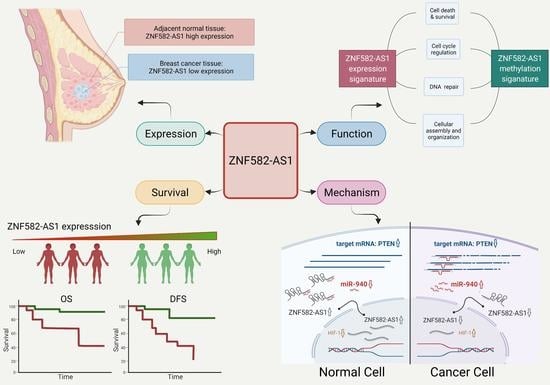
LncRNA ZNF582-AS1 Expression and Methylation in Breast Cancer and Its Biological and Clinical Implications
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Breast Cancer Patients and Tumor Samples in Our Study
2.2. RNA Extraction and ZNF582-AS1 Measurement
2.3. The Cancer Genome Atlas (TCGA) Data
2.4. Gene Expression Omnibus (GEO) Data
2.5. Meta-Analysis of ZNF582-AS1 Expression in Association with Survival
2.6. In Silico Prediction of ZNF582-AS1 Function and Regulation
2.7. Statistical Analysis
3. Results
3.1. ZNF582-AS1 Expression in Breast Cancer
3.2. ZNF582-AS1 Expression and Patient Characteristics
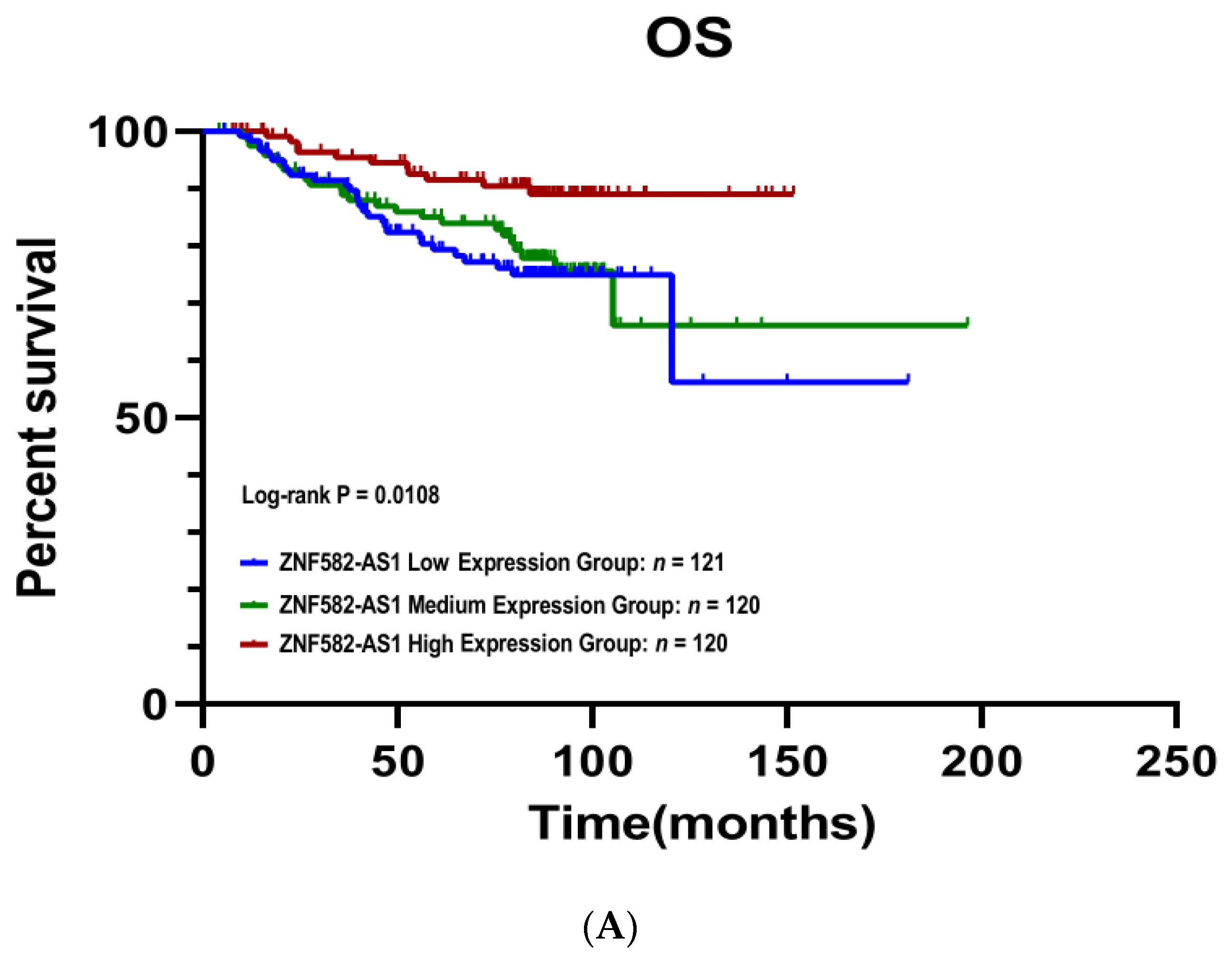
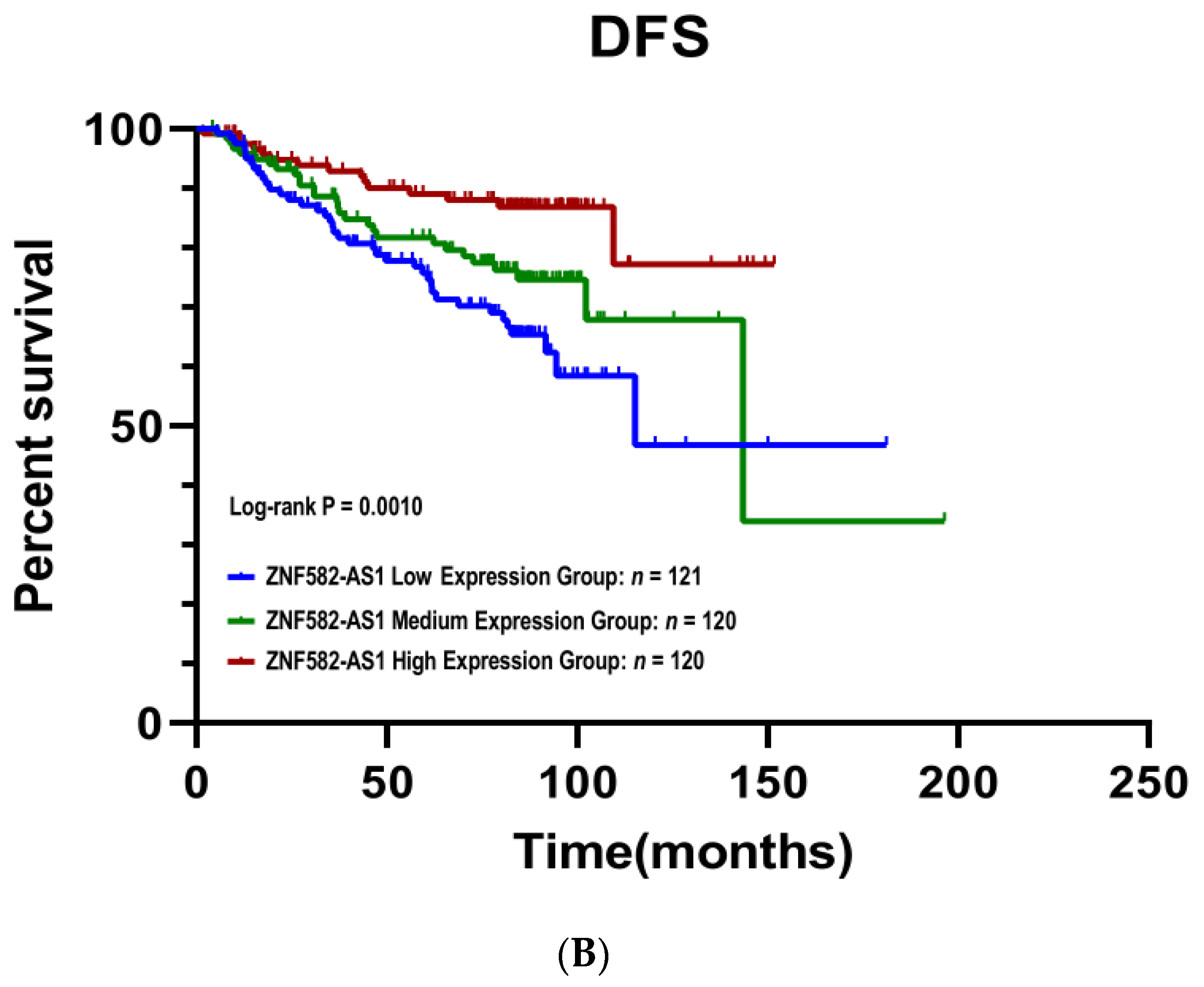
3.3. ZNF582-AS1 Expression and Breast Cancer Survival
3.4. Meta-Analysis of ZNF582-AS1 in Association with Survival
3.5. DNA Methylation and ZNF582-AS1 Expression
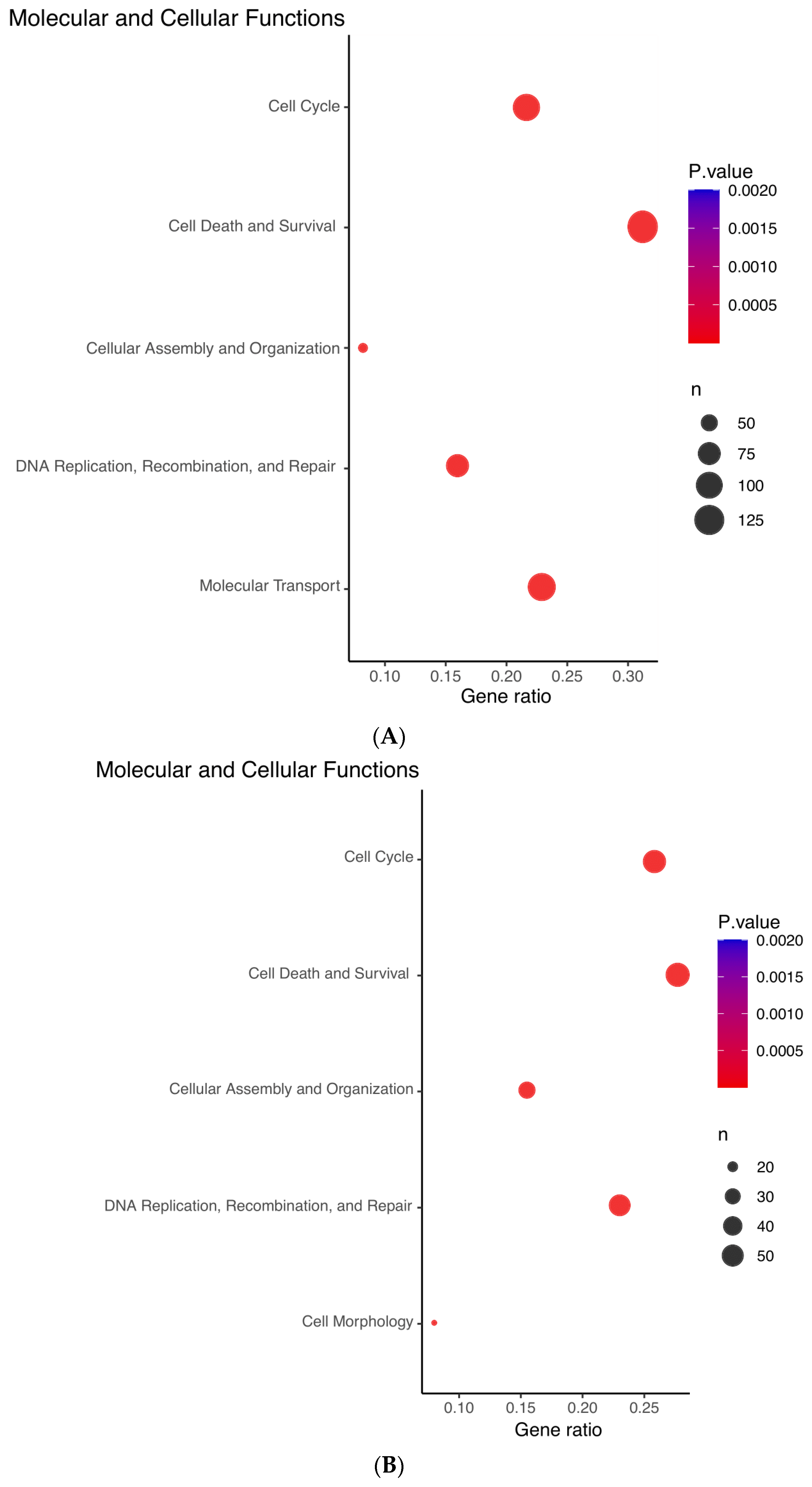
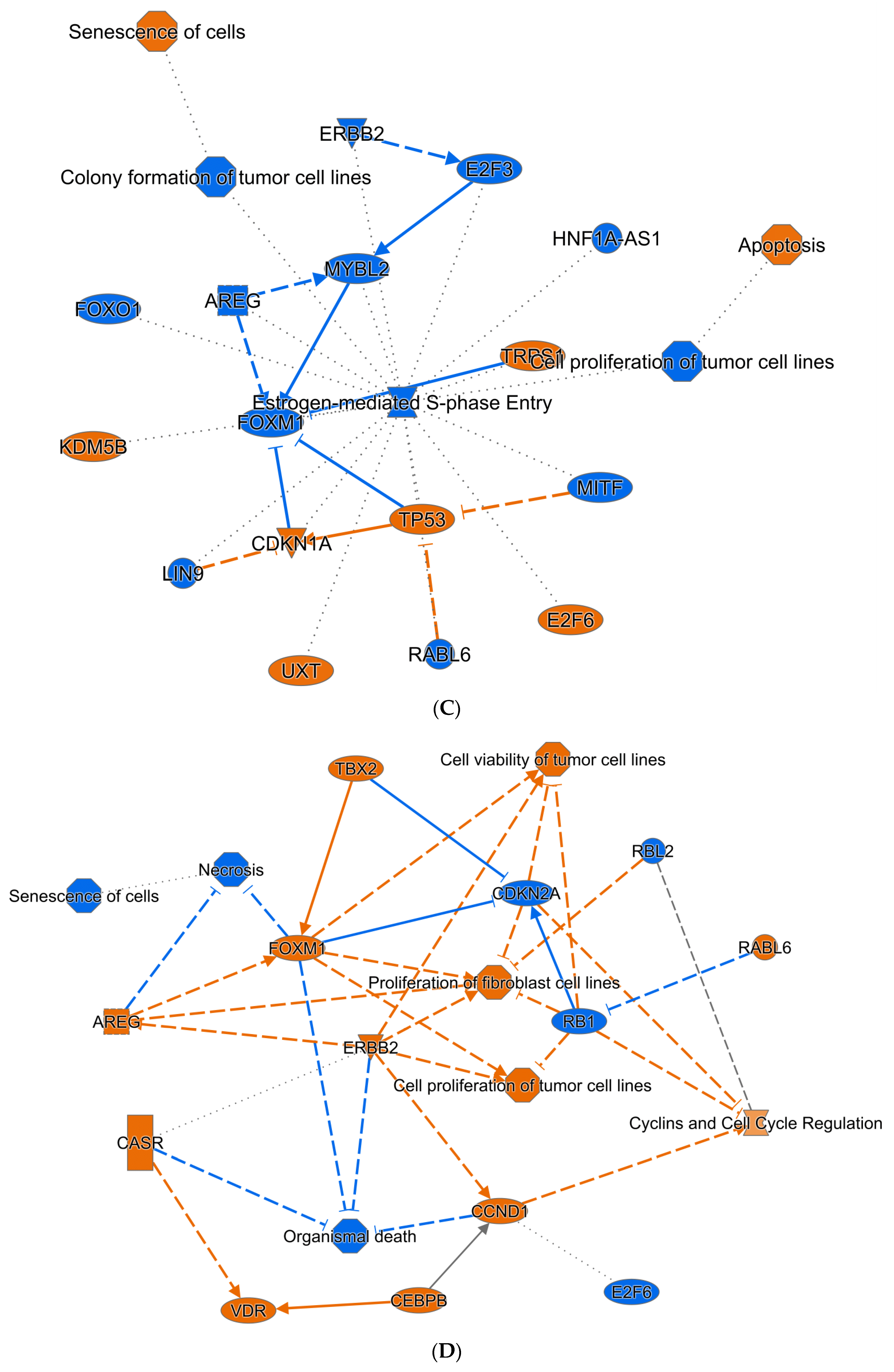
3.6. Bioinformatic Interrogation of ZNF582-AS1 Expression and Methylation Signatures
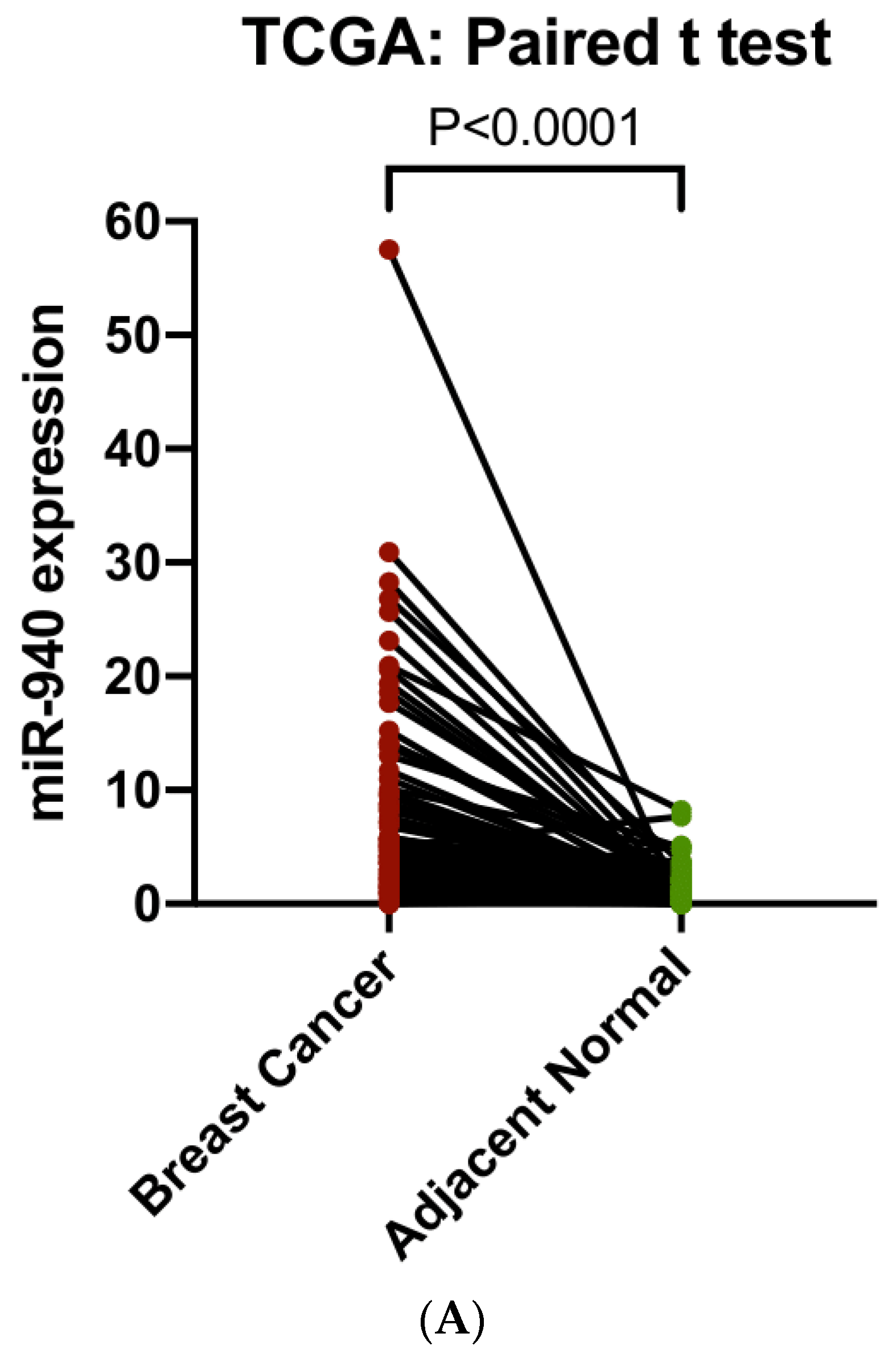
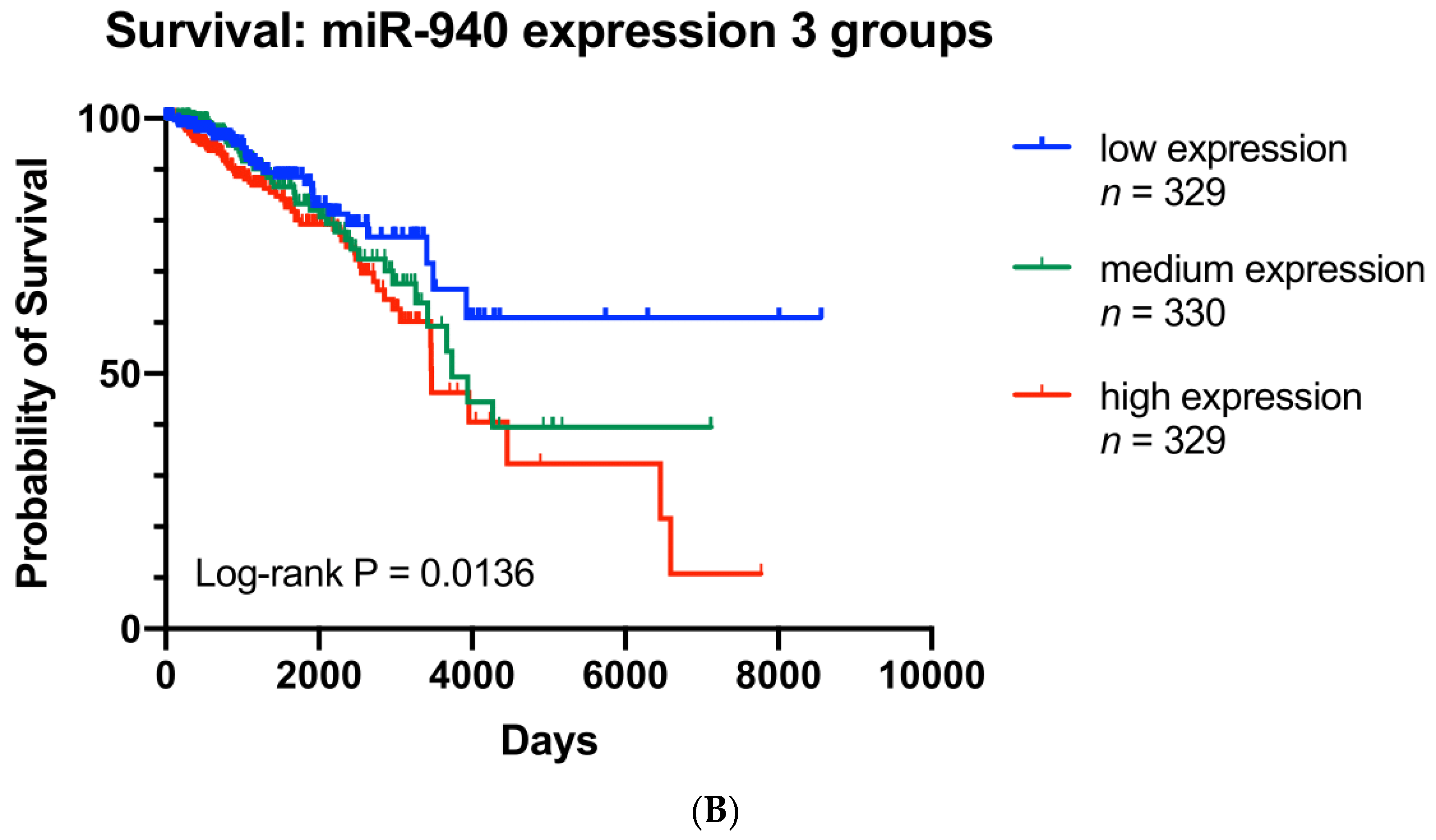
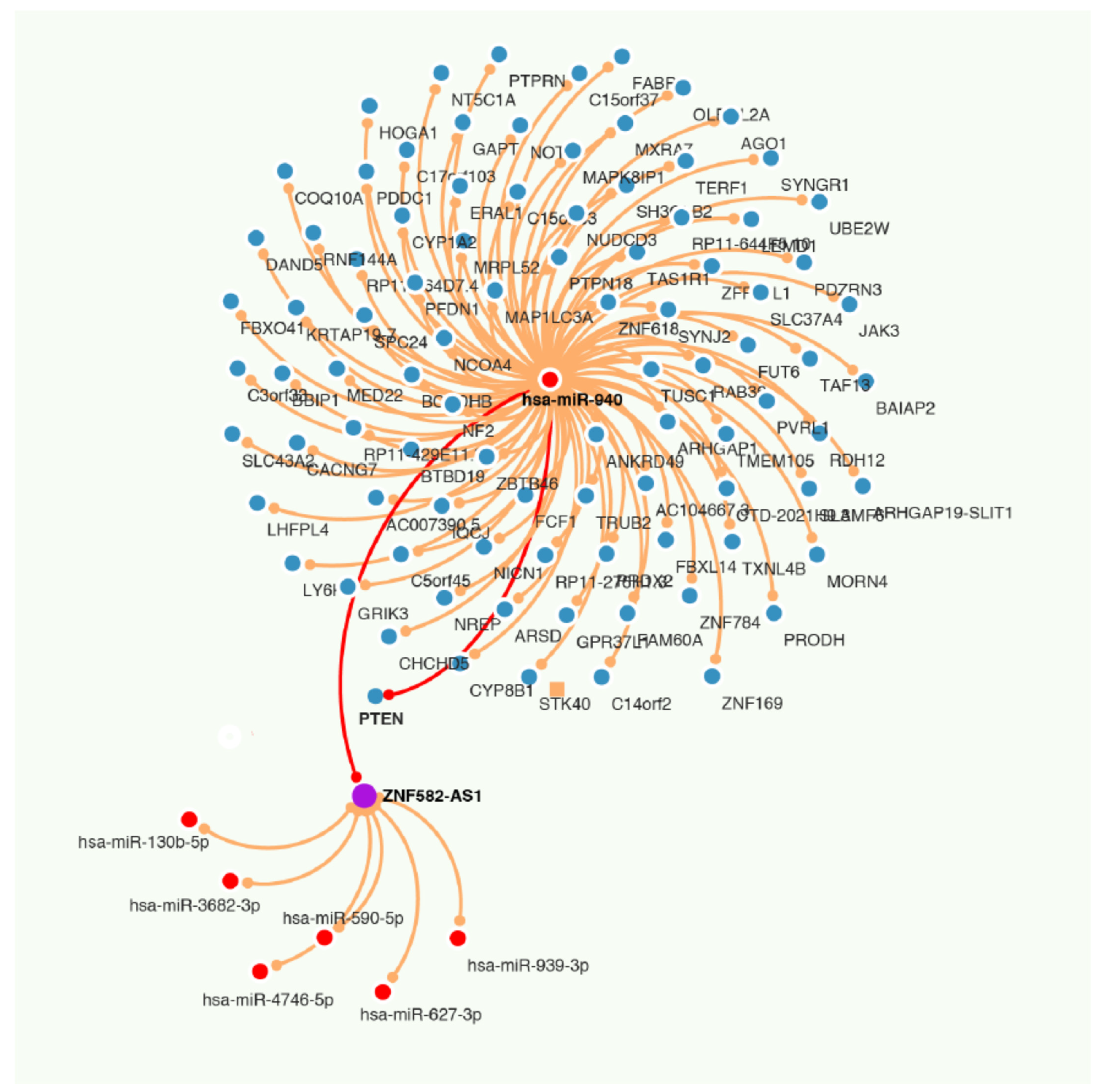
3.7. ZNF582-AS1 and hsa-miR-940
3.8. Regulation of ZNF582-AS1 Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S. Initial impact of the sequencing of the human genome. Nature 2011, 470, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Kapranov, P.; Willingham, A.T.; Gingeras, T.R. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007, 8, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer 2018, 18, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.; Chai, P.; Yu, J.; Xing, Y.; Wang, S.; Fan, J.; Ge, S.; Wang, Y.; Jia, R.; Fan, X. LncRNA CANT1 suppresses retinoblastoma progression by repellinghistone methyltransferase in PI3Kgamma promoter. Cell Death Dis. 2020, 11, 306. [Google Scholar] [CrossRef]
- Bhan, A.; Soleimani, M.; Mandal, S.S. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017, 77, 3965–3981. [Google Scholar] [CrossRef]
- Li, J.; Han, L.; Roebuck, P.; Diao, L.; Liu, L.; Yuan, Y.; Weinstein, J.N.; Liang, H. TANRIC: An Interactive Open Platform to Explore the Function of lncRNAs in Cancer. Cancer Res. 2015, 75, 3728–3737. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Jensen, M.A.; Ferretti, V.; Grossman, R.L.; Staudt, L.M. The NCI Genomic Data Commons as an engine for precision medicine. Blood 2017, 130, 453–459. [Google Scholar] [CrossRef]
- Diez-Villanueva, A.; Mallona, I.; Peinado, M.A. Wanderer, an interactive viewer to explore DNA methylation and gene expression data in human cancer. Epigenetics Chromatin 2015, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Pawitan, Y.; Bjohle, J.; Amler, L.; Borg, A.L.; Egyhazi, S.; Hall, P.; Han, X.; Holmberg, L.; Huang, F.; Klaar, S.; et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: Derived and validated in two population-based cohorts. Breast Cancer Res. 2005, 7, R953–R964. [Google Scholar] [CrossRef] [PubMed]
- Haibe-Kains, B.; Desmedt, C.; Di Leo, A.; Azambuja, E.; Larsimont, D.; Selleslags, J.; Delaloge, S.; Duhem, C.; Kains, J.P.; Carly, B.; et al. Genome-wide gene expression profiling to predict resistance to anthracyclines in breast cancer patients. Genom. Data 2013, 1, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zou, L.; Li, Q.; Haibe-Kains, B.; Tian, R.; Li, Y.; Desmedt, C.; Sotiriou, C.; Szallasi, Z.; Iglehart, J.D.; et al. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat. Med. 2010, 16, 214–218. [Google Scholar] [CrossRef]
- Kao, K.J.; Chang, K.M.; Hsu, H.C.; Huang, A.T. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: Implications for treatment optimization. BMC Cancer 2011, 11, 143. [Google Scholar] [CrossRef]
- Sabatier, R.; Finetti, P.; Adelaide, J.; Guille, A.; Borg, J.P.; Chaffanet, M.; Lane, L.; Birnbaum, D.; Bertucci, F. Down-regulation of ECRG4, a candidate tumor suppressor gene, in human breast cancer. PLoS ONE 2011, 6, e27656. [Google Scholar] [CrossRef]
- Clarke, C.; Madden, S.F.; Doolan, P.; Aherne, S.T.; Joyce, H.; O’Driscoll, L.; Gallagher, W.M.; Hennessy, B.T.; Moriarty, M.; Crown, J.; et al. Correlating transcriptional networks to breast cancer survival: A large-scale coexpression analysis. Carcinogenesis 2013, 34, 2300–2308. [Google Scholar] [CrossRef]
- Ivshina, A.V.; George, J.; Senko, O.; Mow, B.; Putti, T.C.; Smeds, J.; Lindahl, T.; Pawitan, Y.; Hall, P.; Nordgren, H.; et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006, 66, 10292–10301. [Google Scholar] [CrossRef]
- Metzger-Filho, O.; Michiels, S.; Bertucci, F.; Catteau, A.; Salgado, R.; Galant, C.; Fumagalli, D.; Singhal, S.K.; Desmedt, C.; Ignatiadis, M.; et al. Genomic grade adds prognostic value in invasive lobular carcinoma. Ann. Oncol. 2013, 24, 377–384. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.R.; Liu, S.; Sun, W.J.; Zheng, L.L.; Zhou, H.; Yang, J.H.; Qu, L.H. ChIPBase v2.0: Decoding transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data. Nucleic Acids Res. 2017, 45, D43–D50. [Google Scholar] [CrossRef] [PubMed]
- Paraskevopoulou, M.D.; Vlachos, I.S.; Karagkouni, D.; Georgakilas, G.; Kanellos, I.; Vergoulis, T.; Zagganas, K.; Tsanakas, P.; Floros, E.; Dalamagas, T.; et al. DIANA-LncBase v2: Indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016, 44, D231–D238. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Kumegawa, K.; Maruyama, R.; Yamamoto, E.; Ashida, M.; Kitajima, H.; Tsuyada, A.; Niinuma, T.; Kai, M.; Yamano, H.O.; Sugai, T.; et al. A genomic screen for long noncoding RNA genes epigenetically silenced by aberrant DNA methylation in colorectal cancer. Sci. Rep. 2016, 6, 26699. [Google Scholar] [CrossRef]
- Cheng, G.; Liu, D.; Liang, H.; Yang, H.; Chen, K.; Zhang, X. A cluster of long non-coding RNAs exhibit diagnostic and prognostic values in renal cell carcinoma. Aging 2019, 11, 9597–9615. [Google Scholar] [CrossRef]
- Yuan, L.Y.; Qin, X.; Li, L.; Zhou, J.; Zhou, M.; Li, X.; Xu, Y.; Wang, X.J.; Xing, H. The transcriptome profiles and methylation status revealed the potential cancer-related lncRNAs in patients with cervical cancer. J. Cell. Physiol. 2019, 234, 9756–9763. [Google Scholar] [CrossRef]
- Yang, W.; Zhang, K.; Li, L.; Xu, Y.; Ma, K.; Xie, H.; Zhou, J.; Cai, L.; Gong, Y.; Gong, K. Downregulation of lncRNA ZNF582-AS1 due to DNA hypermethylation promotes clear cell renal cell carcinoma growth and metastasis by regulating the N(6)-methyladenosine modification of MT-RNR1. J. Exp. Clin. Cancer Res. 2021, 40, 92. [Google Scholar] [CrossRef]
- Cheng, S.J.; Chang, C.F.; Lee, J.J.; Chen, H.M.; Wang, H.J.; Liou, Y.L.; Yen, C.; Chiang, C.P. Hypermethylated ZNF582 and PAX1 are effective biomarkers for detection of oral dysplasia and oral cancer. Oral. Oncol. 2016, 62, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Juan, Y.C.; Su, Y.F.; Zhang, W.B.; Yu, Y.; Yang, H.Y.; Yu, G.Y.; Peng, X. Hypermethylated PAX1 and ZNF582 genes in the tissue sample are associated with aggressive progression of oral squamous cell carcinoma. J. Oral. Pathol. Med. 2020, 49, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Park, S.L.; Li, Y.; Sheng, X.; Hom, V.; Xia, L.; Zhao, K.; Pooler, L.; Setiawan, V.W.; Lim, U.; Monroe, K.R.; et al. Genome-Wide Association Study of Liver Fat: The Multiethnic Cohort Adiposity Phenotype Study. Hepatol. Commun. 2020, 4, 1112–1123. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wang, G.; Tang, J.; Zhuang, W.; Wang, L.P.; Liou, Y.L.; Liu, Y.Z.; Zhou, H.H.; Zhu, Y.S. DNA Methylation Status of PAX1 and ZNF582 in Esophageal Squamous Cell Carcinoma. Int. J. Env. Res. Public Health 2017, 14, 216. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Liou, Y.L.; Wan, Z.R.; Tang, J.; Zhou, Y.; Zhuang, W.; Wang, G. Aberrant DNA methylation of PAX1, SOX1 and ZNF582 genes as potential biomarkers for esophageal squamous cell carcinoma. Biomed. Pharmacother. 2019, 120, 109488. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.Y.; Zhang, X.; Chu, T.; Zhu, S.; Deng, Y.; Zhou, Y.; Wang, Y.; Zhao, X.; Liu, L.; Fang, C.; et al. High methylation of ZNF582 in cervical adenocarcinoma affects radiosensitivity and prognosis. Ann. Transl. Med. 2019, 7, 328. [Google Scholar] [CrossRef]
- Harty, P.S.; Sieglinger, B.; Heymsfield, S.B.; Shepherd, J.A.; Bruner, D.; Stratton, M.T.; Tinsley, G.M. Novel body fat estimation using machine learning and 3-dimensional optical imaging. Eur. J. Clin. Nutr. 2020, 74, 842–845. [Google Scholar] [CrossRef]
- Mitri, Z.; Constantine, T.; O’Regan, R. The HER2 Receptor in Breast Cancer: Pathophysiology, Clinical Use, and New Advances in Therapy. Chemother. Res. Pract. 2012, 2012, 743193. [Google Scholar] [CrossRef]
- Park, S.Y.; Boushey, C.J.; Wilkens, L.R.; Haiman, C.A.; Le Marchand, L. High-Quality Diets Associate With Reduced Risk of Colorectal Cancer: Analyses of Diet Quality Indexes in the Multiethnic Cohort. Gastroenterology 2017, 153, 386–394.e2. [Google Scholar] [CrossRef]
- Salomon, D.S.; Normanno, N.; Ciardiello, F.; Brandt, R.; Shoyab, M.; Todaro, G.J. The role of amphiregulin in breast cancer. Breast Cancer Res. Treat. 1995, 33, 103–114. [Google Scholar] [CrossRef]
- O’Regan, R.M.; Nahta, R. Targeting forkhead box M1 transcription factor in breast cancer. Biochem. Pharmacol. 2018, 154, 407–413. [Google Scholar] [CrossRef]
- Myatt, S.S.; Lam, E.W. The emerging roles of forkhead box (Fox) proteins in cancer. Nat. Rev. Cancer 2007, 7, 847–859. [Google Scholar] [CrossRef] [PubMed]
- Koo, C.Y.; Muir, K.W.; Lam, E.W. FOXM1: From cancer initiation to progression and treatment. Biochim. Biophys. Acta 2012, 1819, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Francis, R.E.; Myatt, S.S.; Krol, J.; Hartman, J.; Peck, B.; McGovern, U.B.; Wang, J.; Guest, S.K.; Filipovic, A.; Gojis, O.; et al. FoxM1 is a downstream target and marker of HER2 overexpression in breast cancer. Int. J. Oncol. 2009, 35, 57–68. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, Y.Y.; Fu, S.; Wang, X.P.; Wang, H.Y.; Zeng, M.S.; Shao, J.Y. Down-regulation of c9orf86 in human breast cancer cells inhibits cell proliferation, invasion and tumor growth and correlates with survival of breast cancer patients. PLoS ONE 2013, 8, e71764. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, J.; Wang, W. Construction and investigation of breast-cancer-specific ceRNA network based on the mRNA and miRNA expression data. IET Syst. Biol. 2014, 8, 96–103. [Google Scholar] [CrossRef]
- Paci, P.; Colombo, T.; Farina, L. Computational analysis identifies a sponge interaction network between long non-coding RNAs and messenger RNAs in human breast cancer. BMC Syst. Biol. 2014, 8, 83. [Google Scholar] [CrossRef]
- Zhang, H.; Peng, J.; Lai, J.; Liu, H.; Zhang, Z.; Li, X.; Liang, B.; Chen, X.; Zou, B.; Lin, S.; et al. MiR-940 promotes malignant progression of breast cancer by regulating FOXO3. Biosci. Rep. 2020, 40, BSR20201337. [Google Scholar] [CrossRef]
- De Francesco, E.M.; Maggiolini, M.; Musti, A.M. Crosstalk between Notch, HIF-1alpha and GPER in Breast Cancer EMT. Int. J. Mol. Sci. 2018, 19, 2011. [Google Scholar] [CrossRef]
- Das, J.K.; Felty, Q.; Poppiti, R.; Jackson, R.M.; Roy, D. Nuclear Respiratory Factor 1 Acting as an Oncoprotein Drives Estrogen-Induced Breast Carcinogenesis. Cells 2018, 7, 234. [Google Scholar] [CrossRef]
- Tharakaraman, K.; Bodenreider, O.; Landsman, D.; Spouge, J.L.; Marino-Ramirez, L. The biological function of some human transcription factor binding motifs varies with position relative to the transcription start site. Nucleic Acids Res. 2008, 36, 2777–2786. [Google Scholar] [CrossRef]
- Tabach, Y.; Brosh, R.; Buganim, Y.; Reiner, A.; Zuk, O.; Yitzhaky, A.; Koudritsky, M.; Rotter, V.; Domany, E. Wide-scale analysis of human functional transcription factor binding reveals a strong bias towards the transcription start site. PLoS ONE 2007, 2, e807. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, T.W.; Wang, J.; Collins, P.J.; Partridge, E.C.; Aldred, S.F.; Trinklein, N.D.; Myers, R.M.; Weng, Z. Functional analysis of transcription factor binding sites in human promoters. Genome Biol. 2012, 13, R50. [Google Scholar] [CrossRef] [PubMed]


| ZNF582-AS1 Expression | |||||
|---|---|---|---|---|---|
| Variables | Total (n = 361) No. (%) | Low No. (%) | Mid No. (%) | High No. (%) | p-Value |
| Age | |||||
| Age < 58.14 | 181 (50.14) | 61 (33.70) | 59 (32.60) | 61 (33.70) | 0.965 |
| Age ≥ 58.14 | 180 (49.86) | 60 (33.33) | 61 (33.89) | 59 (32.78) | |
| Disease stage | |||||
| Stage I | 114 (33.33) | 28 (24.56) | 39 (34.21) | 47 (41.23) | 0.055 |
| Stage II | 174 (50.88) | 59 (33.91) | 59 (33.91) | 56 (32.18) | |
| Stage III and IV | 54 (15.79) | 27 (50.00) | 16 (29.63) | 11 (20.37) | |
| Tumor grade | |||||
| Grade 1 | 40 (11.33) | 8 (20.00) | 11 (27.50) | 21 (52.50) | 5.86 × 10−5 |
| Grade 2 | 135 (38.24) | 30 (22.22) | 47 (34.81) | 58 (42.96) | |
| Grade 3 | 178 (50.42) | 80 (44.94) | 58 (32.58) | 40 (22.47) | |
| ER status | |||||
| Positive | 245 (68.63) | 68 (27.76) | 85 (34.69) | 92 (37.55) | 0.003 |
| Negative | 112 (31.37) | 53 (47.32) | 32 (28.57) | 27 (24.11) | |
| PR status | |||||
| Positive | 212 (59.38) | 60 (28.30) | 73 (34.43) | 79 (37.26) | 0.053 |
| Negative | 145 (40.62) | 61 (42.07) | 44 (30.34) | 40 (27.59) | |
| ZNF582-AS1 Expression | Relapse | Death | ||||
|---|---|---|---|---|---|---|
| HR | 95%CI | p-Value | HR | 95%CI | p-Value | |
| Univariate analysis | ||||||
| Low | 1 | 1 | ||||
| Mid | 0.70 | 0.43–1.13 | 0.143 | 0.88 | 0.51–1.51 | 0.646 |
| High | 0.34 | 0.19–0.61 | <0.001 | 0.36 | 0.18–0.73 | 0.005 |
| Multivariate analysis * | ||||||
| Low | 1 | 1 | ||||
| Mid | 0.84 | 0.49–1.42 | 0.507 | 1.15 | 0.62–2.14 | 0.646 |
| High | 0.42 | 0.21–0.61 | 0.012 | 0.71 | 0.33–1.52 | 0.373 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Katsaros, D.; Biglia, N.; Fu, Y.; Benedetto, C.; Loo, L.; Wang, Z.; Yu, H. LncRNA ZNF582-AS1 Expression and Methylation in Breast Cancer and Its Biological and Clinical Implications. Cancers 2022, 14, 2788. https://doi.org/10.3390/cancers14112788
Wang J, Katsaros D, Biglia N, Fu Y, Benedetto C, Loo L, Wang Z, Yu H. LncRNA ZNF582-AS1 Expression and Methylation in Breast Cancer and Its Biological and Clinical Implications. Cancers. 2022; 14(11):2788. https://doi.org/10.3390/cancers14112788
Chicago/Turabian StyleWang, Junlong, Dionyssios Katsaros, Nicoletta Biglia, Yuanyuan Fu, Chiara Benedetto, Lenora Loo, Zhanwei Wang, and Herbert Yu. 2022. "LncRNA ZNF582-AS1 Expression and Methylation in Breast Cancer and Its Biological and Clinical Implications" Cancers 14, no. 11: 2788. https://doi.org/10.3390/cancers14112788
APA StyleWang, J., Katsaros, D., Biglia, N., Fu, Y., Benedetto, C., Loo, L., Wang, Z., & Yu, H. (2022). LncRNA ZNF582-AS1 Expression and Methylation in Breast Cancer and Its Biological and Clinical Implications. Cancers, 14(11), 2788. https://doi.org/10.3390/cancers14112788