Biomedical Relevance of Novel Anticancer Peptides in the Sensitive Treatment of Cancer
Abstract
:1. Introduction
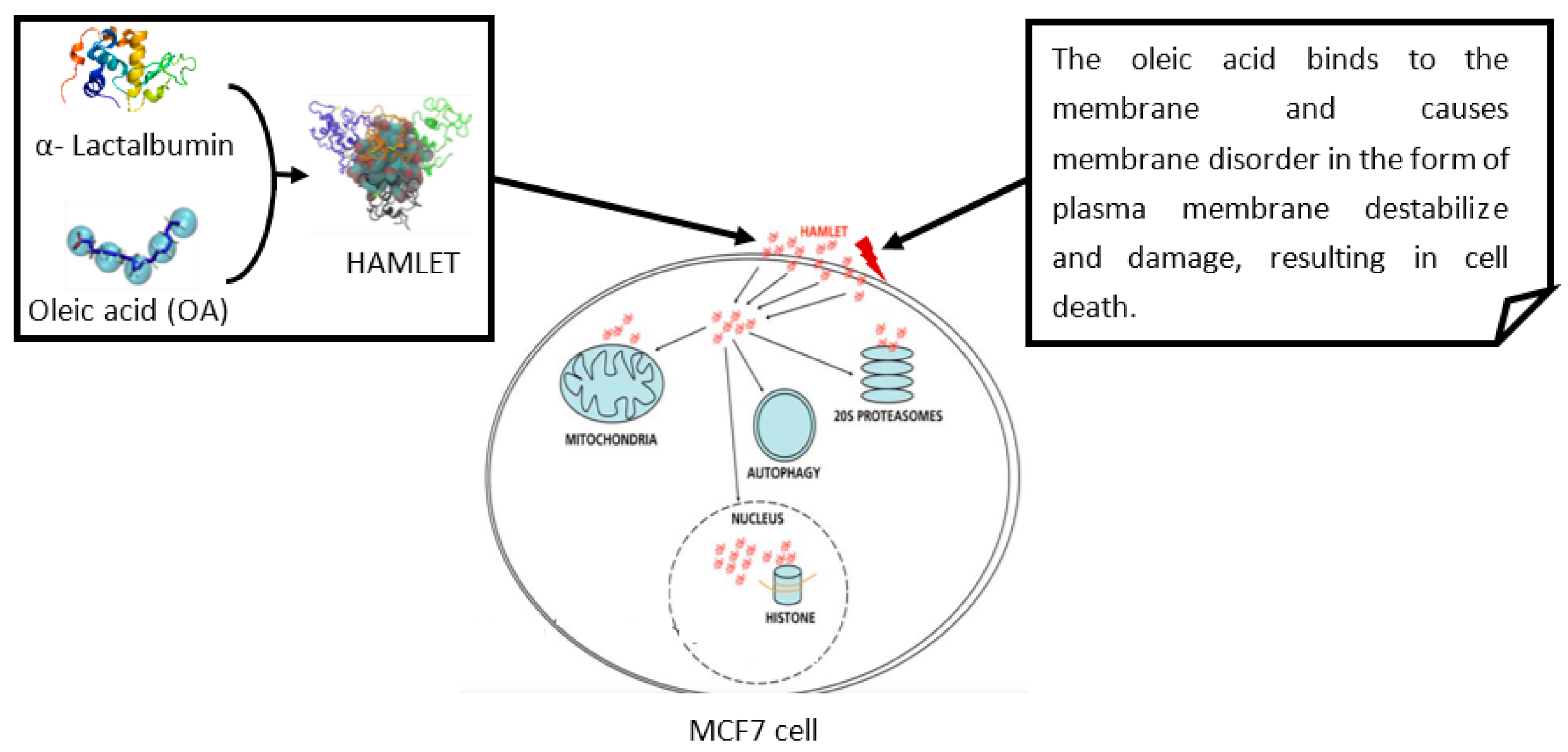
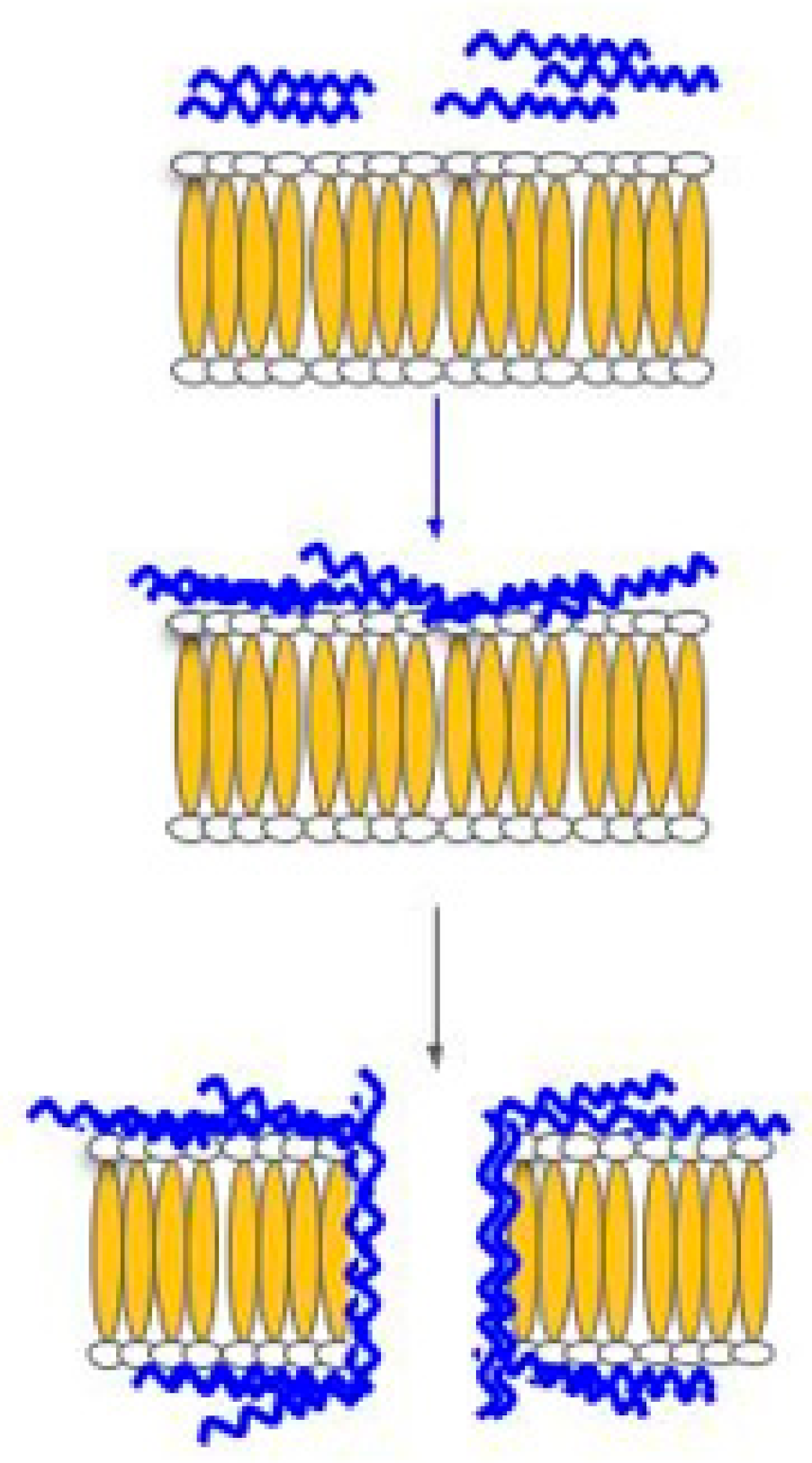
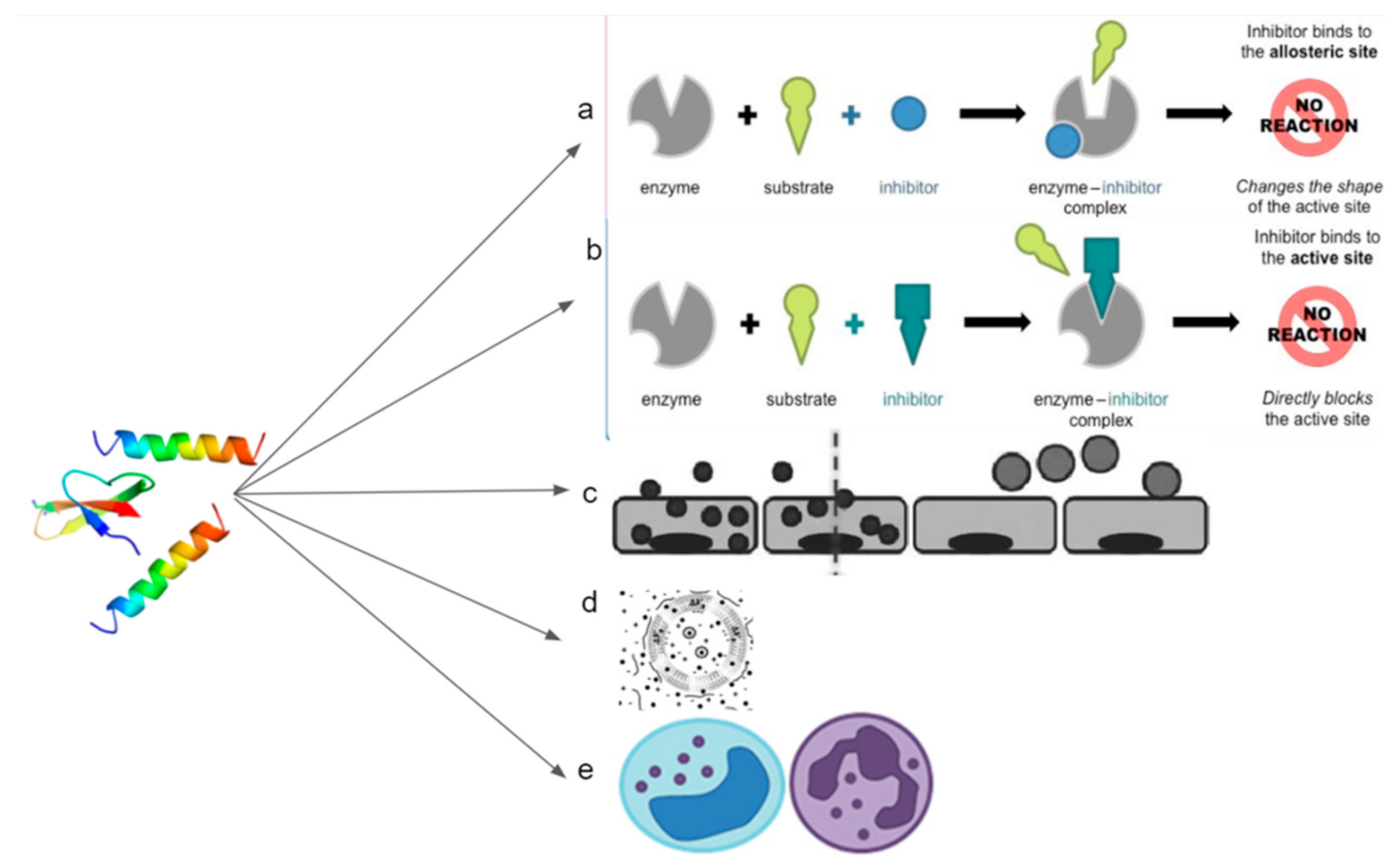
2. Modes of Action of Host-Defense Peptides (HDPs)
| N/B | ACPs | Cancer Type | Mechanism | HDP Sequences | References |
|---|---|---|---|---|---|
| 1 | Leucine-leucine-37 (LL-37) | human oral squamous cell carcinoma (OSCC) cells | Membranolytic activity towards tumor cell using “toroidal pore” mechanism | LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES | [40] |
| 2 | Human defensins or α-defensins (human neutrophil peptides (HNP-1, HNP-2, and HNP-3)) | human myeloid leukemia cell line (U937), human erythroleukemic cell line (K562), and lymphoblastoid B cells (IM-9 and WIL-2). | cytolytic activity | HNP-1: ACYCRIPACIAGERRYGTCIYQGRLWAFCC HNP-2: CYCRIPACIAGERRYGTCIYQGRLWAFCC HNP-3: DCYCRIPACIAGERRYGTCIYQGRLWAFCC | [41] |
| 3 | Human β-defensin-3 (hBD3) | HeLa, Jurkat and U937 cancer cell lines | Binding to cell membrane containing phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] to cause cytolysis | GIINTLQKYYCRVRGGRCAVLSCLPKEEQIGKCSTRGRKCCRRKK | [42] |
| 4 | Bovine lactoferricin (LfcinB) LTX-315 | drug-resistant and drug-sensitive cancer cells | cytolysis and immunogenicity | FKCRRWQWRMKKLGAPSITCVRRAF | [43] |
| 5 | high mobility group box protein 1 (HMGB1) | All cancer types | immature dendritic cells activation and tumor-specific cytotoxic generation | GRRRRSVQWCAVSQPEATKCFQWQRNMRKVRGPPVSCIKRDSPIQCIQA | [34] |
| 6 | Gomesin | murine and human cancer cell lines along with melanoma and leukemia | carpet model for destroying the membrane | QCRRLCYKQRCVTYCRGR | [44] |
| 7 | Mastoparan-C | lung cancer H157, melanocyte MDA-MB-435S, human prostate carcinoma PC-3, human glioblastoma astrocytoma U251MG and human breast cancer MCF-7 cell lines | induce apoptosis and activate phospholipase, inhibition of ATPase activity selectively | INLKALAALAKKIL | [45] |
| 8 | Cecropin B1 | NSCLC cell line | tumor growth inhibition using pore formation and apoptosis | KWKIFKKIEKVGRNIRNGIIKAGPAVAVLGEAKAL | [46] |
| 9 | Magainin 2 | human lung cancer cells A59 and in Ehlrich’s murine ascites cells | formation of pores on cell membranes | GIGKFLHSAKKFGKAFVGEIMNS | [47] |
| 10 | Bufforin IIb | Leukemia, breast, prostate, and colon cancer | Destruction of the membrane using mitochondrial apoptosis | TRSSRAGLQFPVGRVHRLLRK | [48] |
| 11 | Brevinin 2R | T-cell leukemia Jurkat, B-cell lymphoma BJAB, colon carcinoma HT29/219 and SW742, fibrosarcoma L929, breast adenocarcinoma MCF-7, and lung carcinoma A549 cells | Lysosomal death pathway (LDP) and autophagy-like cell death | KLKNFAKGVAQSLLNKASCKLSGQC | [37] |
| 12 | Limnonectes fujianensis brevinvin (LFB) | lung cancer H460, melanoma cell, glioblastoma U251MG, colon cancer HCT116 cell lines | penetrate the lipidic bilayer causing cell death | FLPLAVSLAANFLPKLFCKITKKC | [49] |
| 13 | Phylloseptin-PHa | breast cancer cells MCF-7, breast epithelial cells MCF10A | penetrate the lipidic bilayer causing cell death | FLSLIPAAISAVSALANHF | [50] |
| 14 | Ranatuerin-2PLx | prostate cancer cell PC-3 | cell apoptosis using caspase-3 | GIMDTVKNAAKNLAGQLLDKLKCSITAC | [51] |
| 15 | Dermaseptins (DRS) | prostate cancer cell PC-3 | Pore formation and internalization of the lipid bilayer | GLWSKIKEVGKEAAKAAAKAAGKAALGAVSEAV | [52] |
| 16 | Chrysophsin-1,-2 and-3 | human fibrosarcoma HT-1080, histiocytic lymphoma U937, and cervical carcinoma HeLa cell lines | disrupt the plasma membrane | FFGWLIKGAIHAGKAIHGLIHRRRH | [53] |
| 17 | Ss-arasin | human cervical carcinoma HeLa and colon carcinoma HT-29 | cytotoxicity against cancer cells | SPRVRRRYGRPFGGRPFVGGQFGGRPGCVCIRSPCPCANYG | [52] |
| 18 | Turgencin A and turgencin B | melanoma cancer cells A2058 and the human fibroblast cell line MRC-5 | Pore formation and internalization of the lipid bilayer | Turgencin A: GPKTKAACKMACKLATCGKKPGGWKCKLCELGCDAV Turgencin B: GIKEMLCNMACAQTVCKKSGGPLCDTCQAACKALG | [54] |
| 19 | D-K6L9 | breast and prostate cancer cell lines | reduce neovascularization | LKLLKKLLKKLLKLL | [48] |
| 20 | Dusquetide (SGX942) | neck and head cancer | Binds p62 to cause membrane damage | RIVPA | [37] |
3. Novel Anticancer Peptides (ACPs) Used in Cancer Therapy
4. Mechanism of ACPs for Cancer Treatment
5. Discovery Techniques for the Identification of Sensitive HDPs
6. Molecular Validation Techniques Used for HDPs
7. Challenges for the Use of Anticancer Peptides (ACPs) in Cancer Treatment
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saxena, M.; van der Burg, S.H.; Melief, C.J.; Bhardwaj, N. Therapeutic cancer vaccines. Nat. Rev. Cancer 2021, 21, 360–378. [Google Scholar] [CrossRef]
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2018; Available online: https://gco.iarc.fr/today (accessed on 5 June 2021).
- Black, J.R.; McGranahan, N. Genetic and non-genetic clonal diversity in cancer evolution. Nat. Rev. Cancer 2021, 21, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Marunaka, Y. Roles of interstitial fluid pH and weak organic acids in development and amelioration of insulin resistance. Biochem. Soc. Trans. 2021, 49, 715–726. [Google Scholar] [CrossRef]
- Link, A.; Bornschein, J.; Thon, C. Helicobacter pylori induced gastric carcinogenesis-the best molecular model we have? Best Pract. Res. Clin. Gastroenterol. 2021, 50–51, 101743. [Google Scholar] [CrossRef]
- Calabrese, E.J.; Priest, N.D.; Kozumbo, W.J. Thresholds for carcinogens. Chem. Biol. Interact. 2021, 341, 109464. [Google Scholar] [CrossRef]
- Si, H.; Yang, Q.; Hu, H.; Ding, C.; Wang, H.; Lin, X. Colorectal cancer occurrence and treatment based on changes in intestinal flora. Semin. Cancer Biol. 2021, 70, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Miranda-Galvis, M.; Loveless, R.; Kowalski, L.P.; Teng, Y. Impacts of environmental factors on head and neck cancer pathogenesis and progression. Cells 2021, 10, 389. [Google Scholar] [CrossRef] [PubMed]
- de Martel, C.; Georges, D.; Bray, F.; Ferlay, J.; Clifford, G.M. Global burden of cancer attributable to infections in 2018: A worldwide incidence analysis. Lancet Glob. Health 2020, 8, e180–e190. [Google Scholar] [CrossRef] [Green Version]
- Stelzle, D.; Tanaka, L.F.; Lee, K.K.; Khalil, A.I.; Baussano, I.; Shah, A.S.; McAllister, D.A.; Gottlieb, S.L.; Klug, S.J.; Winkler, A.S. Estimates of the global burden of cervical cancer associated with HIV. Lancet Glob. Health 2021, 9, e161–e169. [Google Scholar] [CrossRef]
- Chen, H.; Wang, J.P.; Liu, H.; Li, H.; Lin, Y.-C.J.; Shi, R.; Yang, C.; Gao, J.; Zhou, C.; Li, Q. Hierarchical transcription factor and chromatin binding network for wood formation in Populus trichocarpa. Plant Cell 2019, 31, 602–626. [Google Scholar] [CrossRef] [Green Version]
- Rosselló-Tortella, M.; Llinàs-Arias, P.; Sakaguchi, Y.; Miyauchi, K.; Davalos, V.; Setien, F.; Calleja-Cervantes, M.E.; Piñeyro, D.; Martínez-Gómez, J.; Guil, S. Epigenetic loss of the transfer RNA-modifying enzyme TYW2 induces ribosome frameshifts in colon cancer. Proc. Natl. Acad. Sci. USA 2020, 117, 20785–20793. [Google Scholar] [CrossRef] [PubMed]
- Riaz, N.; Blecua, P.; Lim, R.S.; Shen, R.; Higginson, D.S.; Weinhold, N.; Norton, L.; Weigelt, B.; Powell, S.N.; Reis-Filho, J.S. Pan-cancer analysis of bi-allelic alterations in homologous recombination DNA repair genes. Nat. Commun. 2017, 8, 857. [Google Scholar] [CrossRef]
- Li, H.-J.; Wu, N.-L.; Pu, C.-M.; Hsiao, C.-Y.; Chang, D.-C.; Hung, C.-F. Chrysin alleviates imiquimod-induced psoriasis-like skin inflammation and reduces the release of CCL20 and antimicrobial peptides. Sci. Rep. 2020, 10, 2932. [Google Scholar] [CrossRef] [Green Version]
- Key, N.S.; Khorana, A.A.; Kuderer, N.M.; Bohlke, K.; Lee, A.Y.; Arcelus, J.I.; Wong, S.L.; Balaban, E.P.; Flowers, C.R.; Francis, C.W. Venous thromboembolism prophylaxis and treatment in patients with cancer: ASCO clinical practice guideline update. J. Clin. Oncol. 2020, 38, 496–520. [Google Scholar] [CrossRef]
- Anwanwan, D.; Singh, S.K.; Singh, S.; Saikam, V.; Singh, R. Challenges in liver cancer and possible treatment approaches. Biochim. Biophys. Acta BBA Rev. Cancer 2020, 1873, 188314. [Google Scholar] [CrossRef]
- Ng, C.X.; Lee, S.H. The Potential Use of Anticancer Peptides (ACPs) in the Treatment of Hepatocellular Carcinoma. Curr. Cancer Drug Targets 2020, 20, 187–196. [Google Scholar] [CrossRef]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef]
- Bosso, A.; Di Maro, A.; Cafaro, V.; Di Donato, A.; Notomista, E.; Pizzo, E. Enzymes as a Reservoir of Host Defence Peptides. Curr. Top. Med. Chem. 2020, 20, 1310–1323. [Google Scholar] [CrossRef]
- Bakare, O.O.; Fadaka, A.O.; Keyster, M.; Pretorius, A. Structural and molecular docking analytical studies of the predicted ligand binding sites of cadherin-1 in cancer prognostics. Adv. Appl. Bioinform. Chem. AABC 2020, 13, 1–9. [Google Scholar] [CrossRef]
- Bakare, O.O.; Fadaka, A.O.; Klein, A.; Pretorius, A. Dietary effects of antimicrobial peptides in therapeutics. All Life 2020, 13, 78–91. [Google Scholar] [CrossRef] [Green Version]
- Arias, M.; Haney, E.F.; Hilchie, A.L.; Corcoran, J.A.; Hyndman, M.E.; Hancock, R.E.; Vogel, H.J. Selective anticancer activity of synthetic peptides derived from the host defence peptide tritrpticin. Biochim. Biophys. Acta BBA Biomembr. 2020, 1862, 183228. [Google Scholar] [CrossRef] [PubMed]
- Németh, T.; Sperandio, M.; Mócsai, A. Neutrophils as emerging therapeutic targets. Nat. Rev. Drug Discov. 2020, 19, 253–275. [Google Scholar] [CrossRef] [PubMed]
- Kunda, N.K. Antimicrobial peptides as novel therapeutics for non-small cell lung cancer. Drug Discov. Today 2020, 25, 238–247. [Google Scholar] [CrossRef]
- Kardani, K.; Bolhassani, A. Antimicrobial/anticancer peptides: Bioactive molecules and therapeutic agents. Immunotherapy 2021, 13, 669–684. [Google Scholar] [CrossRef]
- Al-Smadi, M.; Al-Momani, F. Synthesis, characterization and antimicrobial activity of new 1, 2, 3-selenadiazoles. Molecules 2008, 13, 2740–2749. [Google Scholar] [CrossRef] [Green Version]
- Marqus, S.; Pirogova, E.; Piva, T.J. Evaluation of the use of therapeutic peptides for cancer treatment. J. Biomed. Sci. 2017, 24, 21. [Google Scholar] [CrossRef] [Green Version]
- Szlasa, W.; Zendran, I.; Zalesińska, A.; Tarek, M.; Kulbacka, J. Lipid composition of the cancer cell membrane. J. Bioenerg. Biomembr. 2020, 52, 321–342. [Google Scholar] [CrossRef]
- Chiangjong, W.; Chutipongtanate, S.; Hongeng, S. Anticancer peptide: Physicochemical property, functional aspect and trend in clinical application. Int. J. Oncol. 2020, 57, 678–696. [Google Scholar] [CrossRef]
- Frislev, H.S.; Boye, T.L.; Nylandsted, J.; Otzen, D. Liprotides kill cancer cells by disrupting the plasma membrane. Sci. Rep. 2017, 7, 15129. [Google Scholar] [CrossRef] [PubMed]
- Mamusa, M.; Sitia, L.; Barbero, F.; Ruyra, A.; Calvo, T.D.; Montis, C.; Gonzalez-Paredes, A.; Wheeler, G.N.; Morris, C.J.; McArthur, M. Cationic liposomal vectors incorporating a bolaamphiphile for oligonucleotide antimicrobials. Biochim. Biophys. Acta BBA Biomembr. 2017, 1859, 1767–1777. [Google Scholar] [CrossRef]
- Prasad, S.V.; Fiedoruk, K.; Daniluk, T.; Piktel, E.; Bucki, R. Expression and function of host defense peptides at inflammation sites. Int. J. Mol. Sci. 2020, 21, 104. [Google Scholar] [CrossRef] [Green Version]
- Bhatt, T.; Bhosale, A.; Bajantri, B.; Mathapathi, M.S.; Rizvi, A.; Scita, G.; Majumdar, A.; Jamora, C. Sustained secretion of the antimicrobial peptide S100A7 is dependent on the downregulation of caspase-8. Cell Rep. 2019, 29, 2546–2555. [Google Scholar] [CrossRef]
- Mandke, P.; Vasquez, K.M. Interactions of high mobility group box protein 1 (HMGB1) with nucleic acids: Implications in DNA repair and immune responses. DNA Repair 2019, 83, 102701. [Google Scholar] [CrossRef] [PubMed]
- Oliveira-Junior, N.G.; Freire, M.S.; Almeida, J.A.; Rezende, T.M.; Franco, O.L. Antimicrobial and proinflammatory effects of two vipericidins. Cytokine 2018, 111, 309–316. [Google Scholar] [CrossRef]
- Xhindoli, D.; Pacor, S.; Benincasa, M.; Scocchi, M.; Gennaro, R.; Tossi, A. The human cathelicidin LL-37—A pore-forming antibacterial peptide and host-cell modulator. Biochim. Biophys. Acta BBA Biomembr. 2016, 1858, 546–566. [Google Scholar] [CrossRef]
- Li, B.; Lyu, P.; Xie, S.; Qin, H.; Pu, W.; Xu, H.; Chen, T.; Shaw, C.; Ge, L.; Kwok, H.F. LFB: A novel antimicrobial brevinin-like peptide from the skin secretion of the Fujian large headed frog, Limnonectes fujianensi. Biomolecules 2019, 9, 242. [Google Scholar] [CrossRef] [Green Version]
- Patrzykat, A.; Friedrich, C.L.; Zhang, L.; Mendoza, V.; Hancock, R.E. Sublethal concentrations of pleurocidin-derived antimicrobial peptides inhibit macromolecular synthesis in Escherichia coli. Antimicrob. Agents Chemother. 2002, 46, 605–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexander, L.G.; Sessions, R.B.; Clarke, A.R.; Tatham, A.S.; Shewry, P.R.; Napier, J.A. Characterization and modelling of the hydrophobic domain of a sunflower oleosin. Planta 2002, 214, 546–551. [Google Scholar] [CrossRef]
- Aghazadeh, H.; Memariani, H.; Ranjbar, R.; Pooshang Bagheri, K. The activity and action mechanism of novel short selective LL-37-derived anticancer peptides against clinical isolates of Escherichia coli. Chem. Biol. Drug Des. 2019, 93, 75–83. [Google Scholar] [CrossRef] [Green Version]
- Fruitwala, S.; El-Naccache, D.W.; Chang, T.L. Multifaceted immune functions of human defensins and underlying mechanisms. Semin. Cell Dev. Biol. 2019, 88, 163–172. [Google Scholar] [CrossRef]
- Liu, S.; Zhou, L.; Li, J.; Suresh, A.; Verma, C.; Foo, Y.H.; Yap, E.P.; Tan, D.T.; Beuerman, R.W. Linear analogues of human β-defensin 3: Concepts for design of antimicrobial peptides with reduced cytotoxicity to mammalian cells. Chembiochem 2008, 9, 964–973. [Google Scholar] [CrossRef]
- Zweytick, D. LTX-315–a promising novel antitumor peptide and immunotherapeutic agent. Cell Stress 2019, 3, 328. [Google Scholar] [CrossRef] [Green Version]
- Jeyamogan, S.; Khan, N.A.; Sagathevan, K.; Siddiqui, R. Sera/organ lysates of selected animals living in polluted environments exhibit cytotoxicity against cancer cell lines. Anticancer Agents Med. Chem. 2019, 19, 2251–2268. [Google Scholar] [CrossRef] [PubMed]
- Siddiqua, A.; Khattak, K.; Nwaz, S. Venom proteins; Prospects for anticancer therapy. Pak. J. Biochem. Mol. Biol 2019, 52, 15–26. [Google Scholar]
- Brady, D.; Grapputo, A.; Romoli, O.; Sandrelli, F. Insect cecropins, antimicrobial peptides with potential therapeutic applications. Int. J. Mol. Sci. 2019, 20, 5862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinto, I.B.; dos Santos Machado, L.; Meneguetti, B.T.; Nogueira, M.L.; Carvalho, C.M.E.; Roel, A.R.; Franco, O.L. Utilization of antimicrobial peptides, analogues and mimics in creating antimicrobial surfaces and bio-materials. Biochem. Eng. J. 2019, 150, 107237. [Google Scholar] [CrossRef]
- Zahedifard, F.; Lee, H.; No, J.H.; Salimi, M.; Seyed, N.; Asoodeh, A.; Rafati, S. Anti-leishmanial activity of Brevinin 2R and its Lauric acid conjugate type against L. major: In vitro mechanism of actions and in vivo treatment potentials. PLoS Negl. Trop. Dis. 2019, 13, e0007217. [Google Scholar]
- Liu, Y.; Tavana, O.; Gu, W. p53 modifications: Exquisite decorations of the powerful guardian. J. Mol. Cell Biol. 2019, 11, 564–577. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Zhang, L.; Ma, C.; Zhang, Y.; Xi, X.; Wang, L.; Zhou, M.; Burrows, J.F.; Chen, T. A novel antimicrobial peptide, Ranatuerin-2PLx, showing therapeutic potential in inhibiting proliferation of cancer cells. Biosci. Rep. 2018, 38, BSR20180710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tornesello, A.L.; Borrelli, A.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Antimicrobial peptides as anticancer agents: Functional properties and biological activities. Molecules 2020, 25, 2850. [Google Scholar] [CrossRef]
- Tripathi, A.K.; Kumari, T.; Harioudh, M.K.; Yadav, P.K.; Kathuria, M.; Shukla, P.; Mitra, K.; Ghosh, J.K. Identification of GXXXXG motif in Chrysophsin-1 and its implication in the design of analogs with cell-selective antimicrobial and anti-endotoxin activities. Sci. Rep. 2017, 7, 3384. [Google Scholar] [CrossRef] [PubMed]
- Hansen, I.K.; Isaksson, J.; Poth, A.G.; Hansen, K.Ø.; Andersen, A.J.; Richard, C.S.; Blencke, H.-M.; Stensvåg, K.; Craik, D.J.; Haug, T. Isolation and characterization of antimicrobial peptides with unusual disulfide connectivity from the colonial ascidian Synoicum turgens. Mar. Drugs 2020, 18, 51. [Google Scholar] [CrossRef] [Green Version]
- Matsuzaki, K. Membrane permeabilization mechanisms. In Antimicrobial Peptides; Springer: Berlin/Heidelberg, Germany, 2019; pp. 9–16. [Google Scholar]
- Vitale, D.; Kumar Katakam, S.; Greve, B.; Jang, B.; Oh, E.S.; Alaniz, L.; Götte, M. Proteoglycans and glycosaminoglycans as regulators of cancer stem cell function and therapeutic resistance. FEBS J. 2019, 286, 2870–2882. [Google Scholar] [CrossRef] [Green Version]
- Neundorf, I. Antimicrobial and cell-penetrating peptides: How to understand two distinct functions despite similar physicochemical properties. In Antimicrobial Peptides; Springer: Berlin/Heidelberg, Germany, 2019; pp. 93–109. [Google Scholar]
- Wang, Y.-Y.; Wang, Y.; Li, G.-Z.; Hao, L. Salicylic acid-altering Arabidopsis plant response to cadmium exposure: Underlying mechanisms affecting antioxidation and photosynthesis-related processes. Ecotoxicol. Environ. Saf. 2019, 169, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Blakaj, A.; Bonomi, M.; Gamez, M.E.; Blakaj, D.M. Oral mucositis in head and neck cancer: Evidence-based management and review of clinical trial data. Oral Oncol. 2019, 95, 29–34. [Google Scholar] [CrossRef]
- Rashid, R.; Veleba, M.; Kline, K.A. Focal targeting of the bacterial envelope by antimicrobial peptides. Front. Cell Dev. Biol. 2016, 4, 55. [Google Scholar] [CrossRef]
- Borrelli, A.; Tornesello, A.L.; Tornesello, M.L.; Buonaguro, F.M. Cell penetrating peptides as molecular carriers for anti-cancer agents. Molecules 2018, 23, 295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piotrowska, U.; Sobczak, M.; Oledzka, E. Current state of a dual behaviour of antimicrobial peptides—Therapeutic agents and promising delivery vectors. Chem. Biol. Drug Des. 2017, 90, 1079–1093. [Google Scholar] [CrossRef] [PubMed]
- Bleicken, S.; Assafa, T.E.; Stegmueller, C.; Wittig, A.; Garcia-Saez, A.J.; Bordignon, E. Topology of active, membrane-embedded Bax in the context of a toroidal pore. Cell Death Differ. 2018, 25, 1717–1731. [Google Scholar] [CrossRef] [Green Version]
- Rogers, C.; Erkes, D.A.; Nardone, A.; Aplin, A.E.; Fernandes-Alnemri, T.; Alnemri, E.S. Gasdermin pores permeabilize mitochondria to augment caspase-3 activation during apoptosis and inflammasome activation. Nat. Commun. 2019, 10, 1689. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zou, X.; Qi, G.; Tang, Y.; Guo, Y.; Si, J.; Liang, L. Roles and mechanisms of human cathelicidin LL-37 in cancer. Cell. Physiol. Biochem. 2018, 47, 1060–1073. [Google Scholar] [CrossRef]
- Hsieh, T.-H.; Hsu, C.-Y.; Tsai, C.-F.; Chiu, C.-C.; Liang, S.-S.; Wang, T.-N.; Kuo, P.-L.; Long, C.-Y.; Tsai, E.-M. A novel cell-penetrating peptide suppresses breast tumorigenesis by inhibiting β-catenin/LEF-1 signaling. Sci. Rep. 2016, 6, 19156. [Google Scholar] [CrossRef] [Green Version]
- Haney, E.F.; Straus, S.K.; Hancock, R.E. Reassessing the host defense peptide landscape. Front. Chem. 2019, 7, 43. [Google Scholar] [CrossRef] [Green Version]
- Pan, X.; Xu, J.; Jia, X. Research progress evaluating the function and mechanism of anti-tumor peptides. Cancer Manag. Res. 2020, 12, 397. [Google Scholar] [CrossRef] [Green Version]
- Banerji, U.; Cook, N.; Evans, T.J.; Moreno Candilejo, I.; Roxburgh, P.; Kelly, C.L.; Sabaratnam, N.; Passi, R.; Leslie, S.; Katugampola, S. A Cancer Research UK phase I/IIa trial of BT1718 (a first in class Bicycle Drug Conjugate) given intravenously in patients with advanced solid tumours. J. Clin. Oncol. 2018, 36, TPS2610. [Google Scholar] [CrossRef]
- Shi, Y.; Van der Meel, R.; Chen, X.; Lammers, T. The EPR effect and beyond: Strategies to improve tumor targeting and cancer nanomedicine treatment efficacy. Theranostics 2020, 10, 7921. [Google Scholar] [CrossRef]
- Mai, J.C.; Mi, Z.; Kim, S.-H.; Ng, B.; Robbins, P.D. A proapoptotic peptide for the treatment of solid tumors. Cancer Res. 2001, 61, 7709–7712. [Google Scholar]
- Sabapathy, T.; Deplazes, E.; Mancera, R.L. Revisiting the interaction of melittin with phospholipid bilayers: The effects of concentration and ionic strength. Int. J. Mol. Sci. 2020, 21, 746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanchard, B.J.; Thomas, V.L.; Ingram, V.M. Mechanism of membrane depolarization caused by the Alzheimer Aβ1–42 peptide. Biochem. Biophys. Res. Commun. 2002, 293, 1197–1203. [Google Scholar] [CrossRef]
- Prakash, A.; Jeffryes, M.; Bateman, A.; Finn, R.D. The HMMER web server for protein sequence similarity search. Curr. Protoc. Bioinform. 2017, 60, 3.15.1–3.15.23. [Google Scholar] [CrossRef]
- Grafskaia, E.N.; Polina, N.F.; Babenko, V.V.; Kharlampieva, D.D.; Bobrovsky, P.A.; Manuvera, V.A.; Farafonova, T.E.; Anikanov, N.A.; Lazarev, V.N. Discovery of novel antimicrobial peptides: A transcriptomic study of the sea anemone Cnidopus japonicus. J. Bioinform. Comput. Biol. 2018, 16, 1840006. [Google Scholar] [CrossRef]
- Jäkel, S.; Williams, A. What have advances in transcriptomic technologies taught us about human white matter pathologies? Front. Cell. Neurosci. 2020, 14, 238. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shi, J.; Tong, Z.; Jia, Y.; Yang, B.; Wang, Z. The revitalization of antimicrobial peptides in the resistance era. Pharmacol. Res. 2020, 163, 105276. [Google Scholar] [CrossRef]
- Fields, F.R.; Freed, S.D.; Carothers, K.E.; Hamid, M.N.; Hammers, D.E.; Ross, J.N.; Kalwajtys, V.R.; Gonzalez, A.J.; Hildreth, A.D.; Friedberg, I. Novel antimicrobial peptide discovery using machine learning and biophysical selection of minimal bacteriocin domains. Drug Dev. Res. 2020, 81, 43–51. [Google Scholar] [CrossRef]
- Cardoso, P.; Glossop, H.; Meikle, T.G.; Aburto-Medina, A.; Conn, C.E.; Sarojini, V.; Valery, C. Molecular engineering of antimicrobial peptides: Microbial targets, peptide motifs and translation opportunities. Biophys. Rev. 2021, 13, 1–35. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zhou, Y.; Xu, Y.; Ren, X.; Zhou, S.; Shang, Q.; Jiang, Y.; Luan, Y. Molecular engineering of anti-PD-L1 peptide and photosensitizer for immune checkpoint blockade photodynamic-immunotherapy. Chem. Eng. J. 2020, 400, 125995. [Google Scholar] [CrossRef]
- Aruleba, R.T.; Adekiya, T.A.; Oyinloye, B.E.; Kappo, A.P. Structural studies of predicted ligand binding sites and molecular docking analysis of Slc2a4 as a therapeutic target for the treatment of cancer. Int. J. Mol. Sci. 2018, 19, 386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G. Antimicrobial Peptides: Discovery, Design and Novel Therapeutic Strategies; CABI: Wallingford, UK, 2017. [Google Scholar]
- Tucker, A.T.; Leonard, S.P.; DuBois, C.D.; Knauf, G.A.; Cunningham, A.L.; Wilke, C.O.; Trent, M.S.; Davies, B.W. Discovery of next-generation antimicrobials through bacterial self-screening of surface-displayed peptide libraries. Cell 2018, 172, 618–628.e613. [Google Scholar] [CrossRef] [PubMed]
- Bakare, O.O.; Keyster, M.; Pretorius, A. Identification of biomarkers for the accurate and sensitive diagnosis of three bacterial pneumonia pathogens using in silico approaches. BMC Mol. Cell Biol. 2020, 21, 1–13. [Google Scholar] [CrossRef]
- Williams, M.; Tincho, M.; Gabere, M.; Uys, A.; Meyer, M.; Pretorius, A. Molecular validation of putative antimicrobial peptides for improved human immunodeficiency virus diagnostics via HIV protein p24. J. AIDS Clin. Res. 2016, 7, 2. [Google Scholar] [CrossRef] [Green Version]
- Tincho, M.; Gabere, M.; Pretorius, A. In silico identification and molecular validation of putative antimicrobial peptides for HIV therapy. J. AIDS Clin. Res. 2016, 7, 606. [Google Scholar] [CrossRef] [Green Version]
- Liscano, Y.; Oñate-Garzón, J.; Delgado, J.P. Peptides with dual antimicrobial–anticancer activity: Strategies to overcome peptide limitations and rational design of anticancer peptides. Molecules 2020, 25, 4245. [Google Scholar] [CrossRef]
- Harris, M.A.; Hawkins, C.J.; Miles, M.A. Tetrazolium reduction assays under-report cell death provoked by clinically relevant concentrations of proteasome inhibitors. Mol. Biol. Rep. 2020, 47, 4849–4856. [Google Scholar] [CrossRef]
- Urusov, A.E.; Zherdev, A.V.; Dzantiev, B.B. Towards lateral flow quantitative assays: Detection approaches. Biosensors 2019, 9, 89. [Google Scholar] [CrossRef] [Green Version]
- Mytton, O.T.; McCarthy, N.; Watson, J.; Whiting, P. Interpreting a lateral flow SARS-CoV-2 antigen test. BMJ 2021, 373, n1411. [Google Scholar] [CrossRef] [PubMed]
- Okugawa, Y.; Tanaka, K.; Inoue, Y.; Kawamura, M.; Kawamoto, A.; Hiro, J.; Saigusa, S.; Toiyama, Y.; Ohi, M.; Uchida, K. Brain-derived neurotrophic factor/tropomyosin-related kinase B pathway in gastric cancer. Br. J. Cancer 2013, 108, 121–130. [Google Scholar] [CrossRef] [Green Version]
- Prada-Prada, S.; Flórez-Castillo, J.; Farfán-García, A.; Guzmán, F.; Hernández-Peñaranda, I. Antimicrobial activity of Ib-M peptides against Escherichia coli O157: H7. PLoS ONE 2020, 15, e0229019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, F.; Wu, Q.; Song, S.; She, R.; Zhao, Y.; Yang, Y.; Zhang, M.; Du, F.; Soomro, M.H.; Shi, R. Antimicrobial activity and safety evaluation of peptides isolated from the hemoglobin of chickens. BMC Microbiol. 2016, 16, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Regmi, S.; Choi, Y.H.; Choi, Y.S.; Kim, M.R.; Yoo, J.C. Antimicrobial peptide isolated from Bacillus amyloliquefaciens K14 revitalizes its use in combinatorial drug therapy. Folia Microbiol. 2017, 62, 127–138. [Google Scholar] [CrossRef]
- Steinstraesser, L.; Kraneburg, U.M.; Hirsch, T.; Kesting, M.; Steinau, H.-U.; Jacobsen, F.; Al-Benna, S. Host defense peptides as effector molecules of the innate immune response: A sledgehammer for drug resistance? Int. J. Mol. Sci. 2009, 10, 3951–3970. [Google Scholar] [CrossRef] [Green Version]
- Riedl, S.; Zweytick, D.; Lohner, K. Membrane-active host defense peptides–challenges and perspectives for the development of novel anticancer drugs. Chem. Phys. lipids 2011, 164, 766–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoskin, D.W.; Ramamoorthy, A. Studies on anticancer activities of antimicrobial peptides. Biochim. Biophys. Acta BBA Biomembr. 2008, 1778, 357–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, M.A.; Maloy, W.L.; Zasloff, M.; Jacob, L.S. Anticancer efficacy of Magainin2 and analogue peptides. Cancer Res. 1993, 53, 3052–3057. [Google Scholar] [PubMed]
- Papo, N.; Shai, Y. New lytic peptides based on the D, L-amphipathic helix motif preferentially kill tumor cells compared to normal cells. Biochemistry 2003, 42, 9346–9354. [Google Scholar] [CrossRef] [PubMed]
- Boto, A.; Pérez de la Lastra, J.M.; González, C.C. The road from host-defense peptides to a new generation of antimicrobial drugs. Molecules 2018, 23, 311. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakare, O.O.; Gokul, A.; Wu, R.; Niekerk, L.-A.; Klein, A.; Keyster, M. Biomedical Relevance of Novel Anticancer Peptides in the Sensitive Treatment of Cancer. Biomolecules 2021, 11, 1120. https://doi.org/10.3390/biom11081120
Bakare OO, Gokul A, Wu R, Niekerk L-A, Klein A, Keyster M. Biomedical Relevance of Novel Anticancer Peptides in the Sensitive Treatment of Cancer. Biomolecules. 2021; 11(8):1120. https://doi.org/10.3390/biom11081120
Chicago/Turabian StyleBakare, Olalekan Olanrewaju, Arun Gokul, Ruomou Wu, Lee-Ann Niekerk, Ashwil Klein, and Marshall Keyster. 2021. "Biomedical Relevance of Novel Anticancer Peptides in the Sensitive Treatment of Cancer" Biomolecules 11, no. 8: 1120. https://doi.org/10.3390/biom11081120










