Abstract
Monkeypox is a zoonotic disease caused by a virus member of the genus Orthopoxvirus and is endemic to Central and Western African countries. Previous work has identified two geographically disjuct clades of monkeypox virus based on the analysis of a few genomes coupled with epidemiological and clinical analyses; however, environmental and geographic causes of this differentiation have not been explored. Here, we expand previous phylogenetic studies by analyzing a larger set of monkeypox virus genomes originating throughout Sub-Saharan Africa to identify possible biogeographic barriers associated with genetic differentiation; and projected ecological niche models onto environmental conditions at three periods in the past to explore the potential role of climate oscillations in the evolution of the two primary clades. Analyses supported the separation of the Congo Basin and West Africa clades; the Congo Basin clade shows much shorter branches, which likely indicate a more recent diversification of isolates within this clade. The area between the Sanaga and Cross Rivers divides the two clades and the Dahomey Gap seems to have also served as a barrier within the West African clade. Contraction of areas with suitable environments for monkeypox virus during the Last Glacial Maximum, suggests that the Congo Basin clade of monkeypox virus experienced a severe bottleneck and has since expanded its geographic range.
1. Introduction
Monkeypox (MPX) is a zoonotic disease caused by a member of the Orthopoxvirus genus, which includes other viruses pathogenic to humans (e.g., variola virus, vaccinia virus, and cowpox virus), and produces mild to severe rash illness in infected individuals. The first human case of monkeypox was identified in the Democratic Republic of the Congo (DRC) in 1971 [1] and is currently a major public health concern in that country. Surveillance of human cases of MPX in Central Africa was very active between 1970 and 1986, particularly at the end of the smallpox eradication campaign, with more than 400 human cases reported during this period [2,3,4]. Since then, MPX surveillance has been limited to investigations of outbreaks of the disease [5,6,7,8,9]. In recent years, MPX surveillance in DRC has increased given the growing interest in clarifying the poorly understood natural history of the virus [10,11,12].
Likos et al. [13], based on phylogenetic analyses of monkeypox virus (MPXV) isolates, supported the recognition of two distinct clades of this virus: Western African (WA) and Congo Basin (CB) clades. These two clades are geographically disjunct and have defined epidemiological and clinical differences [7,14,15]. In Likos et al. [13], isolates grouped within the WA clade were obtained from a patient in Liberia [16] and a US soldier returning from Ghana [7]; while the CB clade included two isolates from DRC (formerly Zaire) [3,5] and one from the Republic of the Congo [6].
In the fields of ecology and biogeography, ecological niche modeling (ENM) is commonly used in studies regarding species distributions and to reconstruct their recent evolutionary and biogeographic history [17,18,19,20,21]. Several studies have shown the value of incorporating these methods in the study of the ecology and distribution of various infectious diseases, including MPX [12,22,23,24,25,26]. Ellis et al. [25] successfully produced suitability maps for MPX transmission using ENM; their results suggest the existence of a break in the distribution of suitable environmental conditions for MPX transmission at the Cameroon Highlands. A partition of MPX geographic range at this land feature produces two groups of MPXV that coincide with the WA and CB clades from Likos et al. [13].
Although the phylogenetic analysis of Likos et al. [13] and the ENM from Ellis et al. [25] consistently support the division of MPX into WA and CB clades, the small number of isolates (five) used in the former work does not provide enough resolution to determine whether the Cameroon Highlands is associated with the differentiation of these clades. Particularly, the great geographic distance separating the WA isolates (Liberia and Ghana) and the CB isolates (Republic of the Congo and DRC) makes it difficult to propose the Cameroon Highlands as the only geographic feature involved in the differentiation of MPXV clades because other potential geographic elements are also found between these isolates (e.g., Dahomey Gap, major rivers, etc.).
Climate oscillations during the Quaternary have caused the expansion and contraction of the geographic ranges of biomes (e.g., rainforest, savanna, tundra, etc.), and organisms associated with them, around the world [27,28,29]. In Africa, the rainforest contracted its geographic distribution during glacial maxima and expanded during interglacial periods, potentially impacting the distribution of MPXV and its reservoirs given the tight association between MPXV transmission events and the rainforest in Central and Western Africa [30]. The fragmentation of African rainforest through these climatic cycles could have had a role in the genetic differentiation of MPXV over several thousand years.
Here, we analyze a larger number of genomic sequences from MPXV isolates covering the known geographic distribution of MPXV to add resolution to the phylogenetic analysis and establish relationships among isolates within and between clades. Additionally, we explore the effect that climatic oscillations since the last interglacial period (ca. 135,000 years ago) have had on the distribution of environments suitable for transmission of MPXV and reconstruct its recent history. Finally, we integrate results from ENM with the phylogenetic evidence to identify biogeographic elements and/or geologic events that have potentially influenced the genetic differentiation of MPXV.
2. Materials and Methods
2.1. Genetic Data and Analysis
Table 1 summarizes the MPXV isolates used in our phylogenetic analysis and provides references for the original description of the cases; they include (a) four isolates that correspond to MPXV obtained from outbreaks recorded in laboratories; (b) the five isolates included in the study by Likos et al.; (c) twelve isolates available from human case reports in Sub-Saharan Africa between 1970 and 2010; (d) one isolate obtained from a squirrel from Yambuku, DRC, which is the only MPXV isolate derived directly from wildlife included in this study; and (e) 23 isolates from human cases reported in Sankuru District, DRC between 2006 and 2007 [31]. In all, the 45 analyzed isolates cover the known range for MPX in Central and West Africa (Figure 1). Isolates form Nigeria, Cameroon, and Gabon are of particular interest for the present analysis since they are closer to one of the proposed biogeographic barriers (Cameroon Highlands) than the rest of the isolates from Western Africa or Congo Basin.
Genomes were sequenced using Sanger sequencing or Illumina® (Illumina Inc., San Diego, CA USA) paired end sequencing. Whole genomes of MPXV isolates were aligned using MAFFT v7.017 [32]. Cowpox virus (Grisham 1990, X94355) and horsepox virus (Mongolia 1976, DQ792504) isolates were included as outgroup taxa. The original alignment was 241,258 bp in length. The first 25 kb and last 26 kb are highly variable between CPXV and other Orthopoxviruses since they contain a large number of indels; thus, they were trimmed. Subsequently, all columns containing indels were removed, resulting in an alignment of 173,804 bases from the central conserved region of the genome. A majority-rules consensus tree was estimated from the alignment of all genomes using MrBayes v3.2.2 [33,34]. Settings included a general time reversible model (lset nst = 6) with estimated stationary state frequencies and substitution rates, and a model of gamma-shaped rate variation across sites (rates=gamma). The tree search was carried out over five million generations.
A patristic distances matrix was obtained from the consensus tree and separated into the groups representing the two recognized MPXV clades. We tested the distance values within each group for normality via the Shapiro-Wilk test, compared their variances via the F-test and compared the two groups using a Student’s t-test; all statistical test were performed in R 3.1.1 [35]. MEGA v6.06 [36] was used to calculate within group uncorrelated p-distances for a subset of samples from the Lomela Health Zone in DRC.
2.2. Ecological Niche Models
Environmental data: we used a set of seven diverse and relatively uncorrelated variables [37] from the bioclimatic dataset of WorldClim [38] used in a previous monkeypox ENM study [39]: annual mean temperature, mean diurnal range, maximum temperature of the warmest month, minimum temperature of coldest month, annual precipitation, precipitation of the wettest month, and precipitation of the driest month. These variables were obtained from the WorldClim website (http://www.worldclim.org/) in their digital format at a spatial resolution of ~4 km (2.5 min) for the Last Glacial Maximum (LGM, ca. 21,000 years ago) and Mid-Holocene (MidHol, ca. 6000 years ago) periods; and ~1 km spatial resolution (30 second) for the Last Interglacial period (LIG, ca. 135,000 years ago) [38,40]. A buffer of 5 degrees (~555 km) of radius was created from all monkeypox case localities (see below) and was used to delineate the area in which ENMs is calibrated.
Human case data: we used reports of human MPX from the World Health Organization (WHO) surveillance efforts in DRC between 1970 and 1986. Case localities were georeferenced matching the patient’s village of residence to digital versions of 1:250,000 Joint Operational Graphic (JOG) topographic maps (GNS; http://earth-info.nga.mil/gns.html/) following the georeferencing procedures from MaNIS [41]; details of these procedures are included in a separate publication [39]. The aim of the present study is to identify the environmental conditions required for the disease to be transmitted from its wildlife reservoir to humans; therefore, cases suspected to have resulted from secondary transmission (i.e., the disease acquired by contact with a sick person) were not included in the analysis. In all, 90 unique localities representing the entire geographic distribution of human MPX occurrence were used (white circles in Figure 3A,B). Given that most of the localities were obtained from the active surveillance efforts at the end of the smallpox eradication, this data is biased with better representation of localities in DRC. To account for this bias, we produced 25 subsets of the original dataset by reducing the density of the reported localities in DRC; specifically, we selected subsets of the original dataset in which localities were separated by a distance of 30 min (Approx. 50 km); this process produced datasets containing 41–43 localities.
Ecological niche algorithms: two of the most widely used ecological niche modeling algorithms (GARP and Maxent) have also been used for the study of disease transmission [22,23,26,42,43,44,45,46,47]. Both algorithms search for non-random associations between environmental conditions and the localities where the disease has been recorded; however, models may differ from one algorithm to another in their ability to estimate distributions [48] and to transfer such models into other environmental conditions or geographic areas [49], therefore, independent models were generated with each algorithm.
GARP v1.1.6 produces a model consisting of a set of rules that describe associations between environmental conditions and disease transmission events [50,51]. These models are built via an iterative process of creation, evaluation, modification, and inclusion/exclusion of rules of four basic forms (bioclimatic, atomic, negated, and logistic regression rules). This process stops when the optimization parameter changes by less than 1% from one iteration to the next, or when the maximum number of iterations is reached (1000). For each of the 25 subsets of localities, we ran 100 replicates using 50% of occurrence points to train models on environmental conditions in the present and selected the replicates with the highest performance via the “best subset” consensus approach: retaining the 20% with the lowest omission error and, from this subset, the central 50% in terms of commission error [52]. The models selected by this process (n = 10) were summed and evaluated using the modified ROC (E = 10%, 1000 bootstrap iterations and 50% of points), in which models performing better than random have AUC ratio values higher than 1 [53]. For a particular subset of localities, we selected the areas containing 90% of the training localities to represent the distribution of suitable environments for MPX transmission and created a binary map where 1 = suitable and 0 = unsuitable. Finally, the areas selected from each subset of localities were combined to find areas of agreement by summing the binary maps into one map with values ranging from 0 to 25.
The Maxent algorithm is based on the idea that the best solution for an unknown phenomenon maximizes entropy; thus, it calculates a probability distribution that is closest to a uniform distribution, constrained by the parameters calculated for the locality cases and a regularization parameter (β) that prevents the estimated mean values of the distribution from deviating from the observed mean value [54,55]. We used default settings in Maxent v3.2.1 (i.e., regularization multiplier = 1.0, 1500 maximum iterations, 10,000 background points, convergence limit = 10−5) to create models for each locality subset. We evaluated the models using the modified ROC as described above and selected the areas containing all localities used for training the model for each of the 25 subsets of localities to create a binary map; these maps were combined to find areas of agreement as described above.
ENMs from both algorithms were projected into the environmental conditions during the Mid-Holocene, Last Interglacial and Last Glacial Maximum periods using the environmental datasets described in the previous section. Areas with suitable environments were selected using the methodology described above for each algorithm. The final maps for each period are the combination of the models produced by all 25 locality subsets.
3. Results
3.1. Phylogenetic Analysis
Results of the phylogenetic analysis are shown in Figure 1. The average standard deviation of split frequencies was 0.001,718. Branch lengths between MPXV isolates are shown to scale. The Bayesian analysis grouped 11 isolates in the WA clade and 34 in the CB clade. Geographically, the WA clade isolates correspond to those cases reported from Nigeria, Ghana, Liberia, Cote d’Ivoire, and Sierra Leone, including the four isolates from outbreaks in captive animals; while the CB clade includes all isolates from DRC, Republic of the Congo, Cameroon, and Gabon. Although four polytomies resulted from the phylogenetic analysis, node support was very high (0.99–1.0) throughout most of the tree except for the polytomy that includes groups I-IV, which has support of 0.868. Eight isolates from Lomela Health Zone are included in a polytomy within group III (JX878413, JX878414, JX878415, JX878416, JX878421, JX878422, JX878427, and JX878428), with an average distance between them of 0.002%. Comparison of within clade patristic distances showed that branches in the CB clade are significantly shorter than branches within the WA clade (Shapiro-Wilk p < 0.001 in both clades, F-test p = 0.001, and t-test [unequal variance] p < 0.001). The two Nigerian isolates group together in spite of having high genetic differentiation between them and are from a group that is sister to the one formed by the remaining isolates of the clade; in addition, all isolates obtained from captive animals form a single monophyletic group.
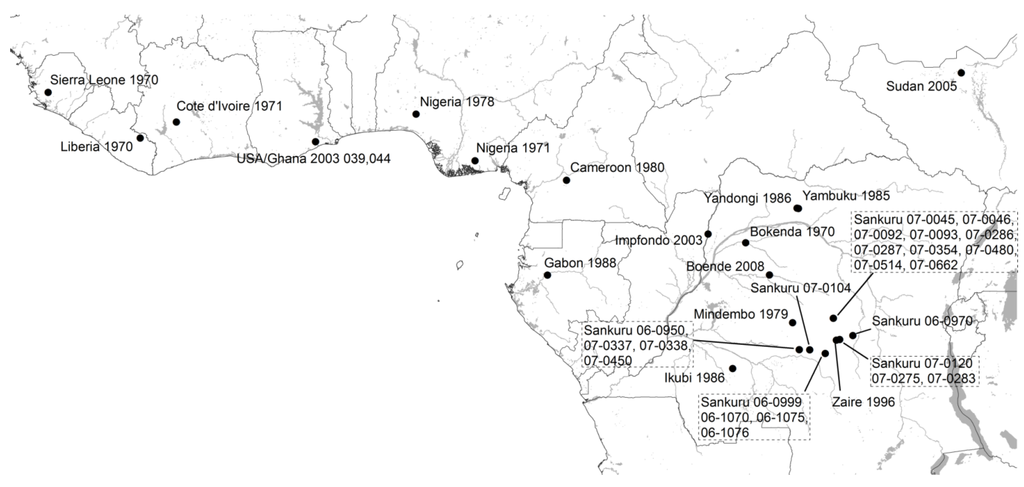
Figure 1.
Isolates included in the phylogenetic analysis (black circles). Isolates from Kugelman et al. [31] are mapped to the centroid of the Health Zone where they were reported. Isolates from Copenhagen, Walter Reed, Paris, and Rotterdam are not mapped because their African origin is unknown. Isolate from Sudan is shown at the outbreak locality, but is thought to have been imported from Northern DRC.
Within the CB clade, five monophyletic groups were formed after the phylogenetic analysis: group I includes the two western most isolates from this clade: Cameroon 1989 and Gabon 1987; group II contains 11 isolates, five from Sankuru District (JX878407, JX878423-25, and JX878429), one from South Sudan (Sudan 2005), one from the Republic of the Congo (Impfondo 2003), and four from other parts of DRC (Boende 2008, Bokenda 1970, Yandongi 1986, and Mindembo 1979); group III is the closest sister group to group II, twelve of the thirteen isolates in this group are from Sankuru District (JX878409-16, JX878421-22, JX878427-28), the remaining one is from Yambuku; six of the seven isolates in group IV are from Sankuru District (JX878408, JX878426, JX878417-20, and Sankuru 1996), the remaining one is Ikubi; group V only has one isolate from Sankuru District (JX878417). The groups are indicated in Figure 2 and Table 1.
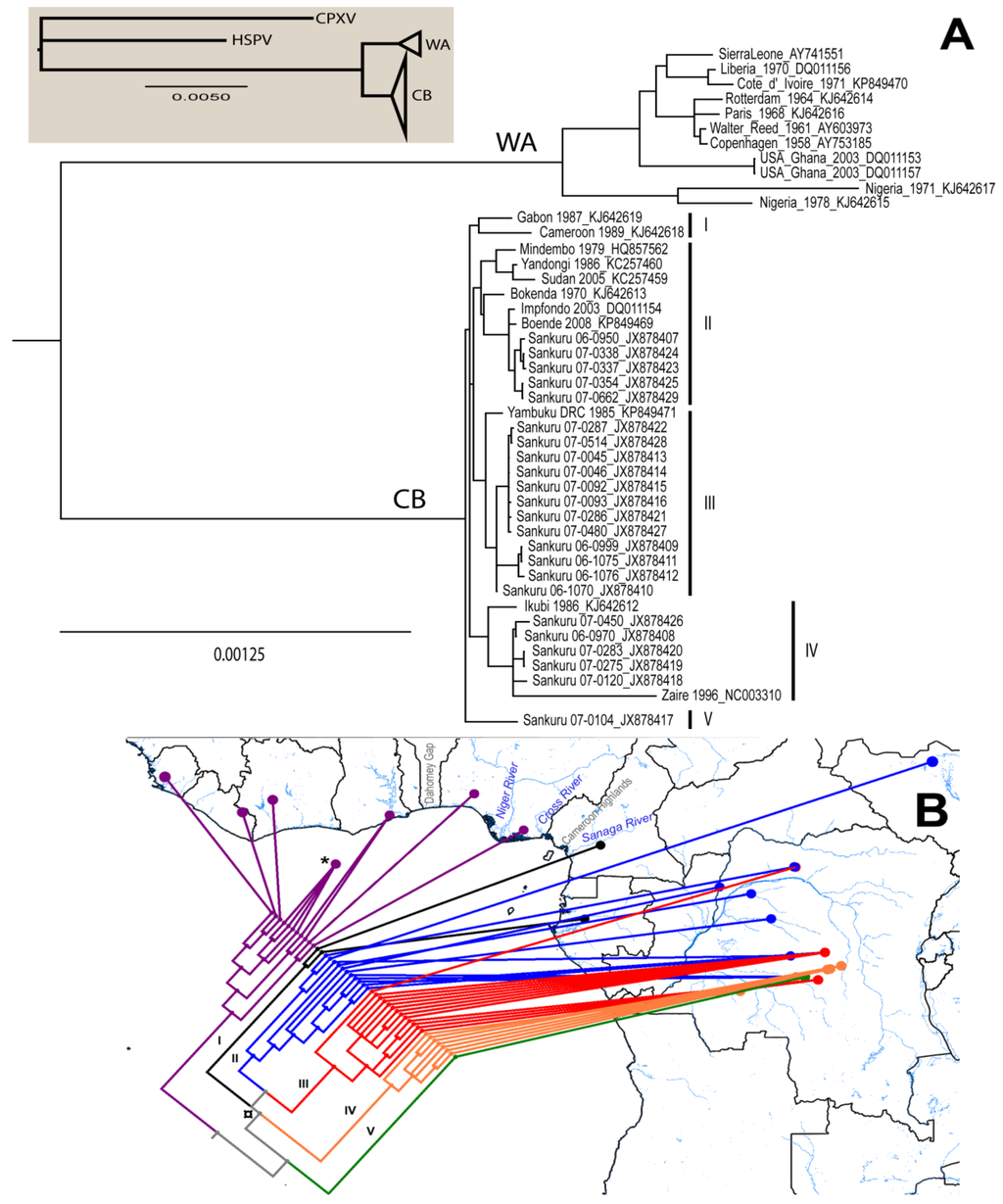
Figure 2.
(A) Majority-rules consensus tree. Clade credibility values for all nodes are 0.99–1.0, except from one polytomy indicated in the text (¤). Branch lengths are shown to scale. CB = Congo Basin Monkeypox clade; WA = West African Monkeypox clade. Groups are indicated by the roman number in parenthesis; (B) Map showing major geographic features of the area and the distribution of isolates used in the phylogenetic analysis, colors indicate groups: I = black, II = blue, III = red, IV = orange, V = green, and WA = purple. Isolates from Copenhagen, Walter Reed, Paris and Rotterdam are mapped to a point in the sea indicated with an asterisk (*) because their African origin is unknown.

Table 1.
Isolates used in the phylogenetic analysis, locality of report, publication source and accession number. ** = Used by Likos et al. [13]. ++ = Used in Kugelman et al. [31]. Roman numerals in parenthesis after the isolate name indicate the Congo Basin group to which they belong based on the phylogenetic analysis.
| Isolate Name | Location | Source | Accession # |
|---|---|---|---|
| Ikubi 1986 (IV) | Ikubi, DRC (Zaire) | [4] | KJ642612 |
| **++ Zaire 1996 (IV) | Akungula, DRC (Zaire) | [5] | NC_003310 |
| Yambuku DRC 1985 (III) | Yambuku, DRC (Zaire) | [56] | KP849471 |
| **++ Mindembo 1979 (II) | Mindembo, DRC (Zaire) | [3] | HQ857562 |
| Yandongi 1986 (II) | Yandongi, DRC | [4] | KC257460 |
| Bokenda 1970 (II) | Bokenda, DRC (Zaire) | [1] | KJ642613 |
| Boende 2008 (II) | Boende, DRC | Unpublished | KP849469 |
| **++ Impfondo 2003 (II) | Impfondo, ROC | [6] | DQ011154 |
| Sudan 2005 (II) | Nuria, South Sudan | [57] | KC257459 |
| Cameroon 1989 (I) | Ekoumdouma, Cameroon | [58] | KJ642618 |
| Gabon 1987 (I) | Gabon | [59] | KJ642619 |
| Nigeria 1971 | Ihie, Nigeria | [16] | KJ642617 |
| Nigeria 1978 | Omifunfun, Nigeria | [3] | KJ642615 |
| ++ USA/Ghana 2003_039 | Ghana | [7] | DQ011157 |
| **++ USA/Ghana 2003_044 | Ghana | [7] | DQ011153 |
| ++ Copenhagen 1958 | Copenhagen | [60] | AY753185 |
| ++ Walter Reed 1961 | Walter Reed | [61] | AY603973 |
| Paris 1968 | Paris | [62] | KJ642616 |
| Rotterdam 1965 | Rotterdam | [63] | KJ642614 |
| Cote d’Ivoire 1971 | Cote d’Ivoire | [64] | KP849470 |
| **++ Liberia 1970 | Liberia | [16] | DQ011156 |
| ++ Sierra Leone 1970 | Sierra Leone | [16] | AY741551 |
| ++ Sankuru 06-0950 (II) | Kole Health Zone, DRC | [31] | JX878407 |
| ++ Sankuru 07-0337 (II) | Kole Health Zone, DRC | [31] | JX878423 |
| ++ Sankuru 07-0338 (II) | Kole Health Zone, DRC | [31] | JX878424 |
| ++ Sankuru 07-0450 (IV) | Kole Health Zone, DRC | [31] | JX878426 |
| ++ Sankuru 06-0999 (III) | Vangakete Health Zone, DRC | [31] | JX878409 |
| ++ Sankuru 06-1075 (III) | Vangakete Health Zone, DRC | [31] | JX878411 |
| ++ Sankuru 06-1076 (III) | Vangakete Health Zone, DRC | [31] | JX878412 |
| ++ Sankuru 06-1070 (III) | Vangakete Health Zone, DRC | [31] | JX878410 |
| ++ Sankuru 07-0045 (III) | Lomela Health Zone, DRC | [31] | JX878413 |
| ++ Sankuru 07-0046 (III) | Lomela Health Zone, DRC | [31] | JX878414 |
| ++ Sankuru 07-0092 (III) | Lomela Health Zone, DRC | [31] | JX878415 |
| ++ Sankuru 07-0093 (III) | Lomela Health Zone, DRC | [31] | JX878416 |
| ++ Sankuru 07-0286 (III) | Lomela Health Zone, DRC | [31] | JX878421 |
| ++ Sankuru 07-0480 (III) | Lomela Health Zone, DRC | [31] | JX878427 |
| ++ Sankuru 07-0514 (III) | Lomela Health Zone, DRC | [31] | JX878428 |
| ++ Sankuru 07-0287 (III) | Lomela Health Zone, DRC | [31] | JX878422 |
| ++ Sankuru 07-0354 (II) | Lomela Health Zone, DRC | [31] | JX878425 |
| ++ Sankuru 07-0662 (II) | Lomela Health Zone, DRC | [31] | JX878429 |
| ++ Sankuru 06-0970 (IV) | Katako Kombe Health Zone, DRC | [31] | JX878408 |
| ++Sankuru 07-0120 (IV) | Djalo-Ndjeka Health Zone, DRC | [31] | JX878418 |
| ++Sankuru 07-0275 (IV) | Djalo-Ndjeka Health Zone, DRC | [31] | JX878419 |
| ++Sankuru 07-0283 (IV) | Djalo-Ndjeka Health Zone, DRC | [31] | JX878420 |
| ++Sankuru 07-0104 (V) | Bena-Dibele Health Zone, DRC | [31] | JX878417 |
3.2. Ecological Niche Model
ENM projections into present day environmental conditions are similar to models presented in previous works [25,39] and identify areas in Central and Western Africa in which MPX is known to occur. Models based on each of the 25 locality subsets performed better than random expectations with modified ROC AUC values between 1.31 and 1.42 for GARP (average = 1.37, standard deviation = 0.03); and between 1.33 and 1.48 for Maxent (average = 1.42, standard deviation 0.04). These models are able to capture those environmental conditions that are suitable for MPX transmission with high confidence. ENM maps presented some differences: (a) GARP models predicted a larger area of suitable environmental conditions in Cameroon and Gabon than the models produced by Maxent; (b) Maxent models show a more uniform distribution of suitable conditions in West Africa, while GARP shows fragmentation; and (c) GARP models show an area of low model agreement in the middle of the Congo River Basin (Figure 3A,B).
GARP model projections onto climatic conditions during the Mid-Holocene show more connection between areas with suitable environments for MPXV in West Africa and also between those suitable areas in Nigeria and Central Africa; the most evident discontinuity of suitable areas separates such areas in costal Nigeria from the rest of West Africa and is located in Benin (Figure 3C). Maxent, however, shows a noticeable reduction of suitable areas in West Africa concentrated in the coasts of Nigeria, Togo and Benin with some smaller suitable areas in Sierra Leone, Liberia and Ghana (Figure 3D). ENM projections onto environmental conditions during the LGM with both algorithms identified smaller areas with suitable environments for MPX transmission. The area with highest model agreement for Maxent is located in western DRC with a few other areas with lower model agreement within this country; however, only few small areas in West Africa were identified as suitable by a few models (Figure 3F). Areas of high model agreement for GARP are also restricted to small areas in northern and western DRC, as well as along the coast throughout West Africa (Figure 3E).
Finally, ENM projections onto environmental conditions during the LIG identified a larger and more continuous area of model agreement for both algorithms than that for the LGM located in central Africa with a few small patches along the coast in West Africa for GARP and Gabon for Maxent (Figure 3G,H).
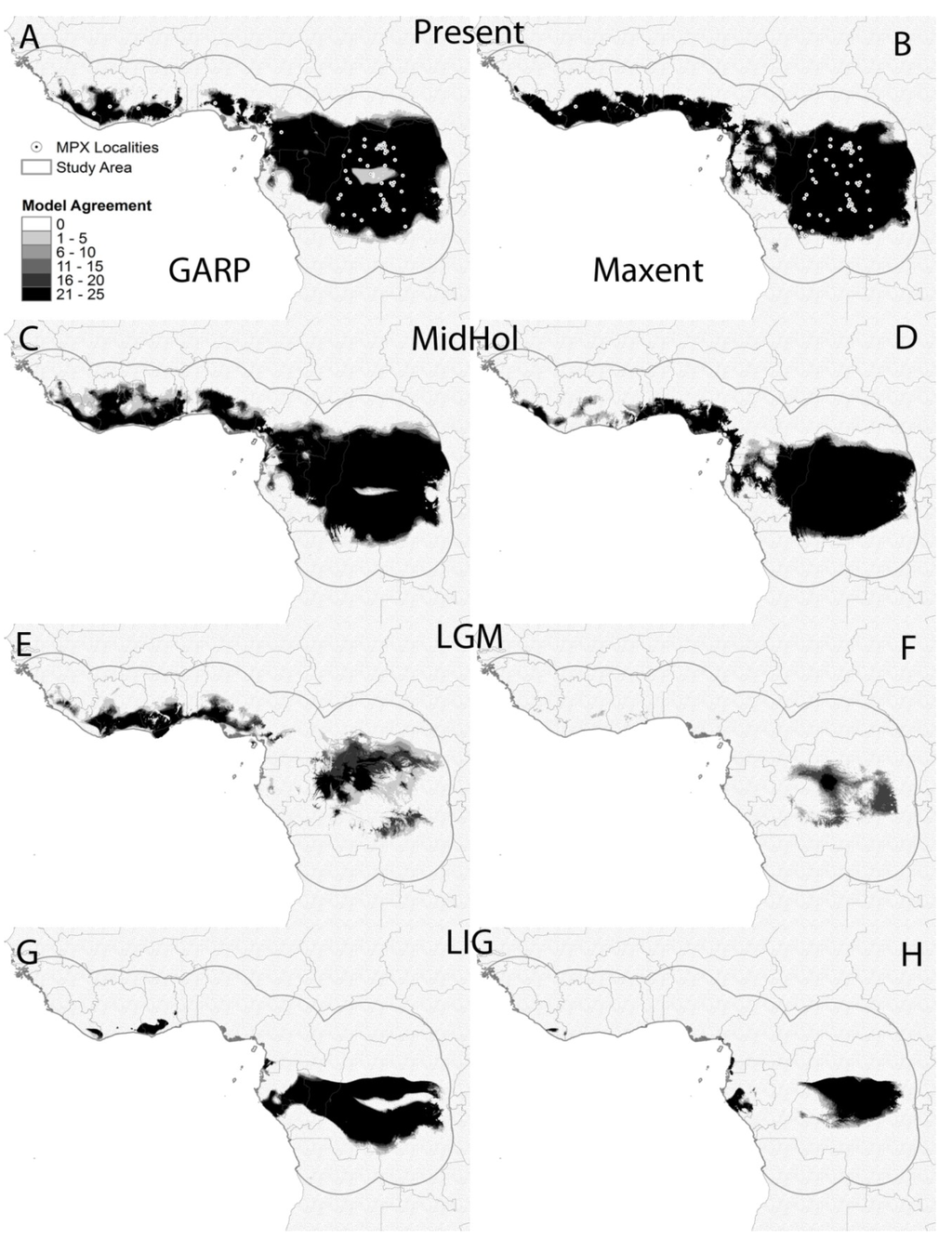
Figure 3.
Model agreement maps from GARP (left column) and Maxent (right column) based on the 25 locality subsets; darker shade = higher agreement. Models projected onto present day environmental conditions (A,B), Mid-Holocene (MidHol: C,D), Last Glacial Maximum (LGM: E,F), and the Last Interglacial period (LIG: G and H). White circles represent MPX localities used for model development.
4. Discussion
Our phylogenetic analysis separates the MPXV isolates into two major groups that correspond to the previously identified WA and CB clades. The eastern most isolate of the WA clade (Ihie, Nigeria) is between the Niger River (west) and the Cross River (east); while the western most isolate of the CB clade (Ekondouma, Cameroon) is south of the Sanaga River (Figure 2B). These rivers (Cross and Sanaga) have been identified as biogeographic barriers for mammal species such as chimpanzees of the genus Pan [65], flying squirrels [66], and mice of the genus Praomys [67]. The two isolates from Nigeria are on opposite sides of the Niger River and they are more genetically divergent than other pairs of isolates from the West African clade, adding support to the idea that rivers could play a role in the differentiation of MPXV.
The Cameroon Highlands are also located between the Cross and Sanaga Rivers; they are recognized as a high biodiversity ecoregion where the dominant vegetation types are tropical and subtropical moist broadleaf forest [68], representing a change in altitude and dominant vegetation coverage from the lowland evergreen forest of the Congo Basin. Although the presence of these geographic features suggest their involvement on the genetic divergence of the two MPXV clades; with the data available to this point, it is not possible to determine which features are currently acting or previously acted as dispersal barriers for MPXV or its reservoir species.
The Dahomey Gap is a savanna corridor that interrupts the West African rain forest in Togo, Benin, and Eastern Ghana [69], which has been hypothesized to act as a barrier to dispersal of mammals [70,71]. Given that most human MPX cases have been associated with the rainforest [3,72,73], we would expect the Dahomey Gap to be a dispersal barrier for MPXV because the dominant land cover and climatic conditions would not be suitable for the reservoir or transmission of the virus based on ENMs. Results of phylogenetic analyses support this by revealing separate groups for isolates located west (Ghana, Cote d’Ivoire, Liberia, and Sierra Leone) and east of the Dahomey Gap (Nigeria). Palynological analyses indicate that an abrupt climatic change into drier conditions favored the establishment of the savanna in this area starting around 4500 years ago [69].
Consistent with the Pleistocene refuge theory [74,75], ecological niche models predicted smaller areas with suitable conditions for MPXV transmission during the LGM (21,000 years ago), especially in the Congo Basin, representing a potential bottleneck for MPXV in the Congo basin. The posterior expansion of the rainforest driven by warmer and more humid conditions in the area, may have allowed MPXV and/or its reservoir(s) to also expand its geographic range, potentially leading to a rapid diversification of the virus, as shown by the relatively short branches of the CB clade and the polytomy that includes groups I to IV in the phylogenetic tree (Figure 2A). As the niche model of MPXV in Western Africa during LGM was less restrictive, it would be expected to maintain a larger portion of its genetic variation.
In the present study, we first identified potential biogeographic barriers for MPXV that could be related to the CB-WA split. Further studies are necessary to determine whether the presence of a river, change in elevation, or change in the dominant vegetation cover is involved in the genetic differentiation of MPXV. The addition of MPXV isolates from the area between the Sanaga and Cross rivers would be ideal; however, cases of human or wildlife MPX have not been reported from this area. Second, we propose that the CB clade is a group with very recent diversification, possibly explained by the colonization of a bigger area with suitable conditions (refuge theory); however, dating the times of differentiation between and within clades is not possible with our current dataset. Tying MPXV cladogenesis to geologic or climatic events is a subject of future efforts. Additional field studies that result in the isolation of MPXV or the finding of serological evidence of infection with this virus in wildlife could be key to better understanding its natural history and biogeography.
Acknowledgments
The authors thank the Emory University Core facility for assistance with genomic sequencing and Chris Upton and Nick Tang in the Department of Biochemistry and Microbiology at the University of Victoria for their assistance with annotation and submission of genomes. Isolation of MPXV from recent DRC cases was the result of research and surveillance efforts led by the Kinshasa School of Public Health, the Institut National de Recherche Biomédicale (INRB), and the Department of Biology at the University of Kinshasa in DRC. Annotation was performed with tools at the Viral Bioinformatics Resource Center (www.virology.ca). This research was supported in part by the appointment of MRM to the Research Participation Program at the CDC, administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the US Department of Energy and the CDC.
Author Contributions
Y.N., M.R.M. and G.L.E. performed phylogenetic and ecological niche model analyses; MGR, RRL and YN compiled can cleaned the locality database of monkeypox cases for their use in ecological niche models; M.G.R., J.-J.M., P.M.K., O.W. and J.M. obtained specimens from monkeypox cases and obtained isolates. Y.L., J.G. and H.Z. completed the sequences of the new isolates included in this work; Y.N., M.R.M., G.L.E., K.L.K., I.K.D. and D.S.C. designed the study, analyzed data and wrote the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Disclaimer
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
References
- Ladnyj, I.D.; Ziegler, P.; Kima, E. A human infection caused by monkeypox virus in Basankusu Territory, Democratic Republic of the Congo. Bull. World Health Organ. 1972, 46, 593–597. [Google Scholar] [PubMed]
- Jezek, Z.; Marennikova, S.S.; Mutumbo, M.; Nakano, J.H.; Paluku, K.M.; Szczeniowski, M. Human monkeypox: A study of 2,510 contacts of 214 patients. J. Infect. Dis. 1986, 154, 551–555. [Google Scholar] [CrossRef] [PubMed]
- Breman, J.G.; Ruti, K.; Steniowski, M.V.; Zanotto, E.; Gromyko, A.I.; Arita, I. Human Monkeypox, 1970–79. Bull. World Health Organ. 1980, 58, 165–182. [Google Scholar] [PubMed]
- Jezek, Z. Smallpox and its post-eradication surveillance. Bull. World. Health. Organ. 1987, 65, 425–434. [Google Scholar] [PubMed]
- Hutin, Y.J.F.; Williams, R.J.; Mailfait, P.; Pebody, R.; Loparev, V.N.; Ropp, S.L.; Rodriguez, M.; Knight, J.C.; Tshioko, F.K.; Khan, A.S.; et al. Outbreak of human monkeypox, Democratic Republic of Congo, 1996–1997. Emerg. Infect. Dis. 2001, 7, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Learned, L.A.; Reynolds, M.G.; Wassa-Wassa, D.; Li, Y.; Olson, V.A.; Karem, K.L.; Stempora, L.L.; Braden, Z.H.; Kline, R.; Likos, A.M.; et al. Extended interhuman transmission of monkeypox in a hospital community in the Republic of the Congo, 2003. Am. J. Trop. Med. Hyg. 2005, 73, 428–434. [Google Scholar] [PubMed]
- Reed, K.D.; Melski, J.W.; Graham, M.B.; Regnery, R.L.; Sotir, M.J.; Wegner, M.V.; Kazmierczak, J.J.; Stratman, E.J.; Li, Y.; Fairley, J.A.; et al. The detection of monkeypox in humans in the Western Hemisphere. N. Engl. J. Med. 2004, 350, 342–350. [Google Scholar] [CrossRef] [PubMed]
- MacNeil, A.; Reynolds, M.G.; Carroll, D.S.; Karem, K.L.; Braden, Z.; Lash, R.R.; Moundeli, A.; Mombouli, J.-V.; Jumaan, A.O.; Schmid, D.S.; et al. Monkeypox or varicella? Lessons from a rash outbreak investigation in the Republic of the Congo. Am. J. Trop. Med. Hyg. 2009, 80, 503–507. [Google Scholar] [PubMed]
- Meyer, H.; Perrichot, M.; Stemmler, M.; Emmerich, P.; Schmitz, H.; Varaine, F.; Shungu, R.; Tshioko, F.; Formenty, P. Outbreaks of disease suspected of being due to monkeypox virus infection in the Democratic Republic of Congo in 2001. J. Clin. Microbiol. 2002, 40, 2919–2921. [Google Scholar] [CrossRef] [PubMed]
- Bass, J.; Tack, D.M.; McCollum, A.M.; Kabamba, J.; Pakuta, E.; Malekani, J.; Nguete, B.; Monroe, B.P.; Doty, J.B.; Karhemere, S.; et al. Enhancing health care worker ability to detect and care for patients with monkeypox in the Democratic Republic of the Congo. Int. Health 2013, 5, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Rimoin, A.W.; Kisalu, N.; Kebela-Ilunga, B.; Mukaba, T.; Wright, L.L.; Formenty, P.; Wolfe, N.D.; Shongo, R.L.; Tshioko, F.; Okitolonda, E.; et al. Endemic human monkeypox, Democratic Republic of Congo, 2001–2004. Emerg. Infect. Dis. 2007, 13, 934–937. [Google Scholar] [CrossRef] [PubMed]
- Fuller, T.; Thomassen, H.A.; Mulembakani, P.M.; Johnston, S.C.; Lloyd-Smith, J.O.; Kisalu, N.K.; Lutete, T.K.; Blumberg, S.; Fair, J.N.; Wolfe, N.D.; et al. Using remote sensing to map the risk of human monkeypox virus in the Congo Basin. EcoHealth 2011, 8, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Likos, A.M.; Sammons, S.A.; Olson, V.A.; France, M.A.; Li, Y.; Olsen-Rasmussen, M.A.; Davidson, W.; Galloway, R.; Khristova, M.L.; Reynolds, M.G.; et al. A tale of two clades: Monkeypox viruses. J. Gen. Virol. 2005, 86, 2661–2672. [Google Scholar] [CrossRef] [PubMed]
- Esposito, J.J.; Knight, J.C. Orthopoxvirus DNA: A comparison of restriction profiles and maps. Virology 1985, 143, 230–251. [Google Scholar] [CrossRef] [PubMed]
- Mackett, M.; Archard, L.C. Conservation and variation in orthopoxvirus genome structure. J. Gen. Virol. 1979, 45, 683–701. [Google Scholar] [CrossRef] [PubMed]
- Foster, S.O.; Brink, E.W.; Hutchins, D.L.; Pifer, J.M.; Lourie, B.; Moser, C.R.; Cummings, E.C.; Kuteyi, O.E.K.; Eke, R.E.A.; Titus, J.B.; et al. Human Monkeypox. Bull. World. Health. Organ. 1972, 46, 569–576. [Google Scholar] [PubMed]
- Waltari, E.; Hijmans, R.J.; Peterson, A.T.; Nyári, Á.S.; Perkins, S.L.; Guralnick, R. Locating Pleistocene refugia: Comparing phylogeographic and ecological niche model predictions. PLoS ONE 2007, 2, e563. [Google Scholar] [CrossRef]
- Bonaccorso, E.; Koch, I.; Peterson, A.T. Pleistocene fragmentation of Amazon species’ ranges. Divers. Distrib. 2006, 12, 157–164. [Google Scholar] [CrossRef]
- Svenning, J.-C.; Fløjgaard, C.; Marske, K.A.; Nogués-Bravo, D.; Normand, S. Applications of species distribution modeling to paleobiology. Quaternary Sci. Rev. 2011, 30, 2930–2947. [Google Scholar] [CrossRef]
- Lima-Ribeiro, M.S.; Diniz-Filho, J.A.F. Modelando a distribuição geográfica das espécies no passado: Uma abordagem promissora em paleoecologia. Rev. Brasileira Paleontol. 2012, 15, 371–385. [Google Scholar] [CrossRef]
- Varela, S.; Lobo, J.M.; Rodríguez, J.; Batra, P. Were the Late Pleistocene climatic changes responsible for the disappearance of the European spotted hyena populations? Hindcasting a spcies geographic distributions across time. Quat. Sci. Rev. 2010, 29, 2027–2035. [Google Scholar] [CrossRef]
- González, C.; Wang, O.; Strutz, S.E.; González-Salazar, C.; Sánchez-Cordero, V.; Sarkar, S. Climate change and risk of leishmaniasis in North America: Predictions from ecological niche models of vector and reservoir species. PLoS Negl. Trop. Dis. 2010, 4, e585. [Google Scholar] [CrossRef] [PubMed]
- Holt, A.C.; Sealkeld, D.J.; Fritz, C.L.; Tucker, J.R.; Gong, P. Spatial analysis of plague in California: Niche modeling predictions of the current distribution and potential response to climate change. Int. J. Health Geogr. 2009, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Komar, O.; Robbins, M.B.; Klenk, K.; Blitvich, B.J.; Marlenee, N.L.; Burkhalter, K.L.; Gubler, D.J.; Gonzálvez, G.; Peña, C.J.; Peterson, A.T.; et al. West Nile virus transmission in resident birds, Dominican Republic. Emerg. Infect. Dis. 2003, 9, 1299–1302. [Google Scholar] [CrossRef] [PubMed]
- Ellis, C.K.; Carroll, D.S.; Lash, R.R.; Peterson, A.T.; Damon, I.K.; Malekani, J.M.; Formenty, P. Ecology and geography of human monkeypox case occurrences across Africa. J. Wildl. Dis. 2012, 48, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, Y.; Williams, R.; Peterson, A.T.; Mead, P.; Staples, E.; Gage, K.L. Climate change effects on plague and tularemia in the United States. Vector-Borne Zoonotic Dis. 2007, 7, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Haffer, J. Speciation in Amazonian forest birds. Science 1969, 195, 131–137. [Google Scholar] [CrossRef]
- Haffer, J. Alternative models of vertebrate speciation in Amazonia: An overview. Biodivers. Conserv. 1997, 6, 451–476. [Google Scholar] [CrossRef]
- Endler, J. Pleistocene forest refuges: Fact or fancy? In Biological Diversification in the Tropics; Prance, G.T., Ed.; Columbia University Press: New York, NY, USA, 1982; pp. 179–200. [Google Scholar]
- Ježek, Z.; Fenner, F. Human Monkeypox; Karger: New York, NY, USA, 1988. [Google Scholar]
- Kugelman, J.R.; Johnson, S.C.; Mulembakani, P.M.; Kisalu, N.; Lee, M.S.; Koroleva, G.; McCarthy, S.E.; Gestole, M.C.; Wolfe, N.D.; Fair, J.N.; et al. Genomic variability of monkeypox virus among humans, Democratic Republic of the Congo. Emerg. Infect. Dis. 2014, 20, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008, 9, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F.; Nielsen, R.; Bollback, J.P. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 2001, 294, 2310–2314. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A language and Environment for Statistical Computing; R Foundation for Statistical Computing, v3.1.1: Vienna, Austria, 2011. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Valverde, A.; Nakazawa, Y.; Lira-Noriega, A.; Peterson, A.T. Environmental correlation structure and ecological niche model projections. Biodivers. Inf. 2009, 6, 28–35. [Google Scholar]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Lash, R.R.; Carroll, D.S.; Hughes, C.M.; Nakazawa, Y.; Karem, K.L.; Damon, I.K.; Peterson, A.T. Effects of georeferencing effort on mapping monkeypox case distributions and transmission risk. Int. J. Health Geogr. 2012, 11, 23. [Google Scholar] [CrossRef] [PubMed]
- Otto-Bliesner, B.L.; Marshall, S.J.; Overpeck, J.T.; Miller, G.H.; Hu, A. CAPE Last Interglacial Project members, Simulating arctic climate warmth and icefield retreat in the Last Interglaciation. Science 2006, 311, 1751–1753. [Google Scholar] [CrossRef] [PubMed]
- Chapman, A.D.; Wieczorek, J. Guide to Best Practices for Georeferencing; Global Biodiversity Information Facility: Copenhagen, Denmark, 2006. [Google Scholar]
- Peterson, A.T.; Sánchez-Cordero, V.; Beard, C.B.; Ramsey, J.M. Ecologic niche modeling and potential reservoirs for Chagas disease, Mexico. Emerg. Infect. Dis. 2002, 8, 662–667. [Google Scholar] [CrossRef] [PubMed]
- Peterson, A.T.; Martínez-Campos, C.; Nakazawa, Y.; Martínez-Meyer, E. Time-specific ecological niche modeling predicts spatial dynamics of vector insects and human dengue cases. Trans. R. Soc. Trop. Med. Hyg. 2005, 99, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Peterson, A.T.; Bauer, J.T.; Mills, J.N. Ecologic and geographic distribution of filovirus disease. Emerg. Infect. Dis. 2004, 10, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, Y.; Williams, R.A.J.; Peterson, A.T.; Mead, P.S.; Kugeler, K.J.; Petersen, J.M. Ecological niche modeling of Francisella tularensis subspecies and clades in the United States. Am. J. Trop. Med. Hyg. 2010, 82, 912–918. [Google Scholar] [CrossRef] [PubMed]
- Peterson, A.T.; Benz, B.W.; Papeş, M. Highly pathogenic H5N1 avian influenza: Entry pathways into North America via bird migration. PLoS ONE 2007, 2, e261. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.A.J.; Peterson, A.T. Ecology and geography of avian influenza (HPAI H5N1) transmission in the Middle East and northeastern Africa. Int. J. Health Geogr. 2009, 8, 47. [Google Scholar] [CrossRef] [PubMed]
- Elith, J.; Graham, C.H.; Anderson, R.P.; Dudik, M.; Ferrier, S.; Guisan, A.; Hijmans, R.J.; Huettman, F.; Leathwick, J.R.; Lehmann, A.; et al. Novel methods improve prediction of species' distributions from occurrence data. Ecography 2006, 29, 129–151. [Google Scholar] [CrossRef]
- Peterson, A.T.; Papeş, M.; Eaton, M. Transferability and model evaluation in ecological niche modeling: A comparison of GARP and Maxent. Ecography 2007, 30, 550–560. [Google Scholar] [CrossRef]
- Stockwell, D. The GARP modelling system: Problems and solutions to automated spatial prediction. Int. J. Geogr. Inf. Sci. 1999, 13, 143–158. [Google Scholar] [CrossRef]
- Stockwell, D.R.B.; Noble, I.R. Introduction of sets of rules from animal distribution data: A robust and informative method of data analysis. Math. Comput. Simulat. 1992, 33, 385–390. [Google Scholar] [CrossRef]
- Anderson, R.P.; Lew, D.; Peterson, A.T. Evaluating predictive models of speciesʼ distributions: Criteria for selecting optimal models. Ecol. Model. 2003, 162, 211–232. [Google Scholar] [CrossRef]
- Peterson, A.T.; Papeş, M.; Soberón, J. Rethinking receiver operating characteristic analysis applications in ecological niche modeling. Ecol. Model. 2008, 213, 63–72. [Google Scholar] [CrossRef]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- Phillips, S.J.; Dudik, M.; Schapire, R.E. A maximum Entropy Approach to Species Distribution Modeling. In Proceedings of the 21st International Conference on Machine Learning, Banff, Canada, July 2004.
- Khodakevich, L.; Ježek, Z.; Kinzanzka, K. Isolation of monkeypox virus from wild squirrel infected in nature. Lancet 1986, 1, 98–99. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, Y.; Emerson, G.L.; Zhao, H.; Li, Y.; Carroll, D.S.; Reynolds, M.G.; Karem, K.L.; Olson, V.A.; Lash, R.R.; Davidson, W.B.; et al. Phylogenetic and ecologic perspectives of a monkeypox outbreak, southern Sudan, 2005. Emerg. Infect. Dis. 2013, 19, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Tchokoteu, P.F.; Kago, I.; Tetanye, E.; Ndoumbe, P.; Pignon, D.; Mbede, J. Variola or a severe case of varicella? A case of human variola due to monkeypox virus in a child from the Cameroon. Ann. Soc. Belg. Med. Trop. 1991, 71, 123–128. [Google Scholar] [PubMed]
- Müller, G.; Meyer, A.; Gras, F.; Emmerich, P.; Kolakowski, T.; Esposito, J.J. Monkeypox virus in liver and spleen of child in Gabon. Lancet 1988, 331, 769–770. [Google Scholar] [CrossRef]
- Arita, I.; Henderson, D.A. Smallpox and monkeypox in non-human primates. Bull. World Health Organ. 1968, 39, 277–283. [Google Scholar] [PubMed]
- McConnell, S.J.; Herman, Y.F.; Mattson, D.E.; Erickson, L. Monkey pox disease in irradiated cynomologous monkeys. Nature 1962, 195, 1128–1129. [Google Scholar] [CrossRef]
- Milhaud, C.; Klein, M.; Virat, J. Analyse dʼun cas de variole du singe (monkeypox) chez le chimpanzé (Pan troglogdytes). Exp. Anim. 1969, 2, 121–135. [Google Scholar]
- Peters, J.C. An epizootic of monkey pox at Rotterdam Zoo. Int. Zoo Yearb. 1966, 6, 274–275. [Google Scholar] [CrossRef]
- Arita, I.; Henderson, D.A. Monkeypox and whitepox viruses in West and Central Africa. Bull. World Health Organ. 1976, 53, 347–353. [Google Scholar] [PubMed]
- Gonder, M.K.; Disotell, T.R.; Oates, J.F. New genetic evidence on the evolution of chimpanzee populations and implications for taxinomy. Int. J. Primatol. 2006, 27, 1103–1127. [Google Scholar] [CrossRef]
- Schunke, A.C.; Hutterer, R. The variance of variation: Geographic patters of coat colouration in Anomalurops and Anomalurus (Mammalia, Rodentia, Anomalouridae). Bonn. Zool. Beitr. 2004, 53, 169–185. [Google Scholar]
- Nicolas, V.; Missoup, A.D.; Denys, C.; Kerbis Peterhans, J.; Katuala, P.; Couloux, A.; Colyn, M. The roles of rivers and Pleistocene refugia in shaping genetic diversity in Praomys misonnei in tropical Africa. J. Biogeogr. 2011, 38, 191–207. [Google Scholar] [CrossRef]
- Olson, D.M.; Dinerstein, E.; Wikramanayake, E.D.; Burgess, N.D.; Powell, G.V.N.; Underwood, E.C.; DʼAmico, J.; Itoua, I.; Strand, H.E.; Morrison, J.C.; et al. Terrestrial ecorregions of the world: A new map of life on Earth. Bioscience 2001, 51, 933–938. [Google Scholar] [CrossRef]
- Salzmann, U.; Hoelzmann, P. The Dahomey Gap: An abrupt climatically induced rain forest fragmentation in West Africa during the late Holocene. Holocene 2005, 15, 190–199. [Google Scholar] [CrossRef]
- Booth, A.H. The Niger, the Volta and teh Dahomey Gap as geographic barriers. Evolution 1958, 12, 48–62. [Google Scholar] [CrossRef]
- Nicolas, V.; Quérouil, S.; Verheyen, E.; Verheyen, W.; Mboumba, J.F.; Dillen, M.; Colyn, M. Mitochondrial phylogeny of African wood mice, genus Hylomyscus (Rodentia, Muridae): Implications for their taxonomy and biogeography. Mol. Phylogenet. Evol. 2006, 38, 779–793. [Google Scholar] [CrossRef] [PubMed]
- Rimoin, A.W.; Mulembakani, P.M.; Johnston, S.C.; Lloyd Smith, J.O.; Kisalu, N.K.; Kinkela, T.L.; Blumberg, S.; Thomassen, H.A.; Pike, B.L.; Fair, J.N.; et al. Major increase in human monkeypox incidence 30 years after smallpox vaccination campaigns cease in the Democratic Republic of Congo. Proc. Natl. Acad. Sci. USA 2010, 107, 16262–16267. [Google Scholar] [CrossRef] [PubMed]
- Jezek, Z.; Fenner, F. Human Monkeypox. In Monographs in Virology; S. Karger AG: Basel, Switzerland, 1988; Volume 14, pp. 1–140. [Google Scholar]
- Mayr, E.; O’Hara, R.J. The biogeographic evidence supporting the Pleistocene forest refuge hypothesis. Evolution 1986, 40, 55–67. [Google Scholar] [CrossRef]
- Plana, V. Mechanisms and tempo of evolution in the African Guineo-Congolian rainforest. Philos. Trans. R. Soc. Lond. B 2004, 359, 1585–1594. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).