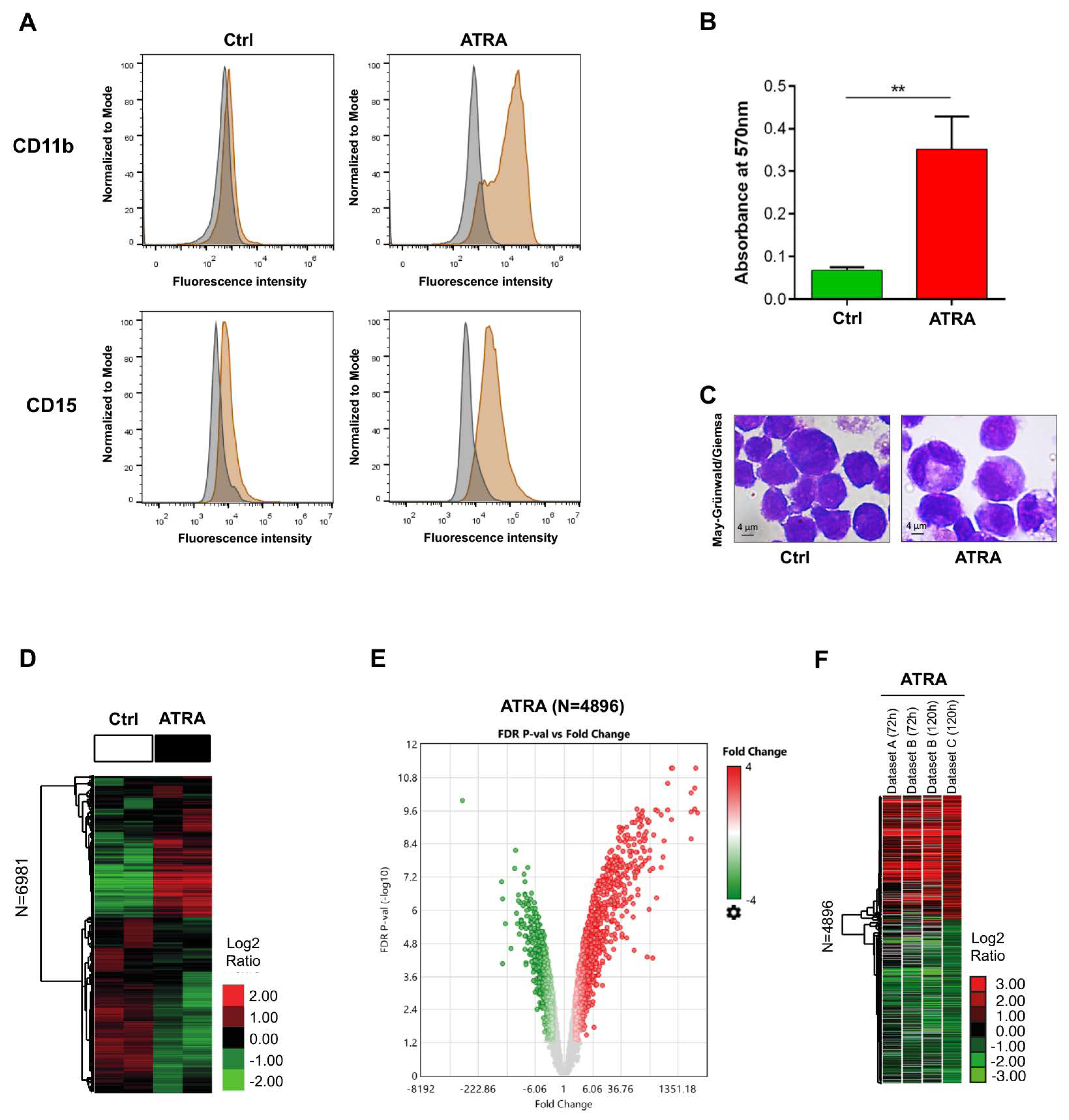
2.2. NBT Assay
2.3. May-Grünwald-Giemsa Staining
2.4. Flow Cytometric Analysis
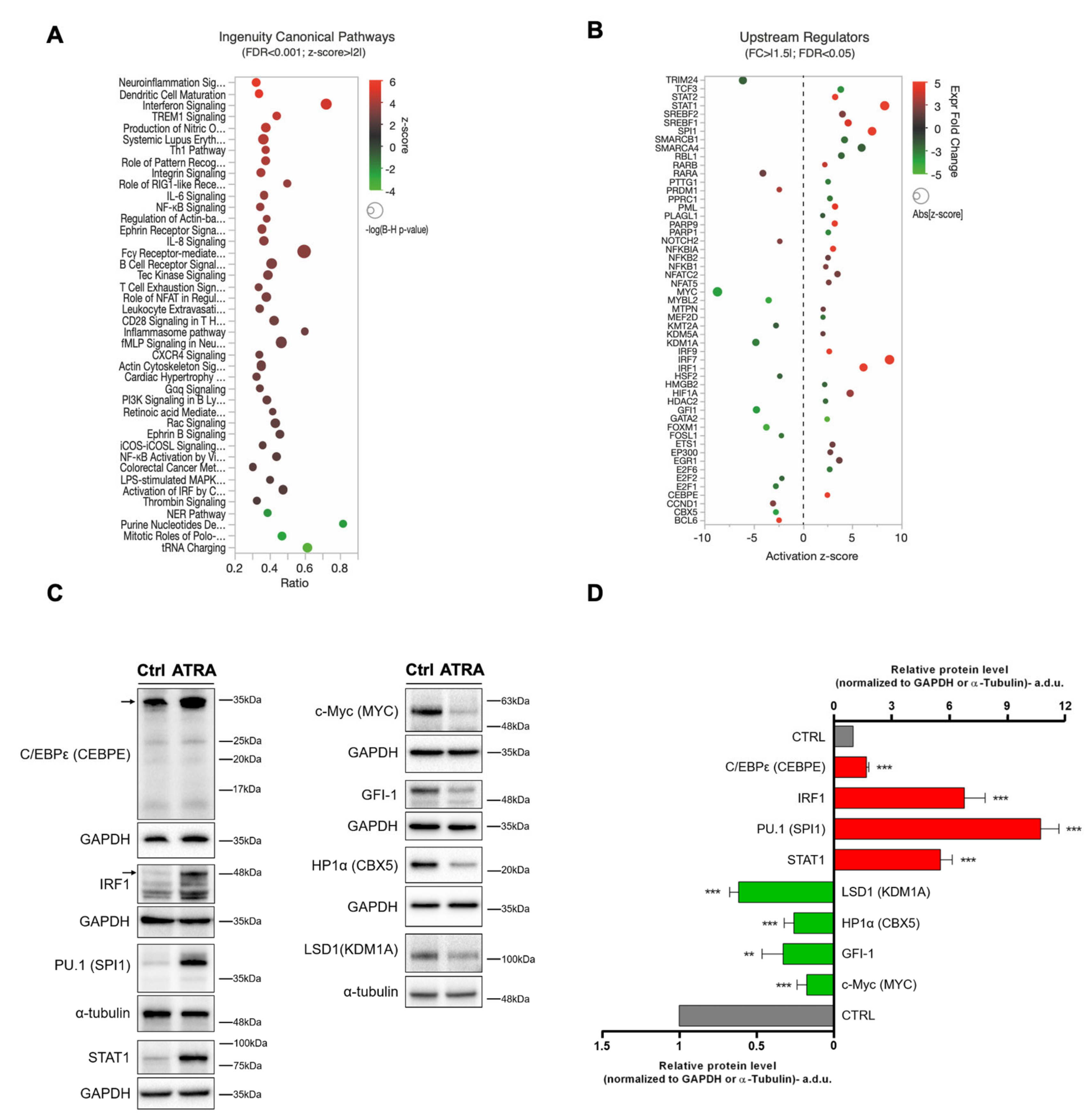
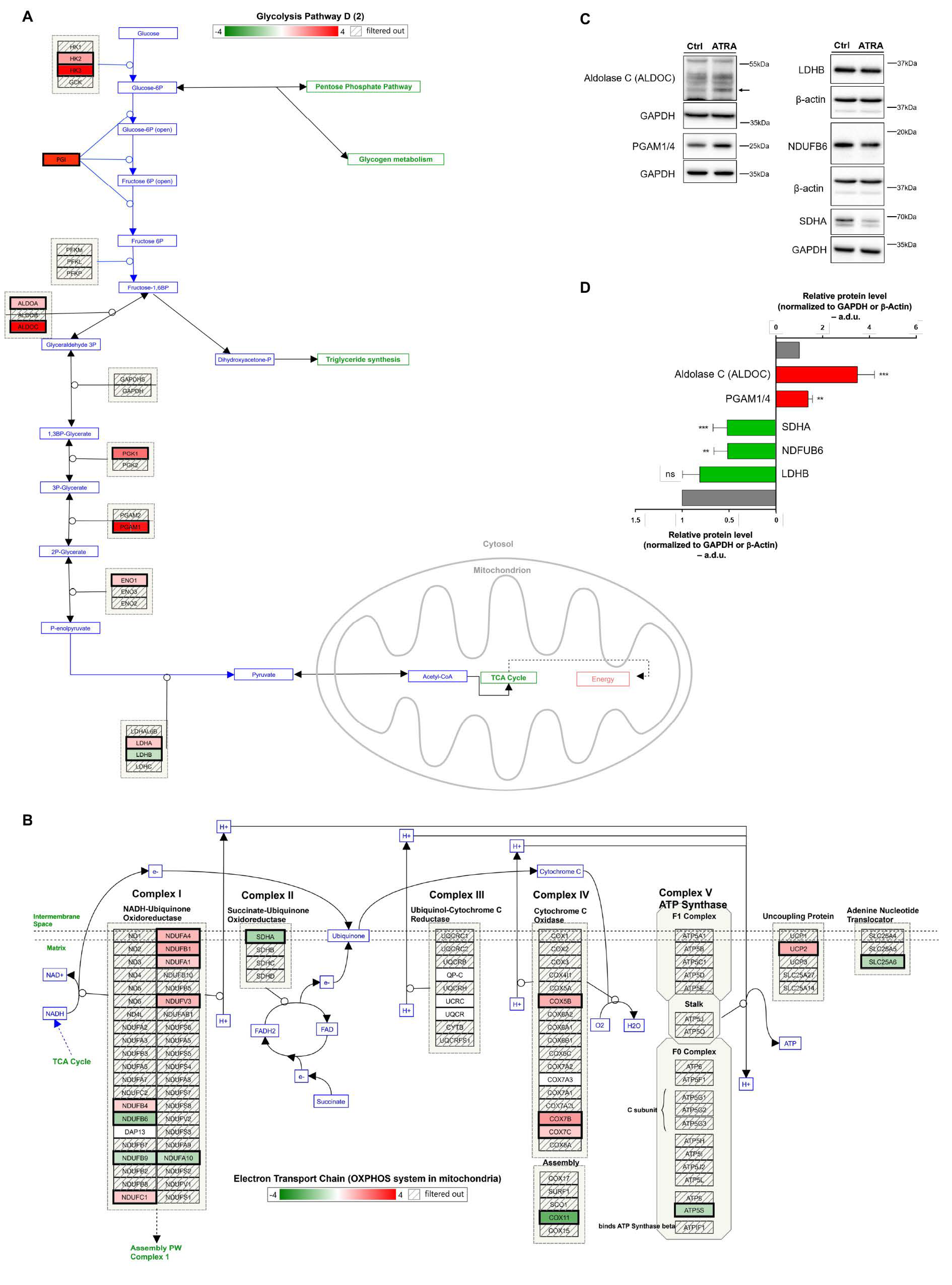
2.5. Western Blot Analysis
2.6. RNA Extraction and Affymetrix Analysis
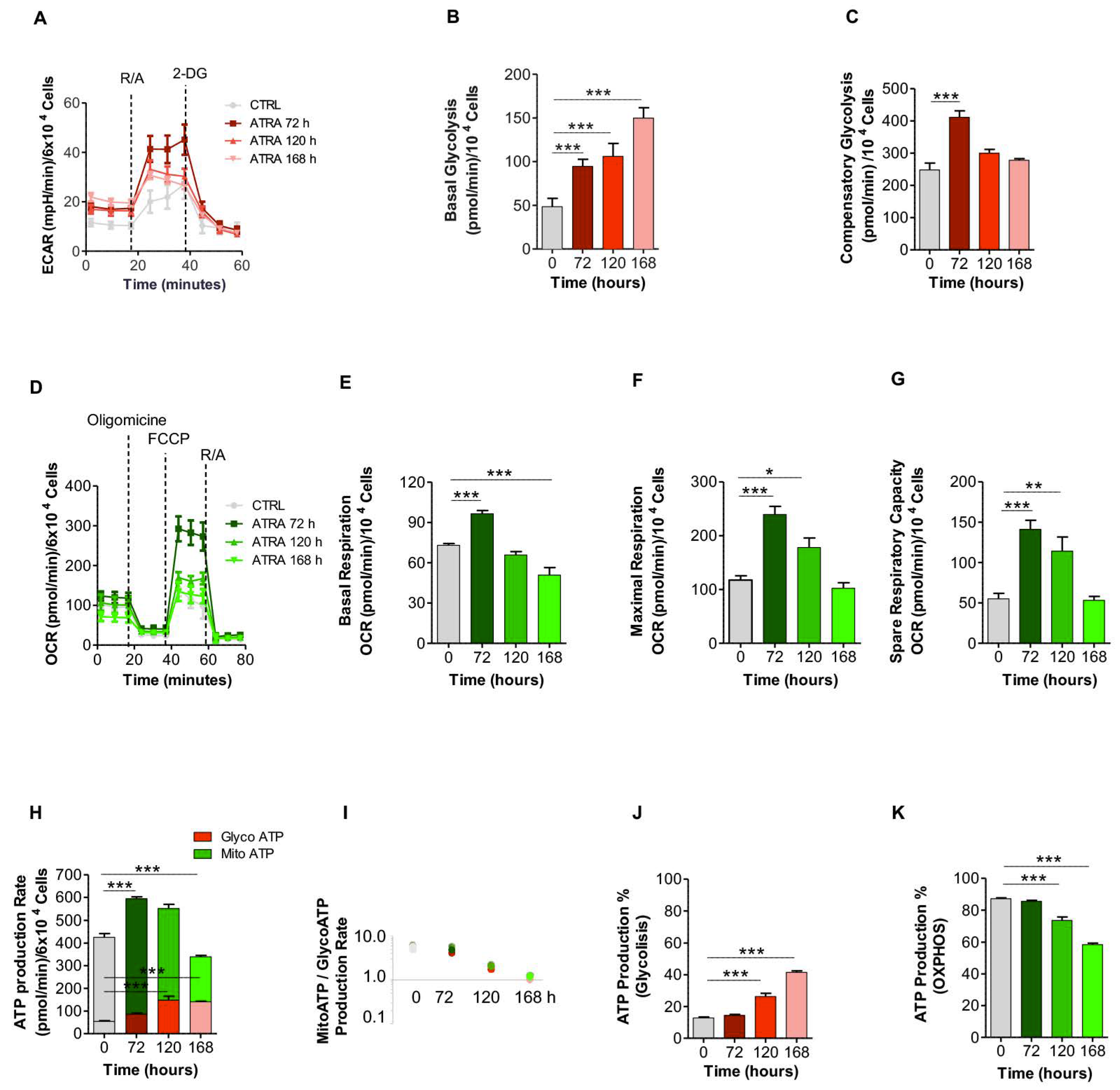
2.7. Metabolic Analyses
2.8. Statistical Analysis
3. Results
3.1. Evaluation of ATRA-Induced NB4 Differentiation
3.2. Genes Differentially Expressed in ATRA-Treated NB4 Cells
3.3. Canonical Pathways and Upstream Regulators from IPA
3.4. Glycolytic and OXPHOS Profiles During ATRA-Induced NB4 Differentiation
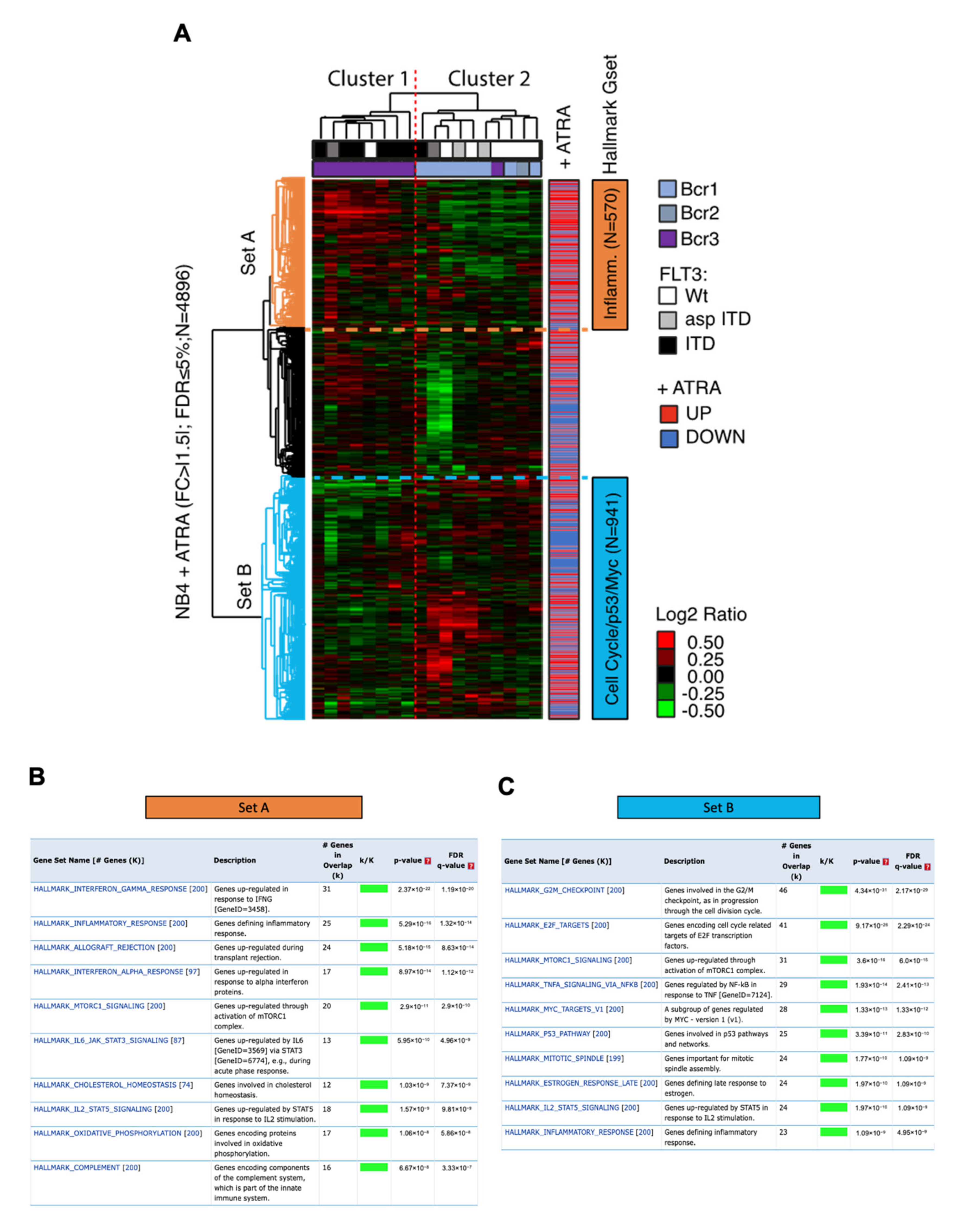
3.5. Impact of ATRA-Signature in A Cohort of Patients with Acute Promyelocytic Leukemia Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pejovic, T.; Schwartz, P.E. Leukemias. Clin Obs. Gynecol. 2002, 45, 866–878. [Google Scholar] [CrossRef] [PubMed]
- Sell, S. Leukemia: Stem cells, maturation arrest, and differentiation therapy. Stem Cell Rev. 2005, 1, 197–206. [Google Scholar] [CrossRef]
- di Masi, A.; Leboffe, L.; De Marinis, E.; Pagano, F.; Cicconi, L.; Rochette-Egly, C.; Lo-Coco, F.; Ascenzi, P.; Nervi, C. Retinoic acid receptors: From molecular mechanisms to cancer therapy. Mol. Aspects Med. 2015, 41, 1–115. [Google Scholar] [CrossRef] [PubMed]
- De Thé, H.; Pandolfi, P.P.; Chen, Z. Acute Promyelocytic Leukemia: A Paradigm for Oncoprotein-Targeted Cure. Cancer Cell 2017, 32, 552–560. [Google Scholar] [CrossRef] [Green Version]
- Cicconi, L.; Fenaux, P.; Kantarjian, H.; Tallman, M.; Sanz, M.A.; Lo-Coco, F. Molecular remission as a therapeutic objective in acute promyelocytic leukemia. Leukemia 2018, 32, 1671–1678. [Google Scholar] [CrossRef]
- De Thé, H.; Lavau, C.; Marchio, A.; Chomienne, C.; Degos, L.; Dejean, A. The PML-RARα fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell 1991, 66, 675–684. [Google Scholar] [CrossRef]
- Pandolfi, P.P.; Alcalay, M.; Fagioli, M.; Zangrilli, D.; Mencarelli, A.; Diverio, D.; Biondi, A.; Lo Coco, F.; Rambaldi, A.; Grignani, F. Genomic variability and alternative splicing generate multiple PML/RAR α transcripts that encode aberrant PML proteins and PML/RAR α isoforms in acute promyelocytic leukaemia. EMBO J. 1992, 11, 1397–1407. [Google Scholar] [CrossRef]
- Cull, E.H.; Altman, J.K. Contemporary treatment of APL. Curr. Hematol. Malig. Rep. 2014, 9, 193–201. [Google Scholar] [CrossRef] [Green Version]
- Noguera, N.I.; Catalano, G.; Banella, C.; Divona, M.; Faraoni, I.; Ottone, T.; Arcese, W.; Voso, M.T. Acute promyelocytic Leukemia: Update on the mechanisms of leukemogenesis, resistance and on innovative treatment strategies. Cancers 2019, 11, 1591. [Google Scholar] [CrossRef] [Green Version]
- Grignani, F.; De Matteis, S.; Nervi, C.; Tomassoni, L.; Gelmetti, V.; Cioce, M.; Fanelli, M.; Ruthardt, M.; Ferrara, F.F.; Zamir, I.; et al. Fusion proteins of the retinoic acid receptor-α recruit histone deacetylase in promyelocytic leukaemia. Nature 1998, 391, 815–818. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.J.; Nagy, L.; Inoue, S.; Shao, W.; Miller, J.; Evans, R.M. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature 1998, 391, 811–814. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.J.; Evans, R.M. Acquisition of oncogenic potential by RAR chimeras in acute promyelocytic leukemia through formation of homodimers. Mol. Cell 2000, 5, 821–830. [Google Scholar] [CrossRef]
- Minucci, S.; Maccarana, M.; Cioce, M.; De Luca, P.; Gelmetti, V.; Segalla, S.; Di Croce, L.; Giavara, S.; Matteucci, C.; Gobbi, A.; et al. Oligomerization of RAR and AML1 transcription factors as a novel mechanism of oncogenic activation. Mol. Cell 2000, 5, 811–820. [Google Scholar] [CrossRef]
- Fazi, F.; Travaglini, L.; Carotti, D.; Palitti, F.; Diverio, D.; Alcalay, M.; McNamara, S.; Miller, W.H.; Lo Coco, F.; Pelicci, P.G.; et al. Retinoic acid targets DNA-methyltransferases and histone deacetylases during APL blast differentiation in vitro and in vivo. Oncogene 2005, 24, 1820–1830. [Google Scholar] [CrossRef] [Green Version]
- Morey, L.; Brenner, C.; Fazi, F.; Villa, R.; Gutierrez, A.; Buschbeck, M.; Nervi, C.; Minucci, S.; Fuks, F.; Di Croce, L. MBD3, a Component of the NuRD Complex, Facilitates Chromatin Alteration and Deposition of Epigenetic Marks. Mol. Cell. Biol. 2008, 28, 5912–5923. [Google Scholar] [CrossRef] [Green Version]
- Noguera, N.I.; Piredda, M.L.; Taulli, R.; Catalano, G.; Angelini, G.; Gaur, G.; Nervi, C.; Voso, M.T.; Lunardi, A.; Pandolfi, P.P.; et al. PML/RARa inhibits PTEN expression in hematopoietic cells by competing with PU.1 transcriptional activity. Oncotarget 2016, 7, 66386–66397. [Google Scholar] [CrossRef] [Green Version]
- Falini, B.; Flenghi, L.; Fagioli, M.; Lo Coco, F.; Cordone, I.; Diverio, D.; Pasqualucci, L.; Biondi, A.; Riganelli, D.; Orleth, A.; et al. Immunocytochemical diagnosis of acute promyelocytic leukemia (M3) with the monoclonal antibody PG-M3 (anti-PML). Blood 1997, 90, 4046–4053. [Google Scholar] [CrossRef] [Green Version]
- De Thé, H.; Le Bras, M.; Lallemand-Breitenbach, V. Acute promyelocytic leukemia, arsenic, and PML bodies. J. Cell Biol. 2012, 198, 11–21. [Google Scholar] [CrossRef] [Green Version]
- Di Masi, A.; Cilli, D.; Berardinelli, F.; Talarico, A.; Pallavicini, I.; Pennisi, R.; Leone, S.; Antoccia, A.; Noguera, N.I.; Lo-Coco, F.; et al. PML nuclear body disruption impairs DNA double-strand break sensing and repair in APL. Cell Death Dis. 2016, 7, e2308-13. [Google Scholar] [CrossRef] [PubMed]
- Leszczyniecka, M.; Roberts, T.; Dent, P.; Grant, S.; Fisher, P.B. Differentiation therapy of human cancer: Basic science and clinical applications. Pharmacol. Ther. 2001, 90, 105–156. [Google Scholar] [CrossRef]
- Gurnari, C.; De Bellis, E.; Divona, M.; Ottone, T.; Lavorgna, S.; Voso, M.T. When Poisons Cure: The Case of Arsenic in Acute Promyelocytic Leukemia. Chemotherapy 2019, 64, 1–10. [Google Scholar] [CrossRef]
- Nervi, C.; Ferrara, F.F.; Fanelli, M.; Rippo, M.R.; Tomassini, B.; Ferrucci, P.F.; Ruthardt, M.; Gelmetti, V.; Gambacorti-Passerini, C.; Diverio, D.; et al. Caspases mediate retinoic acid-induced degradation of the acute promyelocytic leukemia PML/RARα fusion protein. Blood 1998, 92, 2244–2251. [Google Scholar]
- Zhu, J.; Gianni, M.; Kopf, E.; Honoré, N.; Chelbi-Alix, M.; Koken, M.; Quignon, F.; Rochette-Egly, C.; De Thé, H. Retinoic acid induces proteasome-dependent degradation of retinoic acid receptor α (RARα) and oncogenic RARα fusion proteins. Proc. Natl. Acad. Sci. USA 1999, 96, 14807–14812. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.E.; Ye, Y.C.; Chen, S.R.; Zhao, J.C.; Gu, L.J.; Cai, J.R.; Zhao, L.; Xie, J.X.; Shen, Z.X.; Wang, Z.Y. All-trans retinoic acid with or without low dose cytosine arabinoside in acute promyelocytic leukemia. Report of 6 cases. Chin. Med. J. 1987, 100, 949–953. [Google Scholar]
- Huang, M.E.; Ye, Y.C.; Chen, S.R.; Chai, J.R.; Lu, J.X.; Zhoa, L.; Gu, L.J.; Wang, Z.Y. Use of all-trans retinoic acid in the treatment of acute promyelocytic leukemia. Blood 1988, 72, 567–572. [Google Scholar] [CrossRef] [Green Version]
- Castaigne, S.; Chomienne, C.; Daniel, M.T.; Ballerini, P.; Berger, R.; Fenaux, P.; Degos, L. All-trans retinoic acid as a differentiation therapy for acute promyelocytic leukemia. I. Clinical results. Blood 1990, 76, 1704–1709. [Google Scholar] [CrossRef] [Green Version]
- Frankel, S.R.; Eardley, A.; Lauwers, G.; Weiss, M.; Warrell, R.P.J. The “retinoic acid syndrome” in acute promyelocytic leukemia. Ann Intern Med. 1992, 117, 292–296. [Google Scholar] [CrossRef]
- Vahdat, L.; Maslak, P.; Miller, W.H.; Eardley, A.; Heller, G.; Scheinberg, D.A.; Warrell, R.P. Early mortality and the retinoic acid syndrome in acute promyelocytic leukemia: Impact of leukocytosis, low-dose chemotherapy, PMN/RAR-α isoform, and CD13 expression in patients treated with all-trans retinoic acid. Blood 1994, 84, 3843–3849. [Google Scholar] [CrossRef] [Green Version]
- Degos, L.; Dombret, H.; Chomienne, C.; Daniel, M.T.; Micléa, J.M.; Chastang, C.; Castaigne, S.; Fenaux, P. All-trans-retinoic acid as a differentiating agent in the treatment of acute promyelocytic leukemia. Blood 1995, 85, 2643–2653. [Google Scholar] [CrossRef]
- Avvisati, G.; Lo Coco, F.; Diverio, D.; Falda, M.; Ferrara, F.; Lazzarino, M.; Russo, D.; Petti, M.C.; Mandelli, F. AIDA (all-trans retinoic acid + idarubicin) in newly diagnosed acute promyelocytic leukemia: A Gruppo Italiano Malattie Ematologiche Maligne dell’Adulto (GIMEMA) pilot study. Blood 1996, 88, 1390–1398. [Google Scholar] [CrossRef] [Green Version]
- Mandelli, F.; Diverio, D.; Avvisati, G.; Luciano, A.; Barbui, T.; Bernasconi, C.; Broccia, G.; Cerri, R.; Falda, M.; Fioritoni, G.; et al. Molecular remission in PML/RAR alpha-positive acute promyelocytic leukemia by combined all-trans retinoic acid and idarubicin (AIDA) therapy. Gruppo Italiano-Malattie Ematologiche Maligne dell’Adulto and Associazione Italiana di Ematologia ed Oncologia Pe. Blood 1997, 90, 1014–1021. [Google Scholar] [PubMed]
- Sanz, M.A.; Lo-Coco, F. Modern approaches to treating acute promyelocytic leukemia. J. Clin. Oncol. 2011, 29, 495–503. [Google Scholar] [CrossRef]
- Sanz, M.A.; Fenaux, P.; Lo Coco, F. Arsenic trioxide in the treatment of acute promyelocytic leukemia. A review of current evidence. Haematologica 2005, 90, 1231–1235. [Google Scholar]
- Lo-Coco, F.; Avvisati, G.; Vignetti, M.; Breccia, M.; Gallo, E.; Rambaldi, A.; Paoloni, F.; Fioritoni, G.; Ferrara, F.; Specchia, G.; et al. Front-line treatment of acute promyelocytic leukemia with AIDA induction followed by risk-adapted consolidation for adults younger than 61 years: Results of the AIDA-2000 trial of the GIMEMA Group. Blood 2010, 116, 3171–3179. [Google Scholar] [CrossRef]
- Powell, B.L.; Moser, B.; Stock, W.; Gallagher, R.E.; Willman, C.L.; Stone, R.M.; Rowe, J.M.; Coutre, S.; Feusner, J.H.; Gregory, J.; et al. Arsenic trioxide improves event-free and overall survival for adults with acute promyelocytic leukemia: North American Leukemia Intergroup Study C9710. Blood 2010, 116, 3751–3757. [Google Scholar] [CrossRef] [Green Version]
- Iland, H.J.; Bradstock, K.; Supple, S.G.; Catalano, A.; Collins, M.; Hertzberg, M.; Browett, P.; Grigg, A.; Firkin, F.; Hugman, A.; et al. All-trans-retinoic acid, idarubicin, and IV arsenic trioxide as initial therapy in acute promyelocytic leukemia (APML4). Blood 2012, 120, 1570–1580. [Google Scholar] [CrossRef]
- Noguera, N.I.; Pelosi, E.; Angelini, D.F.; Piredda, M.L.; Guerrera, G.; Piras, E.; Battistini, L.; Massai, L.; Berardi, A.; Catalano, G.; et al. High-dose ascorbate and arsenic trioxide selectively kill acute myeloid leukemia and acute promyelocytic leukemia blasts in vitro. Oncotarget 2017, 8, 32550–32565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stahl, M.; Tallman, M.S. Differentiation syndrome in acute promyelocytic leukaemia. Br. J. Haematol. 2019, 187, 157–162. [Google Scholar] [CrossRef]
- Montesinos, P.; González, J.D.; González, J.; Rayón, C.; De Lisa, E.; Amigo, M.L.; Ossenkoppele, G.J.; Peñarrubia, M.J.; Pérez-Encinas, M.; Bergua, J.; et al. Therapy-related myeloid neoplasms in patients with acute promyelocytic leukemia treated with all-trans-retinoic acid and anthracycline-based chemotherapy. J. Clin. Oncol. 2010, 28, 3872–3879. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, L.; Grignani, F.; Scicchitano, B.M.; Jetten, A.M.; Diverio, D.; Lo Coco, F.; Avvisati, G.; Gambacorti-Passerini, C.; Adamo, S.; Levin, A.A.; et al. Retinoid-induced differentiation of acute promyelocytic leukemia involves PML-RARα-mediated increase of type II transglutaminase. Blood 1996, 87, 1939–1950. [Google Scholar] [CrossRef]
- Roussel, M.J.S.; Lanotte, M. Maturation sensitive and resistant t(15;17) NB4 cell lines as tools for APL physiopathology: Nomenclature of cells and repertory of their known genetic alterations and phenotypes. Oncogene 2001, 20, 7287–7291. [Google Scholar] [CrossRef] [Green Version]
- Habib, A.; Hamade, E.; Mahfouz, R.; Nasrallah, M.S.; de Thé, H.; Bazarbachi, A. Arsenic trioxide inhibits ATRA-induced prostaglandin E2 and cyclooxygenase-1 in NB4 cells, a model of acute promyelocytic leukemia. Leukemia 2008, 22, 1125–1130. [Google Scholar] [CrossRef] [Green Version]
- Lanotte, M.; Martin-Thouvenin, V.; Najman, S.; Balerini, P.; Valensi, F.; Berger, R. NB4, a maturation inducible cell line with t(15;17) marker isolated from a human acute promyelocytic leukemia (M3). Blood 1991, 77, 1080–1086. [Google Scholar] [CrossRef] [Green Version]
- Rosenbauer, F.; Tenen, D.G. Transcription factors in myeloid development: Balancing differentiation with transformation. Nat. Rev. Immunol. 2007, 7, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Christodoulou, E.G.; Lee, L.M.; Lee, K.L.; Fung, T.K.; So, E.; Petretto, E.; Ong, S.T.; Rackham, O.J.L. Identification of drugs for leukaemia differentiation therapy by network pharmacology. bioRxiv 2019. Available online: https://www.biorxiv.org/content/10.1101/676106v1.abstract (accessed on 20 August 2020).
- Shao, W.; Benedetti, L.; Lamph, W.W.; Nervi, C.; Miller, W.H.J. A Retinoid-Resistant Acute Promyelocytic Leukemia Subclone Expresses a Dominant Negative PML-RARα Mutation. Blood 1997, 89, 4282–4289. [Google Scholar] [CrossRef]
- NCBI-Genome Reference Consortium Human Build 37. Available online: https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.13/ (accessed on 20 August 2020).
- NCBI-Genome Reference Consortium Human Build 38. Available online: https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.26/ (accessed on 20 August 2020).
- Cluster 3.0. Available online: http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm (accessed on 20 August 2020).
- Java TreeView. Available online: http://jtreeview.sourceforge.net (accessed on 20 August 2020).
- GEO. Available online: https://www.ncbi.nlm.nih.gov/geo/ (accessed on 20 August 2020).
- Orfali, N.; O’Donovan, T.R.; Cahill, M.R.; Benjamin, D.; Nanus, D.M.; McKenna, S.L.; Gudas, L.J.; Mongan, N.P. All-trans retinoic acid (ATRA)-induced TFEB expression is required for myeloid differentiation in acute promyelocytic leukemia (APL). Eur. J. Haematol. 2020, 104, 236–250. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Marasca, R.; Maffei, R.; Zucchini, P.; Castelli, I.; Saviola, A.; Martinelli, S.; Ferrari, A.; Fontana, M.; Ravanetti, S.; Torelli, G. Gene expression profiling of acute promyelocytic leukaemia identifies two subtypes mainly associated with Flt3 mutational status. Leukemia 2006, 20, 103–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- GSEA-MSigDB. Available online: https://www.gsea-msigdb.org/gsea/msigdb/index.jsp (accessed on 20 August 2020).
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viale, A.; De Franco, F.; Orleth, A.; Cambiaghi, V.; Giuliani, V.; Bossi, D.; Ronchini, C.; Ronzoni, S.; Muradore, I.; Monestiroli, S.; et al. Cell-cycle restriction limits DNA damage and maintains self-renewal of leukaemia stem cells. Nature 2009, 457, 51–56. [Google Scholar] [CrossRef]
- Liu, T.X.; Zhang, J.W.; Tao, J.; Zhang, R.B.; Zhang, Q.H.; Zhao, C.J.; Tong, J.H.; Lanotte, M.; Waxman, S.; Chen, S.J.; et al. Gene expression networks underlying retinoic acid-induced differentiation of acute promyelocytic leukemia cells. Blood 2000, 96, 1496–1504. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.H.; Chang, M.Y.; Ahn, J.I.; Yu, D.H.; Jung, S.S.; Choi, J.H.; Noh, Y.H.; Lee, Y.S.; Ahn, M.J. Differential gene expression in retinoic acid-induced differentiation of acute promyelocytic leukemia cells, NB4 and HL-60 cells. Biochem. Biophys. Res. Commun. 2002, 296, 1125–1133. [Google Scholar] [CrossRef]
- Yang, L.; Zhao, H.; Li, S.W.; Ahrens, K.; Collins, C.; Eckenrode, S.; Ruan, Q.G.; McIndoe, R.A.; She, J.X. Gene expression profiling during all-trans retinoic acid-induced cell differentiation of acute promyelocytic leukemia cells. J. Mol. Diagnostics 2003, 5, 212–221. [Google Scholar] [CrossRef] [Green Version]
- Zheng, P.Z.; Wang, K.K.; Zhang, Q.Y.; Huang, Q.H.; Du, Y.Z.; Zhang, Q.H.; Xiao, D.K.; Shen, S.H.; Imbeaud, S.; Eveno, E.; et al. Systems analysis of transcriptome and proteome in retinoic acid/arsenic trioxide-induced cell differentiation apoptosis of promyelocytic leukemia. Proc. Natl. Acad. Sci. USA 2005, 102, 7653–7658. [Google Scholar] [CrossRef] [Green Version]
- Park, M.H.; Cho, S.A.; Yoo, K.H.; Yang, M.H.; Ahn, J.Y.; Lee, H.S.; Lee, K.E.; Mun, Y.C.; Cho, D.H.; Seong, C.M.; et al. Gene expression profile related to prognosis of acute myeloid leukemia. Oncol. Rep. 2007, 18, 1395–1402. [Google Scholar] [CrossRef] [Green Version]
- Huynh, T.T.; Sultan, M.; Vidovic, D.; Dean, C.A.; Cruickshank, B.M.; Lee, K.; Loung, C.Y.; Holloway, R.W.; Hoskin, D.W.; Waisman, D.M.; et al. Retinoic acid and arsenic trioxide induce lasting differentiation and demethylation of target genes in APL cells. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Shivdasani, R.A.; Orkin, S.H. The transcriptional control of hematopoiesis. Blood 1996, 87, 4025–4039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosmarin, A.G.; Yang, Z.; Resendes, K.K. Transcriptional regulation in myelopoiesis: Hematopoietic fate choice, myeloid differentiation, and leukemogenesis. Exp. Hematol. 2005, 33, 131–143. [Google Scholar] [CrossRef]
- Ramirez, R.N.; El-Ali, N.C.; Mager, M.A.; Wyman, D.; Conesa, A.; Mortazavi, A. Dynamic Gene Regulatory Networks of Human Myeloid Differentiation. Cell Syst. 2017, 4, 416–429.e3. [Google Scholar] [CrossRef] [Green Version]
- Slenter, D.N.; Kutmon, M.; Hanspers, K.; Riutta, A.; Windsor, J.; Nunes, N.; Mélius, J.; Cirillo, E.; Coort, S.L.; DIgles, D.; et al. WikiPathways: A multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018, 46, D661–D667. [Google Scholar] [CrossRef]
- Hulse, M.; Caruso, L.B.; Madzo, J.; Tan, Y.; Johnson, S.; Tempera, I. Poly(ADP-ribose) polymerase 1 is necessary for coactivating hypoxia-inducible factor-1-dependent gene expression by Epstein-Barr virus latent membrane protein 1. PLoS Pathog. 2018, 14, 1–27. [Google Scholar] [CrossRef]
- Peng, M.; Yin, N.; Chhangawala, S.; Xu, K.; Leslie, C.S.; Li, M.O. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science 2016, 354, 481–484. [Google Scholar] [CrossRef]
- Hopkinson, B.M.; Desler, C.; Kalisz, M.; Vestentoft, P.S.; Juel Rasmussen, L.; Bisgaard, H.C. Bioenergetic Changes during Differentiation of Human Embryonic Stem Cells along the Hepatic Lineage. Oxid. Med. Cell. Longev. 2017, 2017, 5080128. [Google Scholar] [CrossRef]
- Lapa, B.; Gonçalves, A.C.; Jorge, J.; Alves, R.; Pires, A.S.; Abrantes, A.M.; Coucelo, M.; Abrunhosa, A.; Botelho, M.F.; Nascimento-Costa, J.M.; et al. Acute myeloid leukemia sensitivity to metabolic inhibitors: Glycolysis showed to be a better therapeutic target. Med. Oncol. 2020, 37, 1–13. [Google Scholar] [CrossRef]
- Rombouts, W.J.C.; Broyl, A.; Martens, A.C.M.; Slater, R.; Ploemacher, R.E. Human acute myeloid leukemia cells with internal tandem duplications in the Flt3 gene show reduced proliferative ability in stroma supported long-term cultures. Leukemia 1999, 13, 1071–1078. [Google Scholar] [CrossRef] [Green Version]
- Rombouts, W.J.C.; Blokland, I.; Löwenberg, B.; Ploemacher, R.E. Biological characteristics and prognosis of adult acute myeloid leukemia with internal tandem duplications in the Flt3 gene. Leukemia 2000, 14, 675–683. [Google Scholar] [CrossRef] [Green Version]
- Kottaridis, P.D.; Gale, R.E.; Frew, M.E.; Harrison, G.; Langabeer, S.E.; Belton, A.A.; Walker, H.; Wheatley, K.; Bowen, D.T.; Burnett, A.K.; et al. The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: Analysis of 854 patients from the United King. Blood 2001, 98, 1752–1759. [Google Scholar] [CrossRef]
- Kelly, L.M.; Liu, Q.; Kutok, J.L.; Williams, I.R.; Boulton, C.L.; Gilliland, D.G. FLT3 internal tandem duplication mutations associated with human acute myeloid leukemias induce myeloproliferative disease in a murine bone marrow transplant model. Blood 2002, 99, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Schnittger, S.; Schoch, C.; Dugas, M.; Kern, W.; Staib, P.; Wuchter, C.; Löffler, H.; Sauerland, C.M.; Serve, H.; Büchner, T.; et al. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: Correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood 2002, 100, 59–66. [Google Scholar] [CrossRef] [Green Version]
- Thiede, C.; Steudel, C.; Mohr, B.; Schaich, M.; Schäkel, U.; Platzbecker, U.; Wermke, M.; Bornhäuser, M.; Ritter, M.; Neubauer, A.; et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: Association with FAB subtypes and identification of subgroups with poor prognosis. Blood 2002, 99, 4326–4335. [Google Scholar] [CrossRef] [Green Version]
- Beitinjaneh, A.; Jang, S.; Roukoz, H.; Majhail, N.S. Prognostic significance of FLT3 internal tandem duplication and tyrosine kinase domain mutations in acute promyelocytic leukemia: A systematic review. Leuk. Res. 2010, 34, 831–836. [Google Scholar] [CrossRef]
- Breccia, M.; Loglisci, G.; Loglisci, M.G.; Ricci, R.; Diverio, D.; Latagliata, R.; Foà, R.; Lo-Coco, F. FLT3-ITD confers poor prognosis in patients with acute promyelocytic leukemia treated with AIDA protocols: Long-term follow-up analysis. Haematologica 2013, 98, e161–e163. [Google Scholar] [CrossRef] [Green Version]
- Noguera, N.I.; Breccia, M.; Divona, M.; Diverio, D.; Costa, V.; De Santis, S.; Avvisati, G.; Pinazzi, M.B.; Petti, M.C.; Mandelli, F.; et al. Alterations of the FLT3 gene in acute promyelocytic leukemia: Association with diagnostic characteristics and analysis of clinical outcome in patients treated with the Italian AIDA protocol. Leukemia 2002, 16, 2185–2189. [Google Scholar] [CrossRef] [Green Version]
- Callens, C.; Chevret, S.; Cayuela, J.M.; Cassinat, B.; Raffoux, E.; de Botton, S.; Thomas, X.; Guerci, A.; Fegueux, N.; Pigneux, A.; et al. Prognostic implication of FLT3 and Ras gene mutations in patients with acute promyelocytic leukemia (APL): A retrospective study from the European APL Group. Leukemia 2005, 19, 1153–1160. [Google Scholar] [CrossRef] [Green Version]
- Cicconi, L.; Divona, M.; Ciardi, C.; Ottone, T.; Ferrantini, A.; Lavorgna, S.; Alfonso, V.; Paoloni, F.; Piciocchi, A.; Avvisati, G.; et al. PML-RARα kinetics and impact of FLT3-ITD mutations in newly diagnosed acute promyelocytic leukaemia treated with ATRA and ATO or ATRA and chemotherapy. Leukemia 2016, 30, 1987–1992. [Google Scholar] [CrossRef]
- Esnault, C.; Rahmé, R.; Rice, K.L.; Berthier, C.; Gaillard, C.; Quentin, S.; Maubert, A.L.; Kogan, S.; de Thé, H. FLT3-ITD impedes retinoic acid, but not arsenic, responses in murine acute promyelocytic leukemias. Blood 2019, 133, 1495–1506. [Google Scholar] [CrossRef]
- De Mendonça Oliveira, L.; Teixeira, F.M.E.; Sato, M.N. Impact of Retinoic Acid on Immune Cells and Inflammatory Diseases. Mediators Inflamm. 2018, 2018. [Google Scholar] [CrossRef] [Green Version]
- Tanay, A.; Regev, A. Scaling single-cell genomics from phenomenology to mechanism. Nature 2017, 541, 331–338. [Google Scholar] [CrossRef] [Green Version]
- Hirai, H.; Zhang, P.; Dayaram, T.; Hetherington, C.J.; Mizuno, S.I.; Imanishi, J.; Akashi, K.; Tenen, D.G. C/EBPβ is required for “emergency” granulopoiesis. Nat. Immunol. 2006, 7, 732–739. [Google Scholar] [CrossRef]
- Matatall, K.A.; Shen, C.C.; Challen, G.A.; King, K.Y. Type II interferon promotes differentiation of myeloid-biased hematopoietic stem cells. Stem Cells 2014, 32, 3023–3030. [Google Scholar] [CrossRef] [Green Version]
- Matatall, K.A.; Jeong, M.; Chen, S.; Sun, D.; Chen, F.; Mo, Q.; Kimmel, M.; King, K.Y. Chronic Infection Depletes Hematopoietic Stem Cells through Stress-Induced Terminal Differentiation. Cell Rep. 2016, 17, 2584–2595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietras, E.M.; Mirantes-Barbeito, C.; Fong, S.; Loeffler, D.; Kovtonyuk, L.V.; Zhang, S.; Lakshminarasimhan, R.; Chin, C.P.; Techner, J.M.; Will, B.; et al. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat. Cell Biol. 2016, 18, 607–618. [Google Scholar] [CrossRef]
- Matikainen, S.; Ronni, T.; Hurme, M.; Pine, R.; Julkunen, I. Retinoic acid activates interferon regulatory factor-1 gene expression in myeloid cells. Blood 1996, 88, 114–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matikainen, S.; Ronni, T.; Lehtonen, A.; Sareneva, T.; Melén, K.; Nordling, S.; Levy, D.E.; Julkunen, I. Retinoic acid induces signal transducer and activator of transcription (STAT) 1, STAT2, and p48 expression in myeloid leukemia cells and enhances their responsiveness to interferons. Cell Growth Differ. 1997, 8, 687–698. [Google Scholar]
- Chelbi-Alix, M.K.; Pelicano, L. Retinoic acid and interferon signaling cross talk in normal and RA-resistant APL cells. Leukemia 1999, 13, 1167–1174. [Google Scholar] [CrossRef] [Green Version]
- Pelicano, L.; Brumpt, C.; Pitha, P.M.; Chelbi-Alix, M.K. Retinoic acid resistance in NB4 APL cells is associated with lack of interferon α synthesis Stat1 and p48 induction. Oncogene 1999, 18, 3944–3953. [Google Scholar] [CrossRef] [Green Version]
- Koller, E.; Krieger, O.; Kasparu, H.; Lutz, D. Restoration of all-trans retinoic acid sensitivity by interferon in acute promyelocytic leukaemia. Lancet 1991, 338, 1154–1155. [Google Scholar] [CrossRef]
- Lazzarino, M.; Corso, A.; Regazzi, M.B.; Iacona, I.; Bernasconi, C. Modulation of all-trans retinoid acid pharmacokinetics in acute promyelocytic leukaemia by prolonged interferon-α therapy. Br. J. Haematol. 1995, 90, 928–930. [Google Scholar] [CrossRef]
- Nguyen, H.; Hiscott, J.; Pitha, P.M. The growing family of interferon regulatory factors. Cytokine Growth Factor Rev. 1997, 8, 293–312. [Google Scholar] [CrossRef]
- Marié, I. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998, 17, 6660–6669. [Google Scholar] [CrossRef]
- Jefferies, C.A. Regulating IRFs in IFN driven disease. Front. Immunol. 2019, 10, 325. [Google Scholar] [CrossRef] [Green Version]
- Liebermann, D.A.; Hoffman, B. Myeloid differentiation (MyD)/growth arrest DNA damage (GADD) genes in tumor suppression, immunity and inflammation. Leukemia 2002, 16, 527–541. [Google Scholar] [CrossRef] [Green Version]
- Testa, U.; Stellacci, E.; Pelosi, E.; Sestili, P.; Venditti, M.; Orsatti, R.; Fragale, A.; Petrucci, E.; Pasquini, L.; Belardelli, F.; et al. Impaired myelopoiesis in mice devoid of interferon regulatory factor 1. Leukemia 2004, 18, 1864–1871. [Google Scholar] [CrossRef] [Green Version]
- Coccia, E.M.; Stellacci, E.; Valtieri, M.; Masella, B.; Feccia, T.; Marziali, G.; Hiscott, J.; Testa, U.; Peschle, C.; Battistini, A. Ectopic expression of interferon regulatory factor-1 potentiates granulocytic differentiation. Biochem J. 2001, 360, 285–294. [Google Scholar] [CrossRef]
- Lu, R.; Pitha, P.M. Monocyte Differentiation to Macrophage Requires Interferon Regulatory Factor 7. J. Biol. Chem. 2001, 276, 45491–45496. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Jiang, Z.; Wang, L.; Wang, A.; Liu, J.; Chen, C.; Yu, K.; Zou, F.; Wang, W.; Liu, J.; et al. All-trans retinoic acid exerts selective anti-FLT3-ITD acute myeloid leukemia efficacy through downregulating Chk1 kinase. Cancer Lett. 2020, 473, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Mandal, P.K.; Rossi, D.J. DNA-damage-induced differentiation in hematopoietic stem cells. Cell 2012, 148, 847–848. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Sun, Q.; Morita, Y.; Jiang, H.; Groß, A.; Lechel, A.; Hildner, K.; Guachalla, L.M.; Gompf, A.; Hartmann, D.; et al. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell 2012, 148, 1001–1014. [Google Scholar] [CrossRef] [Green Version]
- Crawford, L.J.; Johnston, C.K.; Irvine, A.E. TRIM proteins in blood cancers. J. Cell Commun. Signal. 2018, 12, 21–29. [Google Scholar] [CrossRef] [Green Version]
- Gandini, D.; De Angel, C.; Aguiari, G.; Manzati, E.; Lanza, F.; Pandolfi, P.P.; Cuneo, A.; Castoldi, G.L.; del Senno, L. Preferential expression of the transcription coactivator HTIF1α gene in acute myeloid leukemia and MDS-related AML. Leukemia 2002, 16, 886–893. [Google Scholar] [CrossRef]
- Pennisi, R.; Albanesi, J.; Ascenzi, P.; Nervi, C.; di Masi, A. Are DNA damage response kinases a target for the differentiation treatment of acute myeloid leukemia? IUBMB Life 2018, 70, 1057–1066. [Google Scholar] [CrossRef] [Green Version]
- Aoki, Y.; Sato, A.; Mizutani, S.; Takagi, M. Hematopoietic myeloid cell differentiation diminishes nucleotide excision repair. Int. J. Hematol. 2014, 100, 260–265. [Google Scholar] [CrossRef]
- Faraoni, I.; Compagnone, M.; Lavorgna, S.; Angelini, D.F.; Cencioni, M.T.; Piras, E.; Panetta, P.; Ottone, T.; Dolci, S.; Venditti, A.; et al. BRCA1, PARP1 and γH2AX in acute myeloid leukemia: Role as biomarkers of response to the PARP inhibitor olaparib. Biochim. Biophys. Acta 2015, 1852, 462–472. [Google Scholar] [CrossRef] [Green Version]
- Gil-Kulik, P.; Dudzińska, E.; Radzikowska-Büchner, E.; Wawer, J.; Jojczuk, M.; Nogalski, A.; Wawer, G.A.; Feldo, M.; Kocki, W.; Cioch, M.; et al. Different regulation of PARP1, PARP2, PARP3 and TRPM2 genes expression in acute myeloid leukemia cells. BMC Cancer 2020, 20, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Faraoni, I.; Giansanti, M.; Voso, M.T.; Lo-Coco, F.; Graziani, G. Targeting ADP-ribosylation by PARP inhibitors in acute myeloid leukaemia and related disorders. Biochem. Pharmacol. 2019, 167, 133–148. [Google Scholar] [CrossRef]
- Shi, Y.; Lan, F.; Matson, C.; Mulligan, P.; Whetstine, J.R.; Cole, P.A.; Casero, R.A.; Shi, Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004, 119, 941–953. [Google Scholar] [CrossRef] [Green Version]
- Lokken, A.A.; Zeleznik-Le, N.J. Breaking the LSD1/KDM1A Addiction: Therapeutic Targeting of the Epigenetic Modifier in AML. Cancer Cell 2012, 21, 451–453. [Google Scholar] [CrossRef] [Green Version]
- Harris, W.J.; Huang, X.; Lynch, J.T.; Spencer, G.J.; Hitchin, J.R.; Li, Y.; Ciceri, F.; Blaser, J.G.; Greystoke, B.F.; Jordan, A.M.; et al. The Histone Demethylase KDM1A Sustains the Oncogenic Potential of MLL-AF9 Leukemia Stem Cells. Cancer Cell 2012, 21, 473–487. [Google Scholar] [CrossRef] [Green Version]
- Christensen, J.; Agger, K.; Cloos, P.A.C.; Pasini, D.; Rose, S.; Sennels, L.; Rappsilber, J.; Hansen, K.H.; Salcini, A.E.; Helin, K. RBP2 Belongs to a Family of Demethylases, Specific for Tri-and Dimethylated Lysine 4 on Histone 3. Cell 2007, 128, 1063–1076. [Google Scholar] [CrossRef] [Green Version]
- Klose, R.J.; Yan, Q.; Tothova, Z.; Yamane, K.; Erdjument-Bromage, H.; Tempst, P.; Gilliland, D.G.; Zhang, Y.; Kaelin, W.G. The Retinoblastoma Binding Protein RBP2 Is an H3K4 Demethylase. Cell 2007, 128, 889–900. [Google Scholar] [CrossRef] [Green Version]
- Shokri, G.; Doudi, S.; Fathi-Roudsari, M.; Kouhkan, F.; Sanati, M.H. Targeting histone demethylases KDM5A and KDM5B in AML cancer cells: A comparative view. Leuk. Res. 2018, 68, 105–111. [Google Scholar] [CrossRef]
- Valk, P.J.M.; Verhaak, R.G.W.; Beijen, M.A.; Erpelinck, C.A.J.; Van Waalwijk Van Doorn-Khosrovani, S.B.; Boer, J.M.; Beverloo, H.B.; Moorhouse, M.J.; Van Der Spek, P.J.; Löwenberg, B.; et al. Prognostically Useful Gene-Expression Profiles in Acute Myeloid Leukemia. N. Engl. J. Med. 2004, 350, 1617–1628. [Google Scholar] [CrossRef] [Green Version]
- Klein, H.U.; Ruckert, C.; Kohlmann, A.; Bullinger, L.; Thiede, C.; Haferlach, T.; Dugas, M. Quantitative comparison of microarray experiments with published leukemia related gene expression signatures. BMC Bioinformatics 2009, 10, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Roma, A.; Spagnuolo, P.A. Estrogen Receptors Alpha and Beta in Acute Myeloid Leukemia. Cancers 2020, 12, 907. [Google Scholar] [CrossRef] [Green Version]
- Corcoran, S.E.; Neill, L.A.J.O.; Corcoran, S.E.; Neill, L.A.J.O. HIF1 a and metabolic reprogramming in inflammation Find the latest version: HIF1 α and metabolic reprogramming in inflammation. J. Clin. Invest. 2016, 126, 3699–3707. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Pore, N.; Behrooz, A.; Ismail-Beigi, F.; Maity, A. Regulation of glut1 mRNA by hypoxia-inducible factor-1: Interaction between H-ras and hypoxia. J. Biol. Chem. 2001, 276, 9519–9525. [Google Scholar] [CrossRef] [Green Version]
- Harris, A.L. Hypoxia - A key regulatory factor in tumour growth. Nat. Rev. Cancer 2002, 2, 38–47. [Google Scholar] [CrossRef]
- Chiavarina, B.; Whitaker-Menezes, D.; Migneco, G.; Martinez-Outschoorn, U.E.; Pavlides, S.; Howell, A.; Tanowitz, H.B.; Casimiro, M.C.; Wang, C.; Pestell, R.G.; et al. HIF1-alpha functions as a tumor promoter in cancer associated fibroblasts, and as a tumor suppressor in breast cancer cells: Autophagy drives compartment-specific oncogenesis. Cell Cycle 2010, 9, 3534–3551. [Google Scholar] [CrossRef] [Green Version]
- Watanuki, S.; Kobayashi, H.; Sorimachi, Y.; Yamamoto, M.; Okamoto, S.; Takubo, K. ATP turnover and glucose dependency in hematopoietic stem/progenitor cells are increased by proliferation and differentiation. Biochem. Biophys. Res. Commun. 2019, 514, 287–294. [Google Scholar] [CrossRef]
- Pearce, E.L.; Pearce, E.J. Metabolic pathways in immune cell activation and quiescence. Immunity 2013, 38, 633–643. [Google Scholar] [CrossRef] [Green Version]
- Ganeshan, K.; Chawla, A. Metabolic regulation of immune responses. Annu. Rev. Immunol. 2014, 32, 609–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albanesi, J.; Noguera, N.I.; Banella, C.; Colangelo, T.; De Marinis, E.; Leone, S.; Palumbo, O.; Voso, M.T.; Ascenzi, P.; Nervi, C.; et al. Transcriptional and Metabolic Dissection of ATRA-Induced Granulocytic Differentiation in NB4 Acute Promyelocytic Leukemia Cells. Cells 2020, 9, 2423. https://doi.org/10.3390/cells9112423
Albanesi J, Noguera NI, Banella C, Colangelo T, De Marinis E, Leone S, Palumbo O, Voso MT, Ascenzi P, Nervi C, et al. Transcriptional and Metabolic Dissection of ATRA-Induced Granulocytic Differentiation in NB4 Acute Promyelocytic Leukemia Cells. Cells. 2020; 9(11):2423. https://doi.org/10.3390/cells9112423
Chicago/Turabian StyleAlbanesi, Jacopo, Nelida Ines Noguera, Cristina Banella, Tommaso Colangelo, Elisabetta De Marinis, Stefano Leone, Orazio Palumbo, Maria Teresa Voso, Paolo Ascenzi, Clara Nervi, and et al. 2020. "Transcriptional and Metabolic Dissection of ATRA-Induced Granulocytic Differentiation in NB4 Acute Promyelocytic Leukemia Cells" Cells 9, no. 11: 2423. https://doi.org/10.3390/cells9112423









