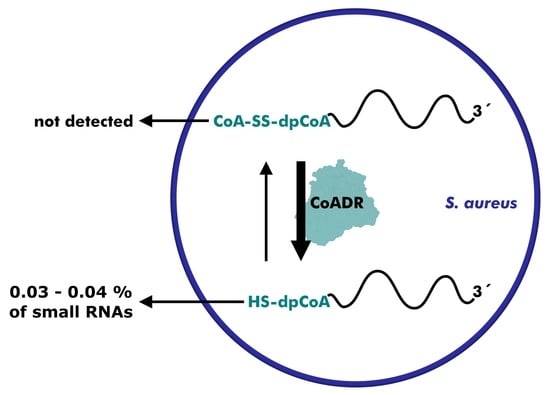
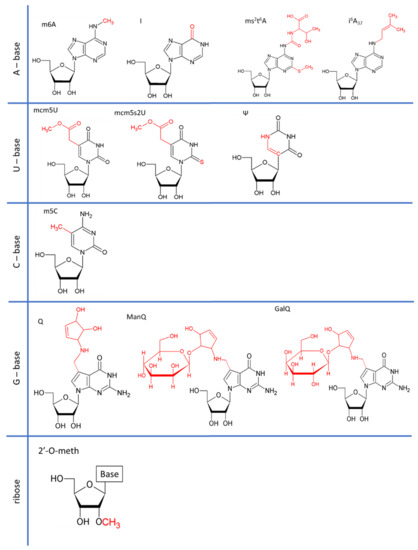
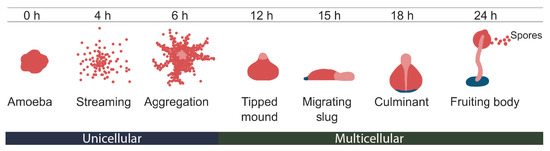
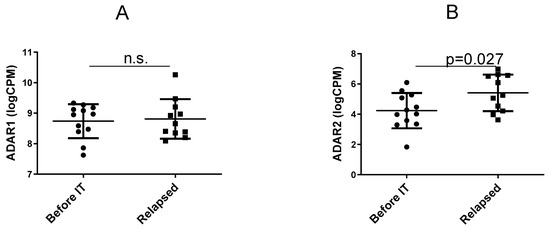
Research on RNA Modification
A topical collection in Non-Coding RNA (ISSN 2311-553X). This collection belongs to the section "RNA Modifications".
Viewed by 23019Editors
Interests: sncRNAs (si, mi and piRNAs); RNA methylation; 2’-0-methylation; epitranscriptomic; chromatin
Topical Collection Information
Dear Colleagues,
RNA modifications, including RNA editing, are today well recognized to play an essential role in the regulation of gene expression through several mechanisms. They have been shown to be involved in many different steps, including regulation of their stability/degradation, localization, transport in and out of the cells, protein interaction, and their role in translational regulation. Post-transcriptional modifications affect virtually all types of RNA (mRNA as well as non-coding RNA such as tRNA, rRNA, sncRNA, lncRNA, circRNA, etc.) in all known kingdoms of life as well as in viral RNAs.
The aim of this Special Issue is to cover all molecular aspects of RNA modification and RNA editing, including biosynthesis of these modified nucleotides, enzymes, and partners involved in their formation, detection methods developed for the analysis of RNA modifications, as well as molecular and cellular mechanisms, for which those modifications are important.
Finally, and excitingly, regarding the nature of RNA and its involvement in epigenetics transmission (e.g., sncRNA), the aspect of RNA modifications on epigenetics mechanisms (epitranscriptomics) will also be covered in this Special Issue.
For this Special Issue, we ask for comprehensive reviews, editorials, original research manuscripts or methodological articles.
Prof. Dr. Clément Carré
Dr. Damien Brégeon
Guest Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 250 words) can be sent to the Editorial Office for assessment.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Non-Coding RNA is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 1800 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- RNA modification
- RNA editing
- Modification biosynthesis
- Modifications detections
- Modifications functions
- Modification diseases
- Epitranscriptomics