Amino Acid and Carotenoid Profiles of Chlorella vulgaris During Two-Stage Cultivation at Different Salinities
Abstract
1. Introduction
2. Materials and Methods
2.1. Microalgae
2.2. Culture Conditions and Experimental Design
2.3. Salinity Treatment
2.4. Analytical Methods: pH and Microalgal Growth Monitoring
2.5. Analytical Methods: Nutrient Concentration Monitoring
2.6. Analytical Methods: Harvesting and Extraction
2.7. Analytical Methods: Amino Acid and Carotenoid Quantification
2.8. Statistical Analysis
3. Results
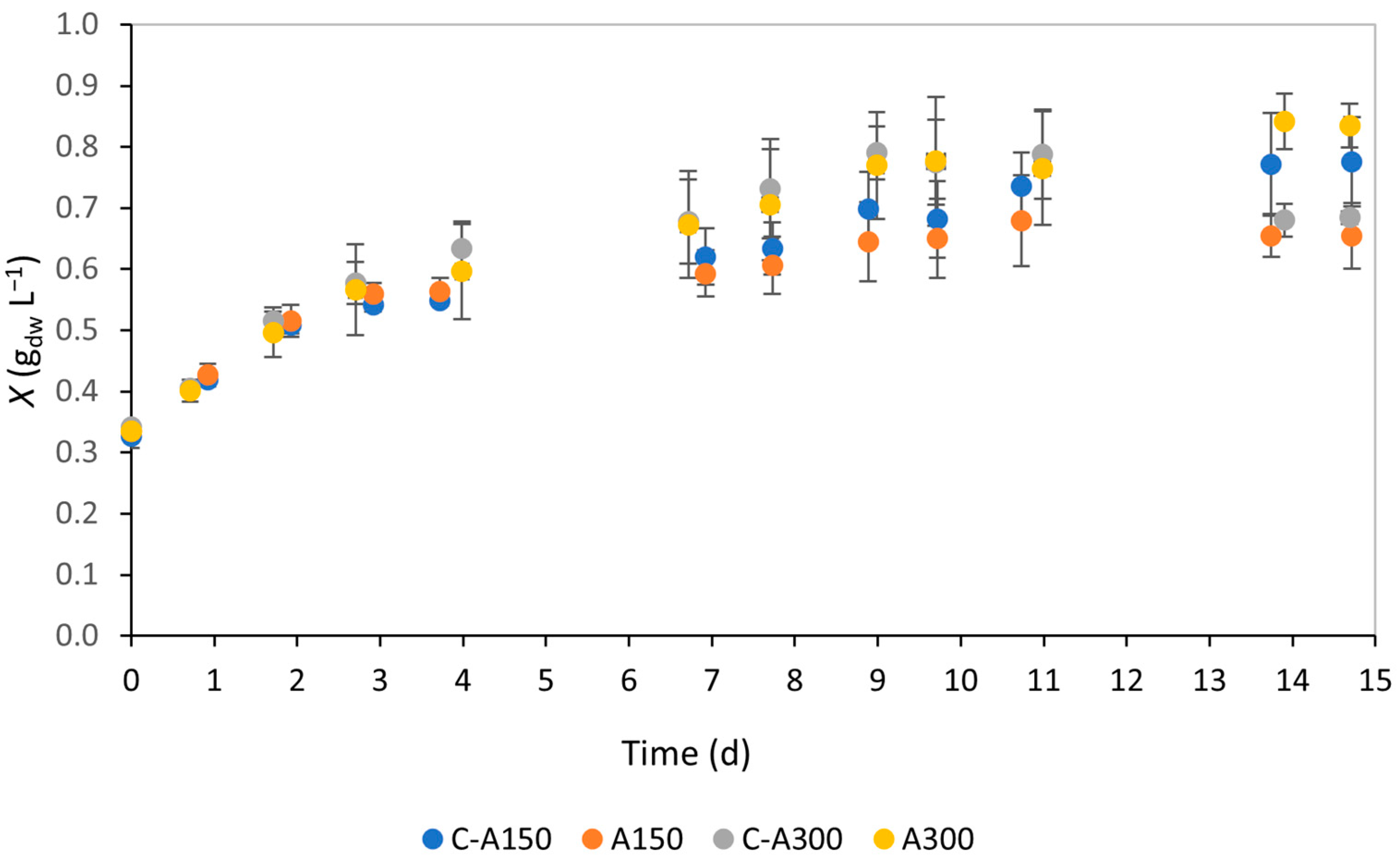
3.1. Biomass Growth and Productivity
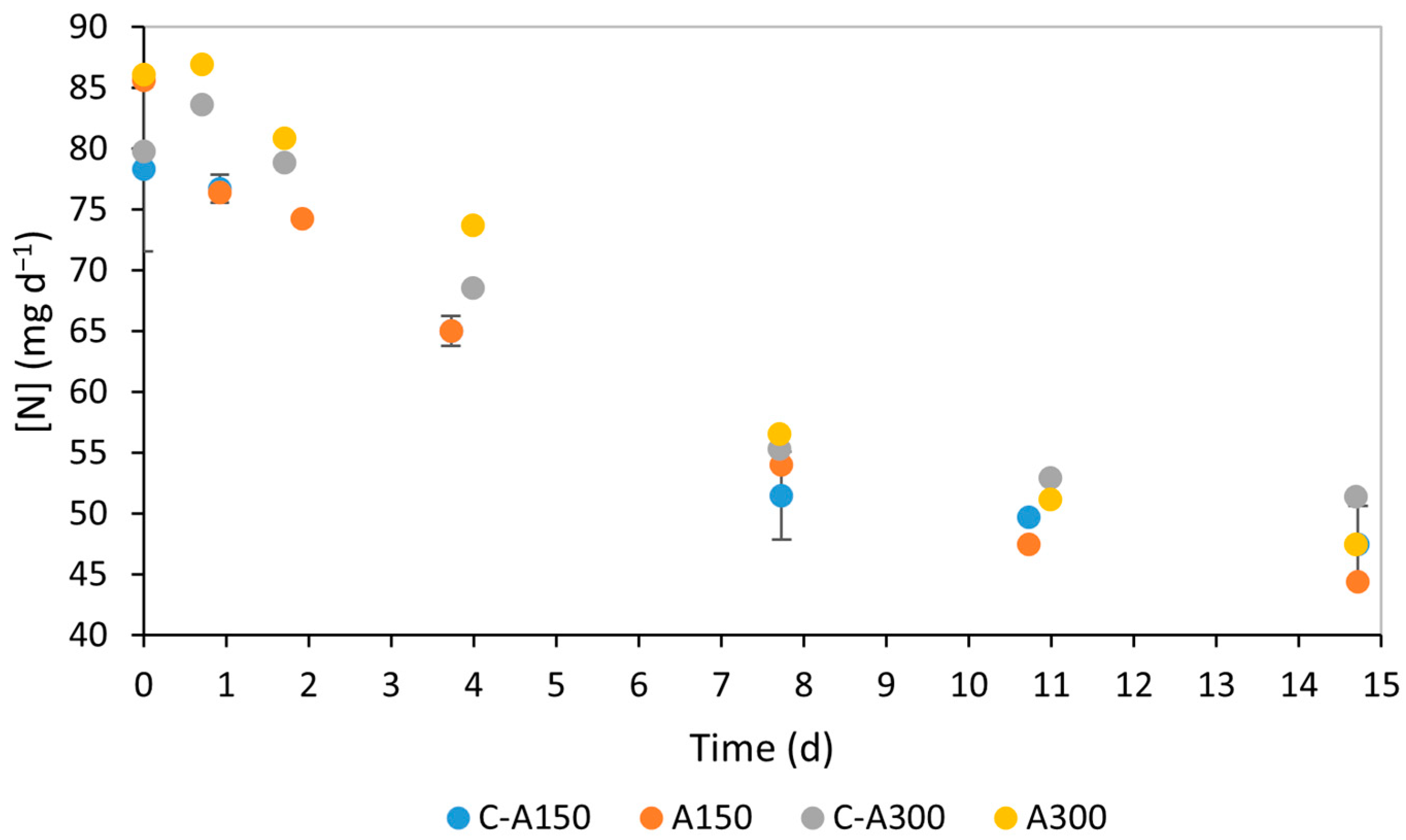
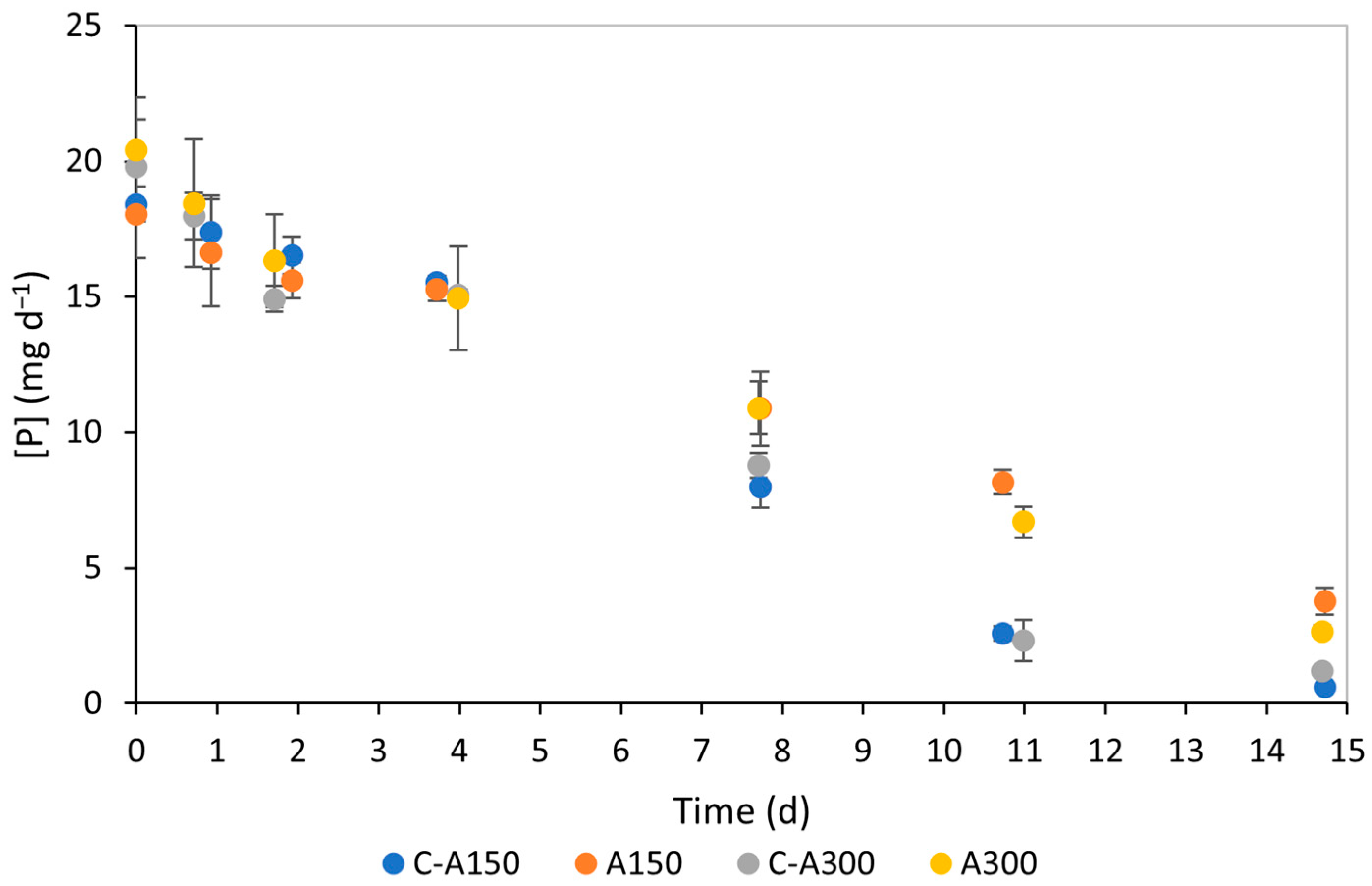
3.2. Nutrient Consumption
3.3. Amino Acid Profile
3.4. Carotenoid Profile
3.5. Principal Component Analysis
4. Discussion
4.1. Biomass Growth and Productivity
4.2. Nutrient Consumption
4.3. Amino Acid Profile
4.4. Carotenoid Profile
4.5. Salinity Impact on Productivity, Nutrient Consumption, and Compound Production
4.6. Principal Component Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Su, Y.Y. Revisiting carbon, nitrogen, and phosphorus metabolisms in microalgae for wastewater treatment. Sci. Total Environ. 2021, 762, 144590. [Google Scholar] [CrossRef]
- Ren, Y.Y.; Sun, H.; Deng, J.Q.; Huang, J.C.; Chen, F. Carotenoid Production from Microalgae: Biosynthesis, Salinity Responses and Novel Biotechnologies. Mar. Drugs 2021, 19, 713. [Google Scholar] [CrossRef]
- Rizwan, M.; Mujtaba, G.; Memon, S.A.; Lee, K.; Rashid, N. Exploring the potential of microalgae for new biotechnology applications and beyond: A review. Renew. Sustain. Energ. Rev. 2018, 92, 394–404. [Google Scholar] [CrossRef]
- Rögner, M. Photosynthesis: Biotechnological Applications with Microalgae; De Gruyter: Berlin, Germany, 2021. [Google Scholar]
- Machado, A.; Pereira, H.; Costa, M.; Santos, T.; Carvalho, B.; Soares, M.; Quelhas, P.; Silva, J.T.; Trovao, M.; Barros, A.; et al. Development of an Organic Culture Medium for Autotrophic Production of Chlorella vulgaris Biomass. Appl. Sci. 2020, 10, 2156. [Google Scholar] [CrossRef]
- Williamson, E.; Ross, I.L.; Wall, B.T.; Hankamer, B. Microalgae: Potential novel protein for sustainable human nutrition. Trends Plant Sci. 2024, 29, 370–382. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, D.; Varjani, S.; Lee, D.J.; Chang, J.S. Sustainable aquaculture and animal feed from microalgae—Nutritive value and techno-functional components. Renew. Sustain. Energ. Rev. 2021, 150, 111549. [Google Scholar] [CrossRef]
- Kumar, R.; Hegde, A.S.; Sharma, K.; Parmar, P.; Srivatsan, V. Microalgae as a sustainable source of edible proteins and bioactive peptides-Current trends and future prospects. Food Res. Int. 2022, 157, 111338. [Google Scholar] [CrossRef]
- Wang, Y.W.; Tibbetts, S.M.; McGinn, P.J. Microalgae as Sources of High-Quality Protein for Human Food and Protein Supplements. Foods 2021, 10, 3002. [Google Scholar] [CrossRef] [PubMed]
- Mohamadnia, S.; Tavakoli, O.; Faramarzi, M.A.; Shamsollahi, Z. Production of fucoxanthin by the microalga Tisochrysis lutea: A review of recent developments. Aquaculture 2020, 516, 734637. [Google Scholar] [CrossRef]
- Odjadjare, E.C.; Mutanda, T.; Olaniran, A.O. Potential biotechnological application of microalgae: A critical review. Crit. Rev. Biotechnol. 2017, 37, 37–52. [Google Scholar] [CrossRef]
- Liu, Z.J.; van den Berg, C.; Weusthuis, R.A.; Dragone, G.; Mussatto, S.I. Strategies for an improved extraction and separation of lipids and carotenoids from oleaginous yeast. Sep. Purif. Technol. 2021, 257, 117946. [Google Scholar] [CrossRef]
- Esteves, A.F.; Gonçalves, A.L.; Vilar, V.J.P.; Pires, J.C.M. Is it possible to shape the microalgal biomass composition with operational parameters for target compound accumulation? Biotechnol. Adv. 2025, 79, 108493. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.M.; Ren, L.J.; Zhao, Q.Y.; Ji, X.J.; Huang, H. Microalgae for the production of lipid and carotenoids: A review with focus on stress regulation and adaptation. Biotechnol. Biofuels 2018, 11, 272. [Google Scholar] [CrossRef]
- Li, T.; Wang, W.N.; Yuan, C.J.; Zhang, Y.; Xu, J.; Zheng, H.L.; Xiang, W.Z.; Li, A.F. Linking lipid accumulation and photosynthetic efficiency in Nannochloropsis sp. under nutrient limitation and replenishment. J. Appl. Phycol. 2020, 32, 1619–1630. [Google Scholar] [CrossRef]
- Zappi, M.E.; Bajpai, R.; Hernandez, R.; Mikolajczyk, A.; Fortela, D.L.; Sharp, W.; Chirdon, W.; Zappi, K.; Gang, D.; Nigam, K.D.P.; et al. Microalgae Culturing to Produce Biobased Diesel Fuels: An Overview of the Basics, Challenges, and a Look toward a True Biorefinery Future. Ind. Eng. Chem. Res. 2019, 58, 15724–15746. [Google Scholar] [CrossRef]
- Shah, S.M.U.; Abdullah, M.A. Effects of Macro/Micronutrients on Green and Brown Microalgal Cell Growth and Fatty Acids in Photobioreactor and Open-Tank Systems (vol 14C, pg 10, 2018). Biocatal. Agr. Biotech. 2021, 31, 10–17. [Google Scholar]
- Sun, X.; Cao, Y.; Xu, H.; Liu, Y.; Sun, J.R.; Qiao, D.R.; Cao, Y. Effect of nitrogen-starvation, light intensity and iron on triacylglyceride/carbohydrate production and fatty acid profile of Neochloris oleoabundans HK-129 by a two-stage process. Bioresour. Technol. 2014, 155, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.E.A.; El-fayoumy, E.A.; Rasmy, W.E.; Soliman, R.M.; Abdullah, M.A. Two-stage cultivation of using light and salt stress conditions for simultaneous production of lipid, carotenoids, and antioxidants. J. Appl. Phycol. 2021, 33, 227–239. [Google Scholar] [CrossRef]
- Sinetova, M.A.; Sidorov, R.A.; Medvedeva, A.A.; Starikov, A.Y.; Markelova, A.G.; Allakhverdiev, S.I.; Los, D.A. Effect of salt stress on physiological parameters of microalgae Vischeria punctata strain IPPAS H-242, a superproducer of eicosapentaenoic acid. J. Biotechnol. 2021, 331, 63–73. [Google Scholar] [CrossRef]
- Pérez-Gálvez, A.; Viera, I.; Roca, M. Carotenoids and Chlorophylls as Antioxidants. Antioxidants 2020, 9, 505. [Google Scholar] [CrossRef]
- Ru, I.T.K.; Sung, Y.Y.; Jusoh, M.; Wahid, M.E.A.; Nagappan, T. Chlorella vulgaris: A perspective on its potential for combining high biomass with high value bioproducts. Appl. Phycol. 2020, 1, 2–11. [Google Scholar] [CrossRef]
- Qiang, S.; Su, A.P.; Li, Y.; Chen, Z.; Hu, C.Y.; Meng, Y.H. Elevated β-Carotene Synthesis by the Engineered Rhodobacter sphaeroides with Enhanced CrtY Expression. J. Agr. Food Chem. 2019, 67, 9560–9568. [Google Scholar] [CrossRef]
- Widomska, J.; Subczynski, W.K. Mechanisms enhancing the protective functions of macular xanthophylls in the retina during oxidative stress. Exp. Eye Res. 2019, 178, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Paliwal, C.; Mitra, M.; Bhayani, K.; Bharadwaj, S.V.V.; Ghosh, T.; Dubey, S.; Mishra, S. Abiotic stresses as tools for metabolites in microalgae. Bioresour. Technol. 2017, 244, 1216–1226. [Google Scholar] [CrossRef]
- Sousa, S.A.; Esteves, A.F.; Ab, E.M.S.; Ab, J.C.M.P. Enhancing urban wastewater treatment: Chlorella vulgaris performance in tertiary treatment and the impact of anaerobic digestate addition. Environ. Technol. Innov. 2024, 34, 103601. [Google Scholar] [CrossRef]
- Lee, B.; Park, S.Y.; Hae, Y.S.; Yea, S.S.; Kim, D.E. Efficient Colorimetric Assay of RNA Polymerase Activity Using Inorganic Pyrophosphatase and Ammonium Molybdate. B Korean Chem. Soc. 2009, 30, 2485–2488. [Google Scholar]
- Collos, Y.; Mornet, F.; Sciandra, A.; Waser, N.; Larson, A.; Harrison, P.J. An optical method for the rapid measurement of micromolar concentrations of nitrate in marine phytoplankton cultures. J. Appl. Phycol. 1999, 11, 179–184. [Google Scholar] [CrossRef]
- Pagels, F.; Pereira, R.N.; Vicente, A.A.; Guedes, A.C. Extraction of Pigments from Microalgae and Cyanobacteria-A Review on Current Methodologies. Appl. Sci. 2021, 11, 5187. [Google Scholar] [CrossRef]
- Kim, D.Y.; Vijayan, D.; Praveenkumar, R.; Han, J.I.; Lee, K.; Park, J.Y.; Chang, W.S.; Lee, J.S.; Oha, Y.K. Cell-wall disruption and lipid/astaxanthin extraction from microalgae: Chlorella and Haematococcus. Bioresour. Technol. 2016, 199, 300–310. [Google Scholar] [CrossRef]
- Mendes-Pinto, M.M.; Raposo, M.F.J.; Bowen, J.; Young, A.J.; Morais, R. Evaluation of different cell disruption processes on encysted cells of Haematococcus pluvialis: Effects on astaxanthin recovery and implications for bio-availability. J. Appl. Phycol. 2001, 13, 19–24. [Google Scholar] [CrossRef]
- Cheng, S.-H.; Khoo, H.E.; Kong, K.W.; Prasad, K.N.; Galanakis, C.M. applications. In Carotenoids: Properties, Processing and Applications; Galanakis, C.M., Ed.; Academic Press: New York, NY, USA, 2020; pp. 259–288. [Google Scholar]
- Hynstova, V.; Sterbova, D.; Klejdus, B.; Hedbavny, J.; Huska, D.; Adam, V. Separation, identification and quantification of carotenoids and chlorophylls in dietary supplements containing Chlorella vulgaris and Spirulina platensis using High Performance Thin Layer Chromatography. J. Pharm. Biomed. 2018, 148, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.A.; Deantonis, K.M. Applications of Amino-Acid Derivatization with 6-Aminoquinolyl-N-Hydroxysuccinimidyl Carbamate—Analysis of Feed Grains, Intravenous Solutions and Glycoproteins. J. Chromatogr. A 1994, 661, 25–34. [Google Scholar] [CrossRef]
- Horwitz, W. Official Methods of Analysis of AOAC International; AOAC International: Gaithersburg, MD, USA, 2000. [Google Scholar]
- Esteves, A.F.; Pardilhó, S.; Gonçalves, A.L.; Vilar, V.J.P.; Pires, J.C.M. Unravelling the impact of light spectra on microalgal growth and biochemical composition using principal component analysis and artificial neural network models. Algal Res. 2025, 85, 103820. [Google Scholar] [CrossRef]
- Bro, R.; Smilde, A.K. Principal component analysis. Anal. Methods 2014, 6, 2812–2831. [Google Scholar] [CrossRef]
- Greenacre, M.; Groenen, P.J.F.; Hastie, T.; D’Enza, A.I.; Markos, A.; Tuzhilina, E. Principal component analysis. Nat. Rev. Methods Primers 2022, 2, 100. [Google Scholar] [CrossRef]
- Gong, Q.T.; Feng, Y.Z.; Kang, L.G.; Luo, M.Y.; Yang, J.H. Effects of Light and pH on Cell Density of Chlorella vulgaris. Energy Proced. 2014, 61, 2012–2015. [Google Scholar] [CrossRef]
- Daliry, S.; Hallajisani, A.; Mohammadi Roshandeh, J.; Nouri, H.; Golzary, A. Investigation of optimal condition for Chlorella vulgaris microalgae growth. Glob. J. Environ. Sci. Manag. 2017, 3, 217–230. [Google Scholar] [CrossRef]
- Salgado, E.M.; Esteves, A.F.; Gonçalves, A.L.; Pires, J.C.M. Microalgal cultures for the remediation of wastewaters with different nitrogen to phosphorus ratios: Process modelling using artificial neural networks. Environ. Res. 2023, 231, 116076. [Google Scholar] [CrossRef]
- Cai, X.; Huang, Q.; Wang, S. Isolation of a novel lutein–protein complex from Chlorella vulgaris and its functional properties. Food Funct. 2015, 6, 1893–1899. [Google Scholar] [CrossRef]
- Church, J.; Hwang, J.H.; Kim, K.T.; McLean, R.; Oh, Y.K.; Nam, B.; Joo, J.C.; Lee, W.H. Effect of salt type and concentration on the growth and lipid content of Chlorella vulgaris in synthetic saline wastewater for biofuel production. Bioresour. Technol. 2017, 243, 147–153. [Google Scholar] [CrossRef]
- Pandit, P.R.; Fulekar, M.H.; Karuna, M.S.L. Effect of salinity stress on growth, lipid productivity, fatty acid composition, and biodiesel properties in Acutodesmus obliquus and Chlorella vulgaris. Environ. Sci. Pollut. Res. 2017, 24, 13437–13451. [Google Scholar] [CrossRef]
- Luangpipat, T.; Chisti, Y. Biomass and oil production by Chlorella vulgaris and four other microalgae—Effects of salinity and other factors. J. Biotechnol. 2017, 257, 47–57. [Google Scholar] [CrossRef] [PubMed]
- El-fayoumy, E.A.; Ali, H.E.A.; Elsaid, K.; Elkhatat, A.; Al-Meer, S.; Rozaini, M.Z.H.; Abdullah, M.A. Co-production of high density biomass and high-value compounds via two-stage cultivation of using light intensity and a combination of salt stressors. Biomass Convers. Bior 2023, 14, 22673–22686. [Google Scholar] [CrossRef]
- Biliani, S.E.; Manariotis, I.D. Sodium chloride and nitrogen effects on Chlorella vulgaris growth and biocommodities production. Desalin Water Treat. 2021, 237, 159–169. [Google Scholar] [CrossRef]
- Li, J.; Li, C.; Lan, C.Q.; Liao, D. Effects of sodium bicarbonate on cell growth, lipid accumulation, and morphology of Chlorella vulgaris. Microb. Cell Factories 2018, 17, 111. [Google Scholar] [CrossRef]
- Alyabyev, A.J.; Loseva, N.L.; Gordon, L.K.; Andreyeva, I.N.; Rachimova, G.G.; Tribunskih, V.I.; Ponomareva, A.A.; Kemp, R.B. The effect of changes in salinity on the energy yielding processes of Chlorella vulgaris and Dunaliella maritima cells. Thermochim. Acta 2007, 458, 65–70. [Google Scholar] [CrossRef]
- Shen, Q.H.; Gong, Y.P.; Fang, W.Z.; Bi, Z.C.; Cheng, L.H.; Xu, X.H.; Chen, H.L. Saline wastewater treatment by Chlorella vulgaris with simultaneous algal lipid accumulation triggered by nitrate deficiency. Bioresour. Technol. 2015, 193, 68–75. [Google Scholar] [CrossRef]
- Ishika, T.; Moheimani, N.R.; Bahri, P.A. Sustainable saline microalgae co-cultivation for biofuel production: A critical review. Renew. Sust. Energ. Rev. 2017, 78, 356–368. [Google Scholar] [CrossRef]
- Lee, T.S. Irrigation Systems and Practices in Challenging Environments; IntechOpen: Rijeka, Croatia, 2012. [Google Scholar] [CrossRef]
- Daneshvar, E.; Santhosh, C.; Antikainen, E.; Bhatnagar, A. Microalgal growth and nitrate removal efficiency in different cultivation conditions: Effect of macro and micronutrients and salinity. J. Environ. Chem. Eng. 2018, 6, 1848–1854. [Google Scholar] [CrossRef]
- Arora, N.; Philippidis, G.P. Unraveling metabolic alterations in Chlorella vulgaris cultivated on renewable sugars using time resolved multi-omics. Sci. Total Environ. 2021, 800, 149504. [Google Scholar] [CrossRef]
- Barera, S.; Forlani, G. The role of proline in the adaptation of eukaryotic microalgae to environmental stress: An underestimated tool for the optimization of algal growth. J. Appl. Phycol. 2023, 35, 1635–1648. [Google Scholar] [CrossRef]
- Ursu, A.V.; Marcati, A.; Sayd, T.; Sante-Lhoutellier, V.; Djelveh, G.; Michaud, P. Extraction, fractionation and functional properties of proteins from the microalgae. Bioresour. Technol. 2014, 157, 134–139. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, P.; Gururani, P.; Rawat, J.; Kumar Jaiswal, K.; Gautam, P.; Nanda, M.; Chauhan, P.K.; Vlaskin, M.S.; Kumar, V. Influence of GABA (Gamma-aminobutyric acid) supplementation on biomass, pigments, lipid and protein content of Pseudochlorella pringsheimii under salinity stress. Curr. Res. Biotechnol. 2024, 7, 100223. [Google Scholar] [CrossRef]
- Shetty, P.; Gitau, M.M.; Maróti, G. Salinity Stress Responses and Adaptation Mechanisms in Eukaryotic Green Microalgae. Cells 2019, 8, 1657. [Google Scholar] [CrossRef]
- Li, X.Y.; Yuan, Y.Z.; Cheng, D.J.; Gao, J.; Kong, L.Z.; Zhao, Q.Y.; Wei, W.; Sun, Y.H. Exploring stress tolerance mechanism of evolved freshwater strain Chlorella sp S30 under 30 g/L salt. Bioresour. Technol. 2018, 250, 495–504. [Google Scholar] [CrossRef]
- Vello, V.; Chu, W.-L.; Lim, P.-E.; Majid, N.A.; Phang, S.-M. Metabolomic profiles of tropical Chlorella species in response to physiological changes during nitrogen deprivation. J. Appl. Phycol. 2018, 30, 3131–3151. [Google Scholar] [CrossRef]
- Aigner, S.; Glaser, K.; Arc, E.; Holzinger, A.; Schletter, M.; Karsten, U.; Kranner, I. Adaptation to Aquatic and Terrestrial Environments in Chlorella vulgaris (Chlorophyta). Front. Microbiol. 2020, 11, 585836. [Google Scholar] [CrossRef]
- Cichonski, J.; Chrzanowski, G. Microalgae as a Source of Valuable Phenolic Compounds and Carotenoids. Molecules 2022, 27, 8852. [Google Scholar] [CrossRef]
- Vello, V.; Umashankar, S.; Phang, S.M.; Chu, W.L.; Lim, P.E.; Majid, N.A.; Liew, K.E.; Swarup, S.; Chew, F.T. Metabolomic profiles of tropical Chlorella and Parachlorella species in response to physiological changes during exponential and stationary growth phase. Algal Res. 2018, 35, 61–75. [Google Scholar] [CrossRef]
- Grudzinski, W.; Krzeminska, I.; Luchowski, R.; Nosalewicz, A.; Gruszecki, W.I. Strong-light-induced yellowing of green microalgae Chlorella: A study on molecular mechanisms of the acclimation response. Algal Res. 2016, 16, 245–254. [Google Scholar] [CrossRef]
- Demmig-Adams, B.; Stewart, J.J.; López-Pozo, M.; Polutchko, S.K.; Adams, W.W. Zeaxanthin, a Molecule for Photoprotection in Many Different Environments. Molecules 2020, 25, 5825. [Google Scholar] [CrossRef] [PubMed]

| Assay | µ (d−1) | PX,avg (mgdw L−1 d−1) |
|---|---|---|
| C-A150 | 0.23 ± 0.02 a | 29 ± 3 a |
| A150 | 0.22 ± 0.02 a | 23 ± 3 b |
| C-A300 | 0.241 ± 0.001 a | 23 ± 1 b |
| A300 | 0.23 ± 0.01 a | 34 ± 3 c |
| Assay | S0 (mg L−1) | Sf (mg L−1) | RE (%) | MR (mg L−1) | RR (mg L−1 d−1) |
|---|---|---|---|---|---|
| C-A150 | 78 ± 7 a | 47 ± 3 a | 39 ± 2 a | 36 ± 4 a,c | 2.6 ± 0.3 a |
| A150 | 85.7 ± 0.5 a | 44 ± 3 a | 49.5 ± 0.4 b | 42.4 ± 0.6 b | 2.88 ± 0.04 b |
| C-A300 | 80 ± 6 a | 51 ± 9 a | 35 ± 1 a | 32 ± 2 a | 2.2 ± 0.2 c |
| A300 | 86 ± 4 a | 47.5 ± 0.3 a | 45 ± 3 c | 39 ± 4 b,c | 2.76 ± 0.05 a,b |
| Assay | S0 (mg L−1) | Sf (mg L−1) | RE (%) | MR (mg L−1) | RR (mg L−1 d−1) |
|---|---|---|---|---|---|
| C-A150 | 18.4 ± 0.6 a | 0.60 ± 0.05 a | 96.7 ± 0.3 a | 17.8 ± 0.6 a | 1.21 ± 0.04 a |
| A150 | 18 ± 2 a | 3.8 ± 0.5 b | 79 ± 2 b | 15.2 ± 0.5 b | 1.03 ± 0.03 b |
| C-A300 | 20 ± 2 a | 1.2 ± 0.1 c | 94 ± 1 a | 19 ± 2 a | 1.3 ± 0.1 a |
| A300 | 20 ± 2 a | 2.6 ± 0.2 d | 87 ± 2 c | 18 ± 2 a | 1.2 ± 0.1 a |
| Amino Acid | Assay | Amino Acid Content (mg gdw−1) | TT (mg gdw−1 d−1) | ||
|---|---|---|---|---|---|
| T8 | T11 | T15 | |||
| ASP–Aspartic acid | C-A150 | 7.23 ± 0.39 a | 7.45 ± 0.33 a | 8.41 ± 0.48 c | 0.17 ± 0.04 |
| A150 | 10.56 ± 0.10 b | 8.88 ± 0.47 c | 6.74 ± 0.37 a,d | −0.55 ± 0.03 | |
| C-A300 | 6.77 ± 0.85 a,d | 6.16 ± 0.49 d | 7.40 ± 0.43 a | 0.09 ± 0.07 | |
| A300 | 6.88 ± 0.20 a,d | 3.60 ± 0.13 e | 3.82 ± 0.19 e | −0.45 ± 0.10 | |
| SER–L-Serine | C-A150 | 4.39 ± 0.18 a,e | 4.57 ± 0.15 a,b | 4.03 ± 0.22 c,e | −0.06 ± 0.03 |
| A150 | 4.93 ± 0.15 b | 4.52 ± 0.31 a,b,e | 3.64 ± 0.12 c | −0.19 ± 0.02 | |
| C-A300 | 4.37 ± 0.53 a,e | 3.62 0.21 c | 4.40 ± 0.12 a,e | 0.01 ± 0.06 | |
| A300 | 4.59 ± 0.06 a,b | 2.34 ± 0.13 d | 2.54 ± 0.12 d | −0.27 ± 0.07 | |
| GLU–Glutamic acid | C-A150 | 8.49 ± 0.46 a,c | 9.48 ± 0.55 d,g | 10.04 ± 0.65 e,g | 0.21 ± 0.06 |
| A150 | 12.42 ± 0.11 b | 10.53 ± 0.51 e | 7.76 ± 0.28 a | −0.67 ± 0.04 | |
| C-A300 | 7.94 ± 0.48 a | 7.87 ± 0.23 a | 9.93 ± 0.49 e,g | 0.30 ± 0.07 | |
| A300 | 9.03 ± 0.25 c,d | 4.43 ± 0.12 f | 5.02 ± 0.19 f | −0.59 ± 0.15 | |
| GLY–L-Glycine | C-A150 | 5.34 ± 0.38 a,c,d | 5.52 ± 0.32 a,c | 4.12 ± 0.17 b,c,d | −3.37 ± 0.99 |
| A150 | 5.17 ± 0.12 a,b,c,d | 4.69 ± 0.15 a,b,c,d | 4.30 ± 0.25 b,c,d | −6.71 ± 1.32 | |
| C-A300 | 6.04 ± 0.16 a | 4.85 ± 0.23 a,b,c,d | 4.89 ± 0.41 a,b,c,d | −3.34 ± 1.33 | |
| A300 | 5.45 ± 0.68 a,c,d | 3.67 ± 0.29 b | 3.85 ± 0.32 b,d | −2.25 ± 0.81 | |
| HIS–L-Histidine | C-A150 | 9.56 ± 0.64 a,f | 9.69 ± 0.60 a,f | 11.54 ± 0.29 d | 2.61 ± 0.53 |
| A150 | 14.39 ± 0.89 b | 11.04 ± 0.83 d | 10.46 ± 0.27 a,d | −1.42 ± 0.36 | |
| C-A300 | 7.37 ± 0.52 c | 6.25 ± 0.49 c,g | 8.91 ± 0.47 f | 1.37 ± 0.63 | |
| A300 | 7.09 ± 0.32 c | 2.12 ± 0.18 e | 5.00 ± 0.15 g | −0.56 ± 0.39 | |
| THR–L-Threonine | C-A150 | 3.79 ± 0.25 a,b | 4.00 ± 0.26 a,b | 3.73 ± 0.20 b | −0.01 ± 0.03 |
| A150 | 4.24 ± 0.09 a | 3.75 ± 0.26 b | 3.10 ± 0.10 c | −0.16 ± 0.02 | |
| C-A300 | 3.71 ± 0.40 b | 3.02 ± 0.22 c | 3.79 ± 0.21 a,b | 0.02 ± 0.05 | |
| A300 | 3.65 ± 0.14 b | 2.10 ± 0.12 d | 2.32 ± 0.16 d | −0.19 ± 0.05 | |
| ARG–Arginine | C-A150 | 17.84 ± 1.90 a | 17.71 ± 1.46 a | 19.19 ± 1.44 a | 0.23 ± 0.17 |
| A150 | 13.87 ± 0.66 b | 13.17 ± 1.80 b | 13.57 ± 0.77 b | −0.01 ± 0.15 | |
| C-A300 | 9.26 ± 0.74 c | 14.60 ± 0.03 b | 25.81 ± 1.83 e | 2.38 ± 0.16 | |
| A300 | 9.84 ± 0.60 c | 6.45 ± 0.52 d | 8.90 ± 0.68 c,d | −0.07 ± 0.18 | |
| ALA–Alanine | C-A150 | 2.57 ± 0.09 a,f | 2.71 ± 0.01 a | 2.74 ± 0.11 a | 0.02 ± 0.01 |
| A150 | 3.42 ± 0.19 b | 3.11 ± 0.15 c | 2.36 ± 0.09 d,f | −0.15 ± 0.02 | |
| C-A300 | 2.70 ± 0.20 a | 2.23 ± 0.10 d | 2.78 ± 0.13 a | 0.01 ± 0.03 | |
| A300 | 2.78 ± 0.11 a | 1.43 ± 0.09 e | 1.70 ± 0.02 g | −0.16 ± 0.05 | |
| PRO–L-Proline | C-A150 | 3.83 ± 0.23 a,e | 3.92 ± 0.25 a,e | 3.64 ± 0.21 e | −0.03 ± 0.02 |
| A150 | 6.89 ± 0.12 b | 6.09 ± 0.40 d | 4.27 ± 0.39 a,c,e | −0.39 ± 0.04 | |
| C-A300 | 4.74 ± 0.46 c,f | 4.32 ± 0.13 a,c | 4.03 ± 0.29 a,e | −0.10 ± 0.03 | |
| A300 | 6.18 ± 0.28 d | 4.78 ± 0.11 c,f | 5.14 ± 0.33 f | −0.14 ± 0.06 | |
| TYR–L-Tyrosine | C-A150 | 5.32 ± 0.39 a | 5.29 ± 0.31 a | 3.85 ± 0.34 d,e | −0.24 ± 0.05 |
| A150 | 4.42 ± 0.69 b,c,e | 4.05 ± 0.01 c,d,e | 3.78 ± 0.36 d,e | −0.09 ± 0.05 | |
| C-A300 | 5.12 ± 0.28 a,b | 4.73 ± 0.31 a,b,c | 4.21 ± 0.09 c,d,e | −0.13 ± 0.03 | |
| A300 | 5.31 ± 0.44 a | 3.61 ± 0.43 d | 3.56 ± 0.23 d | −0.24 ± 0.07 | |
| VAL–Valine | C-A150 | 3.84 ± 0.09 a,c | 3.99 ± 0.24 a,c | 4.01 ± 0.14 a,c | 0.02 ± 0.02 |
| A150 | 4.96 ± 0.26 b | 4.63 ± 0.28 b | 3.80 ± 0.14 a,c | −0.17 ± 0.02 | |
| C-A300 | 4.21 ± 0.49 a | 3.65 ± 0.16 c | 4.53 ± 0.18 b | 0.08 ± 0.05 | |
| A300 | 4.77 ± 0.10 b | 2.46 ± 0.15 d | 2.95 ± 0.11 d | −0.25 ± 0.09 | |
| MET–Methionine | C-A150 | 1.64 ± 0.15 a,d | 1.80 ± 0.09 a,b | 1.26 ± 0.19 c,d | −0.06 ± 0.03 |
| A150 | 2.13 ± 0.20 b | 1.95 ± 0.18 a,b | 1.66 ± 0.15 a,d | −0.07 ± 0.02 | |
| C-A300 | 1.90 ± 0.33 a,b | 1.69 ± 0.08 a | 1.68 ± 0.14 a | −0.03 ± 0.03 | |
| A300 | 2.01 ± 0.22 a,b | 0.98 ± 0.09 c | 0.97 ± 0.24 c | −0.14 ± 0.03 | |
| ILE–L-Isoleucine | C-A150 | 0.39 ± 0.04 a,c | 0.42 ± 0.05 c | 0.31 ± 0.03 b,f | −0.01 ± 0.01 |
| A150 | 0.35 ± 0.04 a,b | 0.31 ± 0.03 b,f | 0.28 ± 0.03 d,f | −0.01 ± 0.01 | |
| C-A300 | 0.30 ± 0.03 b,d,f | 0.24 ± 0.01 d | 0.26 ± 0.02 d,f | 0.00 ± 0.00 | |
| A300 | 0.29 ± 0.03 b,d,f | 0.00 ± 0.00 e | 0.17 ± 0.03 g | −0.02 ± 0.01 | |
| LYS–Lysine | C-A150 | 7.66 ± 0.29 a | 7.84 ± 0.22 a | 8.19 ± 0.32 a | 0.08 ± 0.03 |
| A150 | 9.86 ± 0.82 b | 9.03 ± 0.29 b | 7.66 ± 0.28 a | −0.32 ± 0.05 | |
| C-A300 | 7.75 ± 0.46 a | 6.58 ± 0.45 c | 8.22 ± 0.33 a | 0.10 ± 0.09 | |
| A300 | 8.35 ± 0.26 a | 4.12 ± 0.35 d | 5.01 ± 0.10 e | −0.50 ± 0.16 | |
| LEU–L-Leucine | C-A150 | 5.61 ± 0.28 a,d | 5.73 ± 0.33 a,d | 5.46 ± 0.22 a,d | −0.02 ± 0.03 |
| A150 | 7.10 ± 0.39 b | 6.62 ± 0.37 b | 5.31 ± 0.23 a,d | −0.26 ± 0.04 | |
| C-A300 | 6.78 ± 0.87 b | 5.28 ± 0.19 a | 6.09 ± 0.32 d | −0.09 ± 0.08 | |
| A300 | 6.99 ± 0.16 b | 3.55 ± 0.21 c | 4.05 ± 0.16 c | −0.41 ± 0.13 | |
| PHE–Phenylalanine | C-A150 | 4.89 ± 0.26 a,b | 5.07 ± 0.11 a,b | 3.42 ± 0.16 d | −0.22 ± 0.05 |
| A150 | 4.73 ± 0.52 a,c | 4.06 ± 0.19 c,d | 5.31 ± 0.23 a,b | −0.15 ± 0.04 | |
| C-A300 | 5.48 ± 0.23 b | 4.93 ± 0.39 a,b | 4.09 ± 0.07 c,d | −0.20 ± 0.04 | |
| A300 | 5.52 ± 0.59 b | 3.48 ± 0.45 d | 3.49 ± 0.29 d | −0.28 ± 0.09 | |
| Carotenoid | Assay | Carotenoid Content (mg gdw−1) | TT (mg gdw−1 d−1) | ||
|---|---|---|---|---|---|
| T8 | T11 | T15 | |||
| Neoxanthin | C-A150 | 0.58 ± 0.02 a | 0.30 ± 0.05 b | 0.23 ± 0.02 f | −0.044 ± 0.007 |
| A150 | 0.48 ± 0.05 b | 0.19 ± 0.02 e,f | 0.14 ± 0.02 e,g | −0.048 ± 0.006 | |
| C-A300 | 0.39 ± 0.03 c | 0.28 ± 0.03 d | 0.12 ± 0.01 g | −0.039 ± 0.003 | |
| A300 | 0.37 ± 0.05 c | 0.16 ± 0.02 e,g | 0.13 ± 0.02 e,g | −0.026 ± 0.006 | |
| Violaxantin | C-A150 | 0.26 ± 0.02 a,d | 0.21 ± 0.02 a,c,e | 0.20 ± 0.03 c,e,g | −0.009 ± 0.002 |
| A150 | 0.25 ± 0.03 a,d,e | 0.17 ± 0.02 c,g | 0.11 ± 0.02 f | −0.021 ± 0.002 | |
| C-A300 | 0.33 ± 0.06 b | 0.29 ± 0.03 d | 0.15 ± 0.01 f,g | −0.027 ± 0.004 | |
| A300 | 0.36 ± 0.05 b | 0.16 ± 0.01 c,f,g | 0.15 ± 0.01 f,g | −0.022 ± 0.005 | |
| Lutein | C-A150 | 1.55 ± 0.22 a,d | 1.66 ± 0.08 a,d | 1.37 ± 0.13 d | −0.029 ± 0.015 |
| A150 | 1.56 ± 0.16 a,d | 1.11 ± 0.21 c | 0.89 ± 0.10 c | −0.094 ± 0.014 | |
| C-A300 | 2.18 ± 0.24 b | 1.75 ± 0.21 a | 0.87 ± 0.05 c | −0.193 ± 0.021 | |
| A300 | 2.01 ± 0.30 b | 0.96 ± 0.11 c | 0.99 ± 0.18 c | −0.106 ± 0.035 | |
| Zeaxanthin | C-A150 | 0.20 ± 0.02 a,d,e | 0.29 ± 0.02 b | 0.29 ± 0.04 b | 0.013 ± 0.003 |
| A150 | 0.29 ± 0.05 b | 0.16 ± 0.03 a,c,e | 0.24 ± 0.02 b,d | −0.006 ± 0.005 | |
| C-A300 | 0.21 ± 0.01 a,d | 0.22 ± 0.04 a,d | 0.14 ± 0.01 c,e | −0.011 ± 0.003 | |
| A300 | 0.12 ± 0.02 c | 0.12 ± 0.03 c | 0.17 ± 0.05 a,c,e | 0.010 ± 0.005 | |
| β-carotene | C-A150 | 0.12 ± 0.02 a | 0.09 ± 0.01 c | 0.05 ± 0.01 d | −0.010 ± 0.001 |
| A150 | 0.13 ± 0.03 a | 0.07 ± 0.01 c,d | 0.05 ± 0.01 d | −0.011 ± 0.002 | |
| C-A300 | 0.21 ± 0.01 b | 0.14 ± 0.02 a | 0.07 ± 0.01 c,d | −0.020 ± 0.002 | |
| A300 | 0.21 ± 0.02 b | 0.12 ± 0.01 a | 0.07 ± 0.01 c,d | −0.017 ± 0.002 | |
| Compound | PC1 | PC2 | PC3 | PC4 |
|---|---|---|---|---|
| ASP–Aspartic acid | −0.939 | −0.058 | −0.034 | 0.295 |
| SER–L-Serine | −0.897 | −0.410 | 0.131 | 0.061 |
| GLU–Glutamic acid | −0.961 | −0.076 | 0.034 | 0.183 |
| GLY–L-Glycine | −0.484 | −0.823 | 0.151 | −0.034 |
| HIS–L-Histidine | −0.875 | 0.117 | 0.042 | 0.451 |
| THR–L-Threonine | −0.887 | −0.333 | 0.210 | 0.183 |
| ARG–Arginine | −0.536 | 0.203 | 0.769 | 0.006 |
| ALA–Alanine | −0.967 | −0.201 | −0.035 | 0.126 |
| PRO–L-Proline | −0.349 | −0.077 | −0.883 | −0.172 |
| TYR–L-Tyrosine | −0.261 | −0.909 | 0.250 | 0.000 |
| VAL–Valine | −0.950 | −0.255 | −0.070 | −0.090 |
| MET–Methionine | −0.800 | −0.489 | −0.128 | −0.072 |
| ILE–L-Isoleucine | −0.718 | −0.358 | 0.343 | 0.370 |
| LYS–Lysine | −0.973 | −0.157 | −0.005 | 0.128 |
| LEU–L-Leucine | −0.865 | −0.447 | −0.142 | −0.080 |
| PHE–Phenylalanine | −0.287 | −0.936 | 0.008 | −0.080 |
| Neoxanthin | −0.272 | −0.747 | −0.067 | 0.358 |
| Violaxantin | −0.113 | −0.929 | −0.199 | −0.006 |
| Lutein | −0.135 | −0.936 | −0.102 | 0.174 |
| Zeaxanthin | −0.326 | −0.005 | 0.164 | 0.891 |
| β-carotene | 0.079 | −0.887 | −0.334 | −0.229 |
| Eigenvalue | 12.31 | 4.64 | 1.87 | 1.03 |
| Variance (%) | 58.62 | 22.07 | 8.89 | 4.88 |
| Cumulative variance (%) | 58.62 | 80.69 | 89.59 | 94.47 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinto, A.S.; Maia, C.; Sousa, S.A.; Tavares, T.; Pires, J.C.M. Amino Acid and Carotenoid Profiles of Chlorella vulgaris During Two-Stage Cultivation at Different Salinities. Bioengineering 2025, 12, 284. https://doi.org/10.3390/bioengineering12030284
Pinto AS, Maia C, Sousa SA, Tavares T, Pires JCM. Amino Acid and Carotenoid Profiles of Chlorella vulgaris During Two-Stage Cultivation at Different Salinities. Bioengineering. 2025; 12(3):284. https://doi.org/10.3390/bioengineering12030284
Chicago/Turabian StylePinto, Ana S., Carolina Maia, Sara A. Sousa, Tânia Tavares, and José C. M. Pires. 2025. "Amino Acid and Carotenoid Profiles of Chlorella vulgaris During Two-Stage Cultivation at Different Salinities" Bioengineering 12, no. 3: 284. https://doi.org/10.3390/bioengineering12030284
APA StylePinto, A. S., Maia, C., Sousa, S. A., Tavares, T., & Pires, J. C. M. (2025). Amino Acid and Carotenoid Profiles of Chlorella vulgaris During Two-Stage Cultivation at Different Salinities. Bioengineering, 12(3), 284. https://doi.org/10.3390/bioengineering12030284









