The Phosphoglycerate Kinase (PGK) Gene Family of Maize (Zea mays var. B73)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of PGK-Encoding Genes in the Maize Genome
2.2. Bioinformatic Analysis
2.3. Modeling of PGK Protein Structure
2.4. Promoter Analysis
2.5. Phylogenetic Analysis
2.6. RNA Isolation and Reverse Transcription
2.7. Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
2.8. Enzyme Activity Measurements
3. Results
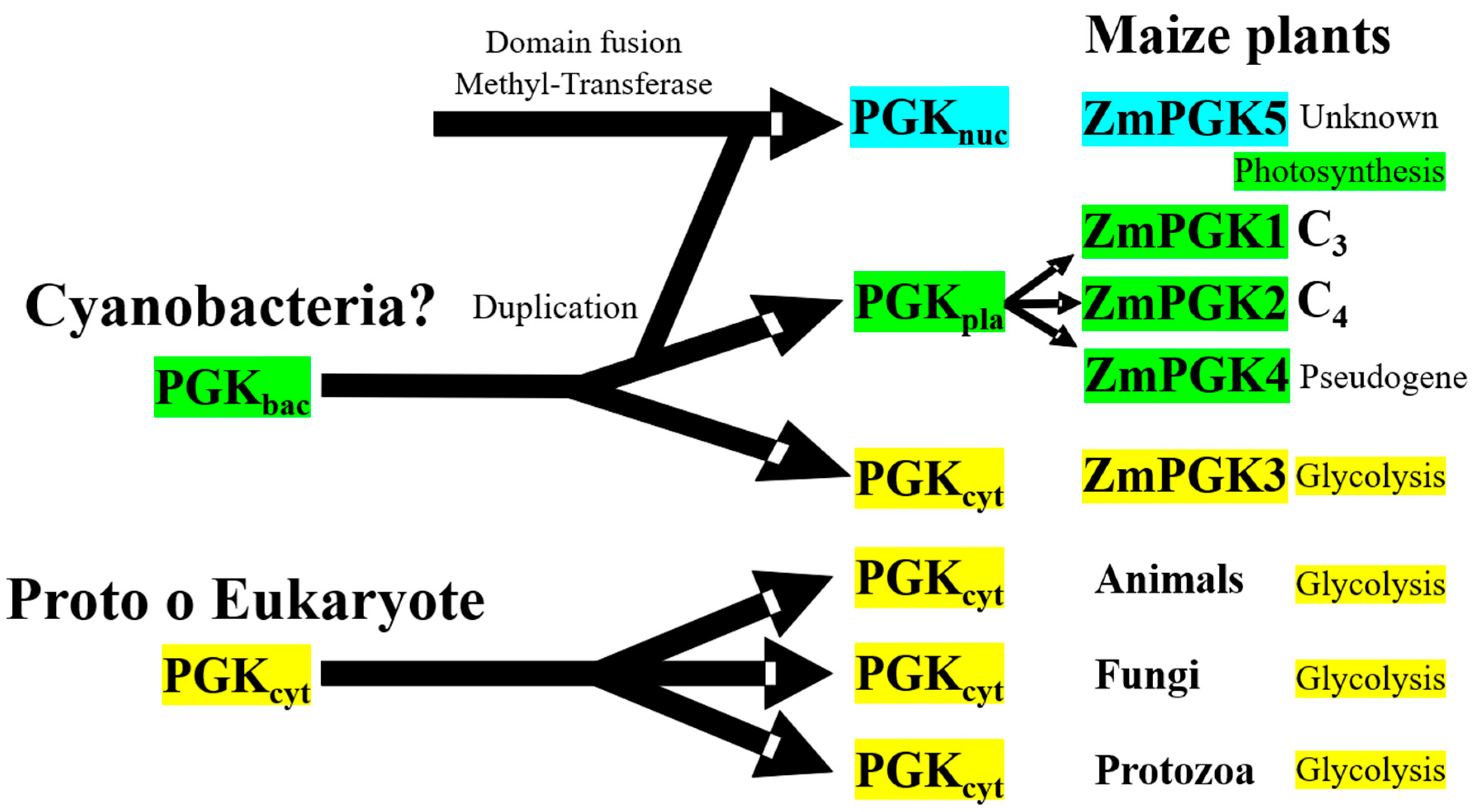
3.1. Identification of PGK Isozymes in Maize Genome (ZmPGKs)
3.2. Analysis of the ZmPGK Protein Sequences
3.3. In Silico Structural ZmPGK Models and Possible Interactions with Their Substrates
3.4. Differential Expression of ZmPgk Genes
3.5. Identification of Cis-Regulatory Elements Correlated with Tissue-Specific Expression of ZmPGKs
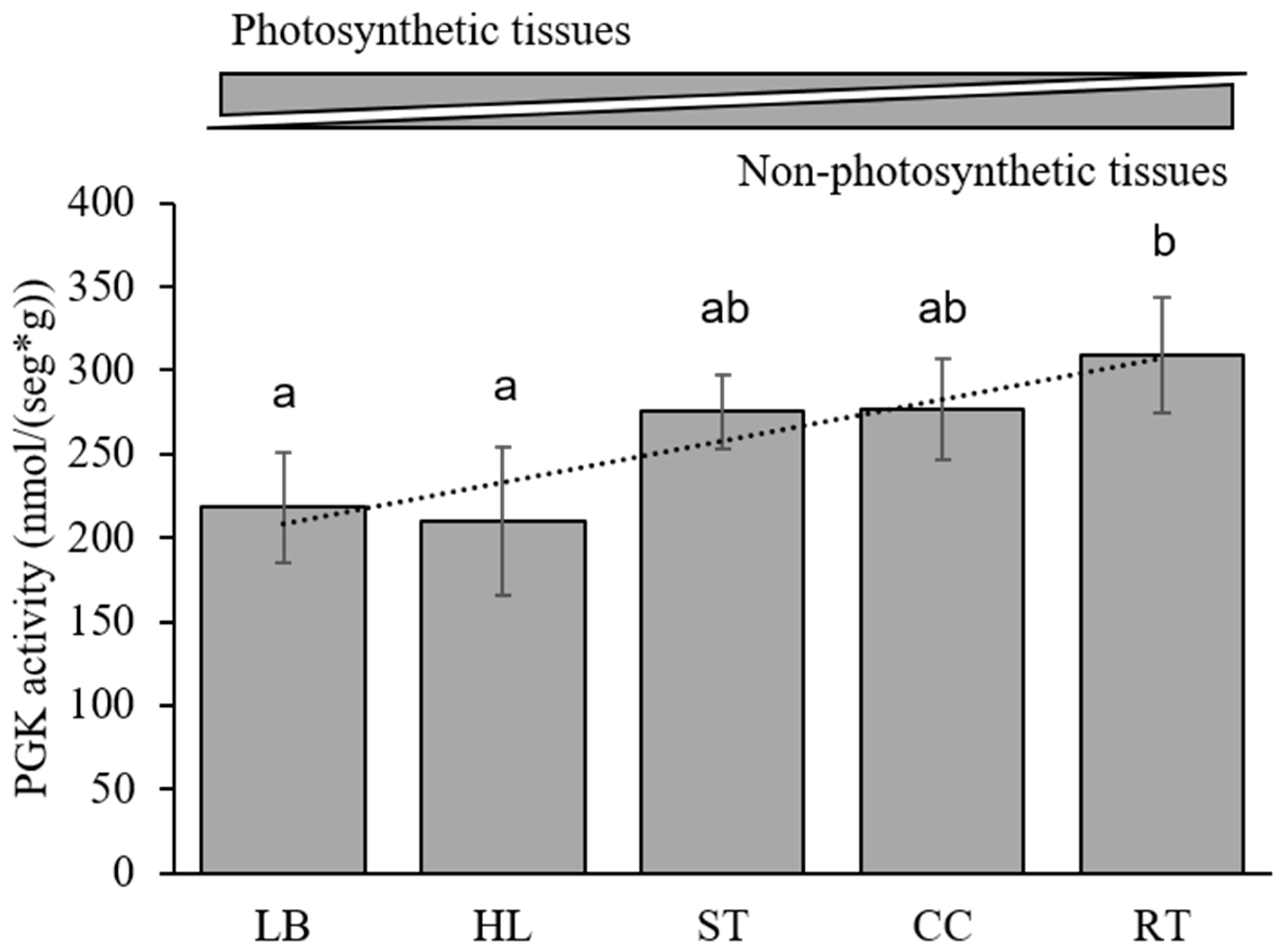
3.6. Kinetic Analysis in Maize
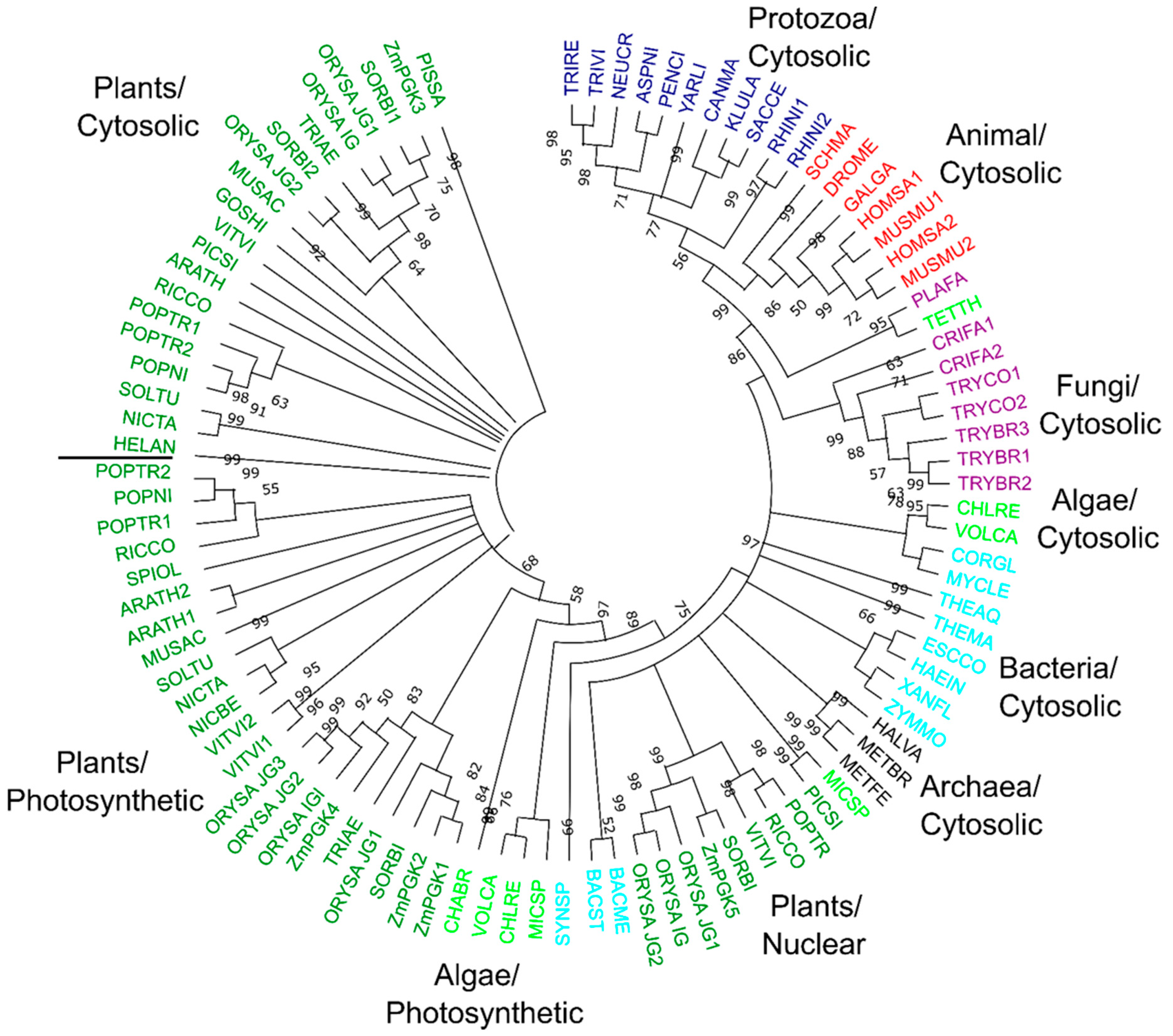
3.7. Phylogenetic Relationships of Plant PGK Isozymes
4. Discussion
4.1. Cytosolic ZmPGK3 is Predicted to be Involved in Glycolysis
4.2. ZmPGK4 is a Pseudogene
4.3. Nuclear ZmPGK5 Isozyme Is a Bacterial Protein Chimera with an Additional Domain
4.4. Molecular Elements Driving Differential ZmPgk Gene Expression
4.5. Evolutionary Diversification of Plant PGKs
4.6. Distinct Roles of Plant PGKs across Species and Isozymes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Martin, W.; Schnarrenberger, C. The evolution of the Calvin cycle from prokaryotic to eukaryotic chromosomes: A case study of functional redundancy in ancient pathways through endosymbiosis. Curr. Genet. 1997, 32, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Raines, C.A. The Calvin cycle revisited. Photosynth. Res. 2003, 75, 1–10. [Google Scholar] [CrossRef]
- Blake, C.C.; Rice, D.W. Phosphoglycerate kinase. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1981, 293, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Tiessen, A.; Perez-Rodriguez, P.; Delaye-Arredondo, L.J. Mathematical modeling and comparison of protein size distribution in different plant, animal, fungal and microbial species reveals a negative correlation between protein size and protein number, thus providing insight into the evolution of proteomes. BMC Res. Notes 2012, 5, 85. [Google Scholar] [CrossRef] [Green Version]
- Ramírez-Sánchez, O.; Pérez-Rodríguez, P.; Delaye, L.; Tiessen, A. Plant Proteins Are Smaller Because They Are Encoded by Fewer Exons than Animal Proteins. Genom. Proteom. Bioinform. 2016, 14, 357–370. [Google Scholar] [CrossRef]
- Banks, R.D.; Blake, C.C.; Evans, P.R.; Haser, R.; Rice, D.W.; Hardy, G.W.; Merrett, M.; Phillips, A.W. Sequence, structure and activity of phosphoglycerate kinase: A possible hinge-bending enzyme. Nature 1979, 279, 773–777. [Google Scholar] [CrossRef] [PubMed]
- Szabo, J.; Varga, A.; Flachner, B.; Konarev, P.V.; Svergun, D.I.; Zavodszky, P.; Vas, M. Communication between the nucleotide site and the main molecular hinge of 3-phosphoglycerate kinase. Biochemistry 2008, 47, 6735–6744. [Google Scholar] [CrossRef] [PubMed]
- Sternberg, M.J.E.; Cohen, F.E.; Taylor, W.R.; Feldmann, R.J.; Smit, J.D.G.; Phillips, D.C.; Blake, C.C.F.; Watson, H.C. Analysis and prediction of structural motifs in the glycolytic enzymes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1981, 293, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Michelson, A.M.; Blake, C.C.; Evans, S.T.; Orkin, S.H. Structure of the human phosphoglycerate kinase gene and the intron-mediated evolution and dispersal of the nucleotide-binding domain. Proc. Natl. Acad. Sci. USA 1985, 82, 6965–6969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zerrad, L.; Merli, A.; Schroder, G.F.; Varga, A.; Graczer, E.; Pernot, P.; Round, A.; Vas, M.; Bowler, M.W. A spring-loaded release mechanism regulates domain movement and catalysis in phosphoglycerate kinase. J. Biol. Chem. 2011, 286, 14040–14048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, H.; Filippova, E.V.; Tkaczuk, K.L.; Dworzynski, P.; Chruszcz, M.; Porebski, P.J.; Wawrzak, Z.; Onopriyenko, O.; Kudritska, M.; Grimshaw, S.; et al. Crystal structures of putative phosphoglycerate kinases from B. anthracis and C. jejuni. J. Struct. Funct. Genom. 2012, 13, 15–26. [Google Scholar] [CrossRef] [Green Version]
- Irani, M.; Maitra, P.K. Isolation and characterization of Escherichia coli mutants defective in enzymes of glycolysis. Biochem. Biophys. Res. Commun. 1974, 56, 127–133. [Google Scholar] [CrossRef]
- Irani, M.H.; Maitra, P.K. Properties of Escherichia coli mutants deficient in enzymes of glycolysis. J. Bacteriol. 1977, 132, 398–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michelson, A.M.; Bruns, G.A.; Morton, C.C.; Orkin, S.H. The human phosphoglycerate kinase multigene family. HLA-associated sequences and an X-linked locus containing a processed pseudogene and its functional counterpart. J. Biol. Chem. 1985, 260, 6982–6992. [Google Scholar] [PubMed]
- Danshina, P.V.; Geyer, C.B.; Dai, Q.; Goulding, E.H.; Willis, W.D.; Kitto, G.B.; McCarrey, J.R.; Eddy, E.M.; O’Brien, D.A. Phosphoglycerate kinase 2 (PGK2) is essential for sperm function and male fertility in mice. Biol. Reprod. 2010, 82, 136–145. [Google Scholar] [CrossRef] [Green Version]
- Cavell, S.; Scopes, K. Isolation and characterization of the “photosynthetic” phosphoglycerate kinase from Beta vulgaris. Eur. J. Biochem. 1976, 63, 483–490. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.; Bradbeer, J.W. The occurrence of chloroplastic and cytosolic isoenzymes of phosphoglycerate kinase in a range of plant species. Planta 1994, 193, 232–237. [Google Scholar] [CrossRef]
- Anderson, L.E.; Bryant, J.A.; Carol, A.A. Both chloroplastic and cytosolic phosphoglycerate kinase isozymes are present in the pea leaf nucleus. Protoplasma 2004, 223, 103–110. [Google Scholar] [CrossRef]
- Troncoso-Ponce, M.A.; Rivoal, J.; Venegas-Caleron, M.; Dorion, S.; Sanchez, R.; Cejudo, F.J.; Garces, R.; Martinez-Force, E. Molecular cloning and biochemical characterization of three phosphoglycerate kinase isoforms from developing sunflower (Helianthus annuus L.) seeds. Phytochemistry 2012, 79, 27–38. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, R.L.B. Grain Fill Stages in Corn Silking Stage. Agron. News 2016, 1, 1–10. [Google Scholar]
- Krogh, A.; Brown, M.; Mian, I.S.; Sjolander, K.; Haussler, D. Hidden Markov models in computational biology. Applications to protein modeling. J. Mol. Biol. 1994, 235, 1501–1531. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Small, I.; Peeters, N.; Legeai, F.; Lurin, C. Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 2004, 4, 1581–1590. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emanuelsson, O.; Nielsen, H.; von Heijne, G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999, 8, 978–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [Green Version]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. ISBN 978-1-59259-890-8. [Google Scholar]
- Stothard, P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, J.; Wen, L.; Gao, X.; Jin, C.; Xue, Y.; Yao, X. DOG 1.0: Illustrator of protein domain structures. Cell Res. 2009, 19, 271–273. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kersey, P.J.; Allen, J.E.; Allot, A.; Barba, M.; Boddu, S.; Bolt, B.J.; Carvalho-Silva, D.; Christensen, M.; Davis, P.; Grabmueller, C.; et al. Ensembl Genomes 2018: An integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res. 2018, 46, D802–D808. [Google Scholar] [CrossRef] [PubMed]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [Green Version]
- Tiessen, A.; Cubedo-Ruiz, E.A.; Winkler, R. Improved Representation of Biological Information by Using Correlation as Distance Function for Heatmap Cluster Analysis. Am. J. Plant Sci. 2017, 08, 502–516. [Google Scholar] [CrossRef] [Green Version]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.L.; Moorman, A.F.M. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.B.; Moorman, A.F.M. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juarez-Colunga, S.; Lopez-Gonzalez, C.; Morales-Elias, N.C.; Massange-Sanchez, J.A.; Trachsel, S.; Tiessen, A. Genome-wide analysis of the invertase gene family from maize. Plant Mol. Biol. 2018, 97, 385–406. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, G.J.; Gamblin, S.J.; Littlechild, J.A.; Dauter, Z.; Wilson, K.S.; Watson, H.C. Structure of the ADP complex of the 3-phosphoglycerate kinase from Bacillus stearothermophilus at 1.65 A. Acta Crystallogr. D Biol. Crystallogr. 1994, 50, 202–209. [Google Scholar] [CrossRef]
- Auerbach, G.; Huber, R.; Grattinger, M.; Zaiss, K.; Schurig, H.; Jaenicke, R.; Jacob, U. Closed structure of phosphoglycerate kinase from Thermotoga maritima reveals the catalytic mechanism and determinants of thermal stability. Structure 1997, 5, 1475–1483. [Google Scholar] [CrossRef]
- Bernstein, B.E.; Hol, W.G. Crystal structures of substrates and products bound to the phosphoglycerate kinase active site reveal the catalytic mechanism. Biochemistry 1998, 37, 4429–4436. [Google Scholar] [CrossRef] [PubMed]
- Tougard, P.; Bizebard, T.; Ritco-Vonsovici, M.; Minard, P.; Desmadril, M. Structure of a circularly permuted phosphoglycerate kinase. Acta Crystallogr. D Biol. Crystallogr. 2002, 58, 2018–2023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekhon, R.S.; Lin, H.; Childs, K.L.; Hansey, C.N.; Buell, C.R.; de Leon, N.; Kaeppler, S.M. Genome-wide atlas of transcription during maize development. Plant J. 2011, 66, 553–563. [Google Scholar] [CrossRef]
- Hoopes, G.M.; Hamilton, J.P.; Wood, J.C.; Esteban, E.; Pasha, A.; Vaillancourt, B.; Provart, N.J.; Buell, C.R. An updated gene atlas for maize reveals organ-specific and stress-induced genes. Plant J. 2019, 97, 1154–1167. [Google Scholar] [CrossRef] [Green Version]
- Downs, G.S.; Bi, Y.-M.; Colasanti, J.; Wu, W.; Chen, X.; Zhu, T.; Rothstein, S.J.; Lukens, L.N. A Developmental Transcriptional Network for Maize Defines Coexpression Modules. Plant Physiol. 2013, 161, 1830–1843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adje, C.A.; Opperdoes, F.R.; Michels, P.A. Molecular analysis of phosphoglycerate kinase in Trypanoplasma borreli and the evolution of this enzyme in kinetoplastida. Gene 1998, 217, 91–99. [Google Scholar] [CrossRef]
- Colasante, C.; Robles, A.; Li, C.-H.; Schwede, A.; Benz, C.; Voncken, F.; Guilbride, D.L.; Clayton, C. Regulated expression of glycosomal phosphoglycerate kinase in Trypanosoma brucei. Mol. Biochem. Parasitol. 2007, 151, 193–204. [Google Scholar] [CrossRef]
- Rosa-Tellez, S.; Anoman, A.D.; Flores-Tornero, M.; Toujani, W.; Alseek, S.; Fernie, A.R.; Nebauer, S.G.; Munoz-Bertomeu, J.; Segura, J.; Ros, R. Phosphoglycerate Kinases Are Co-Regulated to Adjust Metabolism and to Optimize Growth. Plant Physiol. 2018, 176, 1182–1198. [Google Scholar] [CrossRef] [Green Version]
- Podesta, F.E.; Plaxton, W.C. Kinetic and regulatory properties of cytosolic pyruvate kinase from germinating castor oil seeds. Biochem. J. 1991, 279 Pt 2, 495–501. [Google Scholar] [CrossRef] [Green Version]
- Goldschmidt, E.E.; Huber, S.C. Regulation of photosynthesis by end-product accumulation in leaves of plants storing starch, sucrose, and hexose sugars. Plant Physiol. 1992, 99, 1443–1448. [Google Scholar] [CrossRef] [Green Version]
- Friso, G.; Majeran, W.; Huang, M.; Sun, Q.; van Wijk, K.J. Reconstruction of metabolic pathways, protein expression, and homeostasis machineries across maize bundle sheath and mesophyll chloroplasts: Large-scale quantitative proteomics using the first maize genome assembly. Plant Physiol. 2010, 152, 1219–1250. [Google Scholar] [CrossRef] [Green Version]
- Al-Rashdi, J.; Bryant, J.A. Purification of a DNA-binding protein from a multi-protein complex associated with DNA polymerase-α in pea (Pisum sativum). J. Exp. Bot. 1994, 45, 1867–1871. [Google Scholar] [CrossRef]
- Bryant, J.A.; Brice, D.C.; Fitchett, P.N.; Anderson, L.E. A novel DNA-binding protein associated with DNA polymerase-alpha in pea stimulates polymerase activity on infrequently primed templates. J. Exp. Bot. 2000, 51, 1945–1947. [Google Scholar] [CrossRef] [Green Version]
- Ravel, C.; Fiquet, S.; Boudet, J.; Dardevet, M.; Vincent, J.; Merlino, M.; Michard, R.; Martre, P. Conserved cis-regulatory modules in promoters of genes encoding wheat high-molecular-weight glutenin subunits. Front. Plant Sci. 2014, 5, 621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piechulla, B.; Merforth, N.; Rudolph, B. Identification of tomato Lhc promoter regions necessary for circadian expression. Plant Mol. Biol. 1998, 38, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Gowik, U.; Burscheidt, J.; Akyildiz, M.; Schlue, U.; Koczor, M.; Streubel, M.; Westhoff, P. cis-Regulatory elements for mesophyll-specific gene expression in the C4 plant Flaveria trinervia, the promoter of the C4 phosphoenolpyruvate carboxylase gene. Plant Cell 2004, 16, 1077–1090. [Google Scholar] [CrossRef] [Green Version]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.-H.; Hong, X.; Agarwal, M.; Zhu, J.-K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, S.; Sirikhachornkit, A.; Su, X.; Faris, J.; Gill, B.; Haselkorn, R.; Gornicki, P. Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. USA 2002, 99, 8133–8138. [Google Scholar] [CrossRef] [Green Version]
- Longstaff, M.; Raines, C.A.; McMorrow, E.M.; Bradbeer, J.W.; Dyer, T.A. Wheat phosphoglycerate kinase: Evidence for recombination between the genes for the chloroplastic and cytosolic enzymes. Nucleic Acids Res. 1989, 17, 6569–6580. [Google Scholar] [CrossRef] [Green Version]
- Nowitzki, U.; Gelius-Dietrich, G.; Schwieger, M.; Henze, K.; Martin, W. Chloroplast phosphoglycerate kinase from Euglena gracilis: Endosymbiotic gene replacement going against the tide. Eur. J. Biochem. 2004, 271, 4123–4131. [Google Scholar] [CrossRef]
- Brinkmann, H.; Martin, W. Higher-plant chloroplast and cytosolic 3-phosphoglycerate kinases: A case of endosymbiotic gene replacement. Plant Mol. Biol. 1996, 30, 65–75. [Google Scholar] [CrossRef]
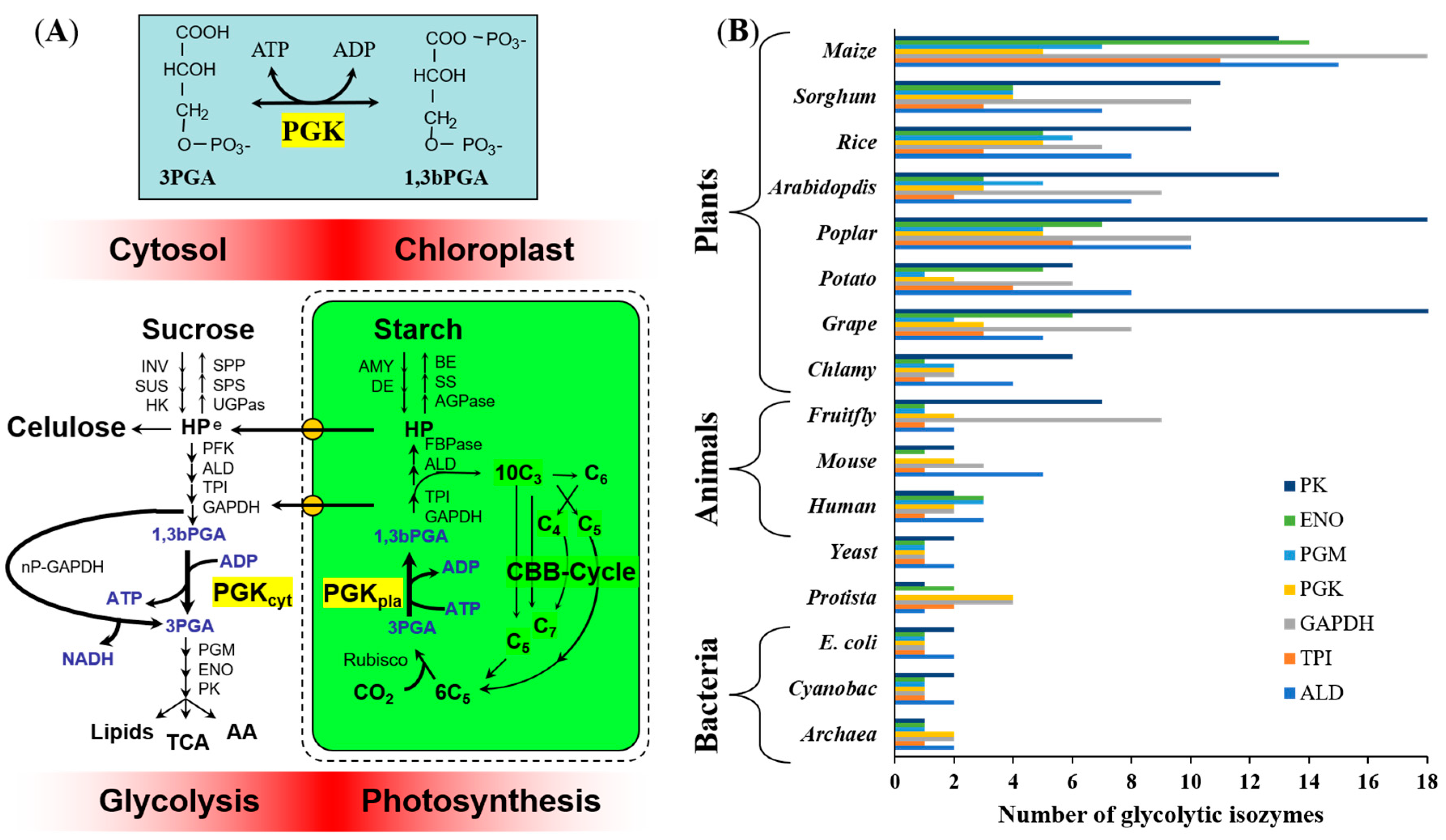
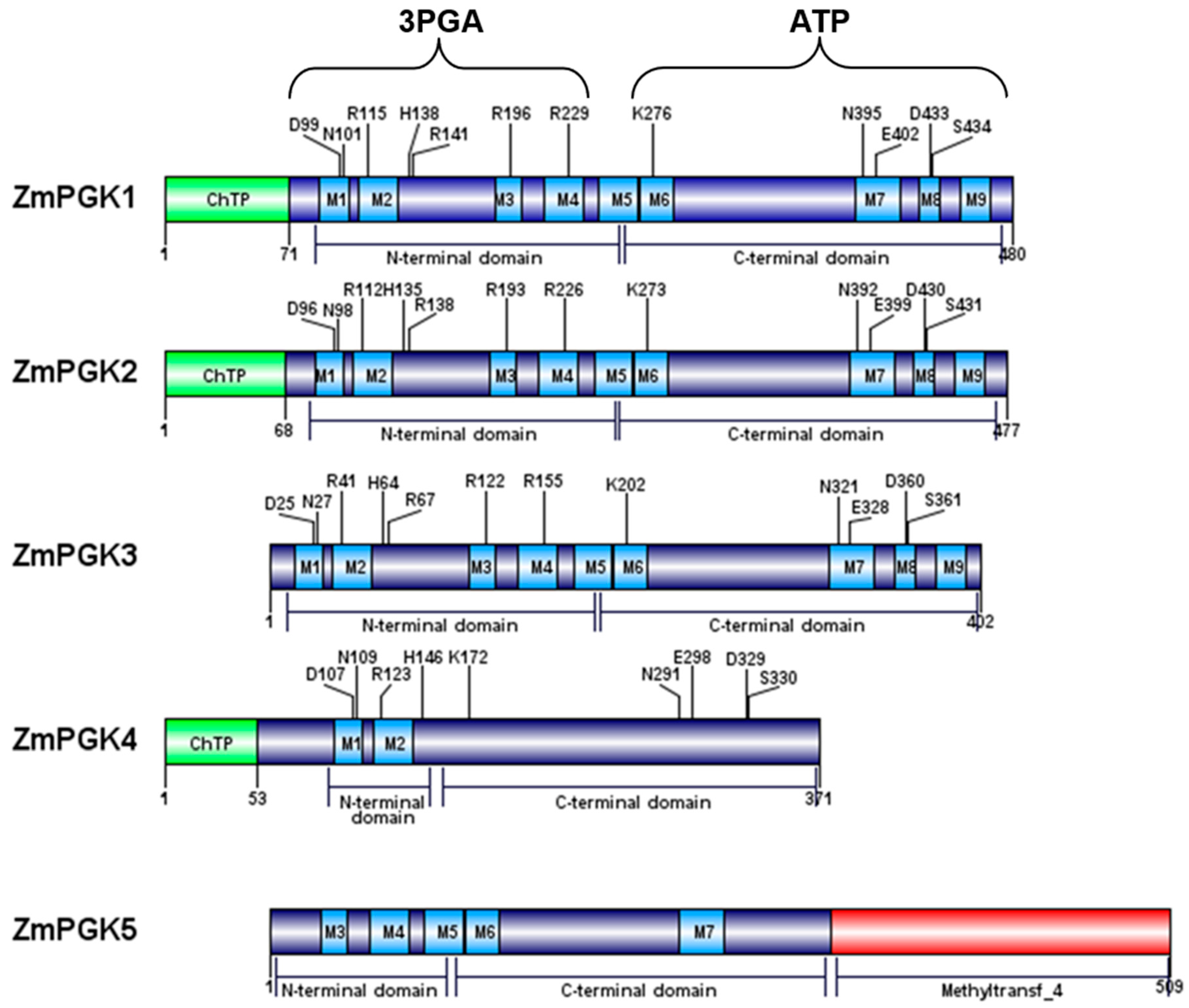

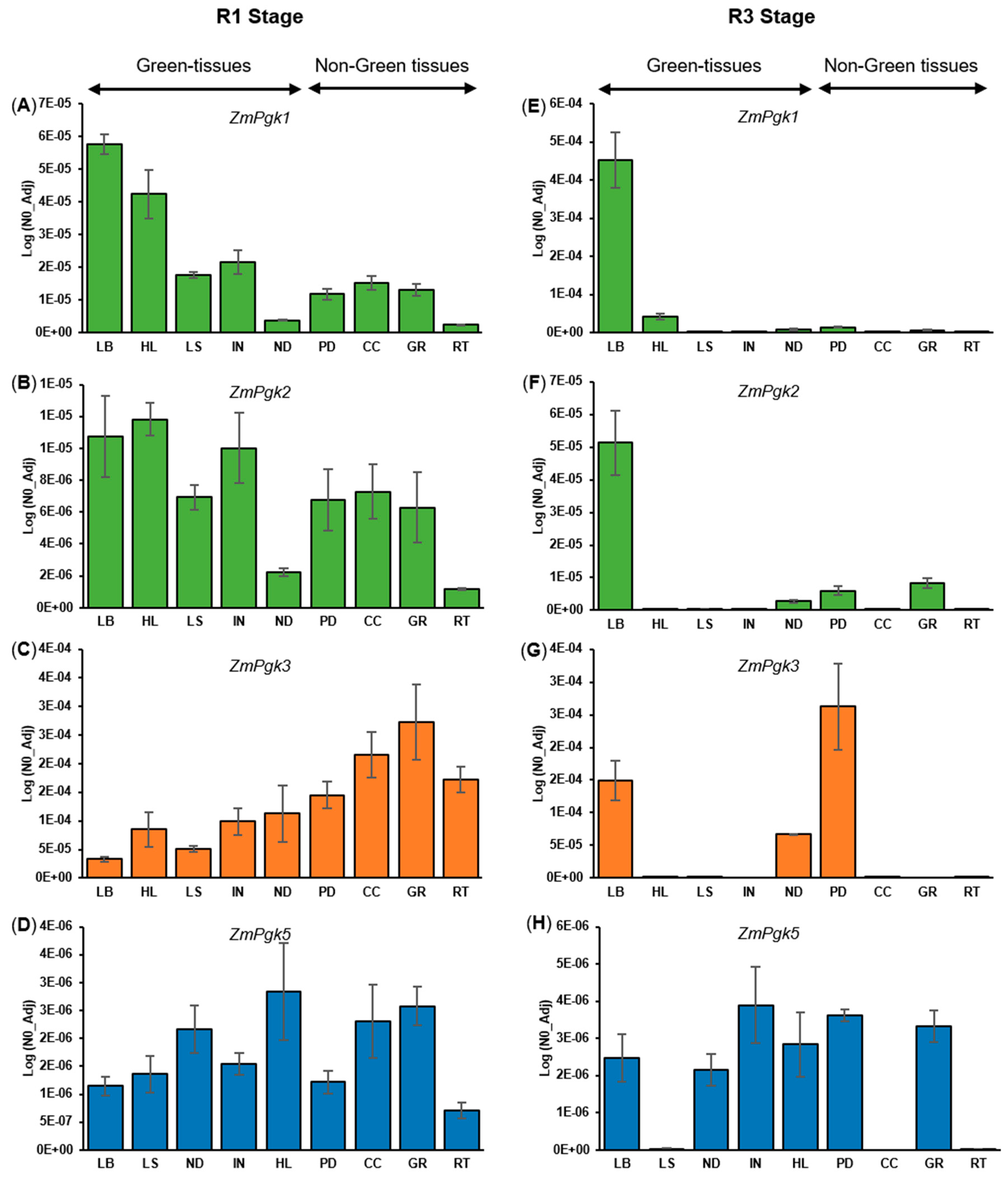
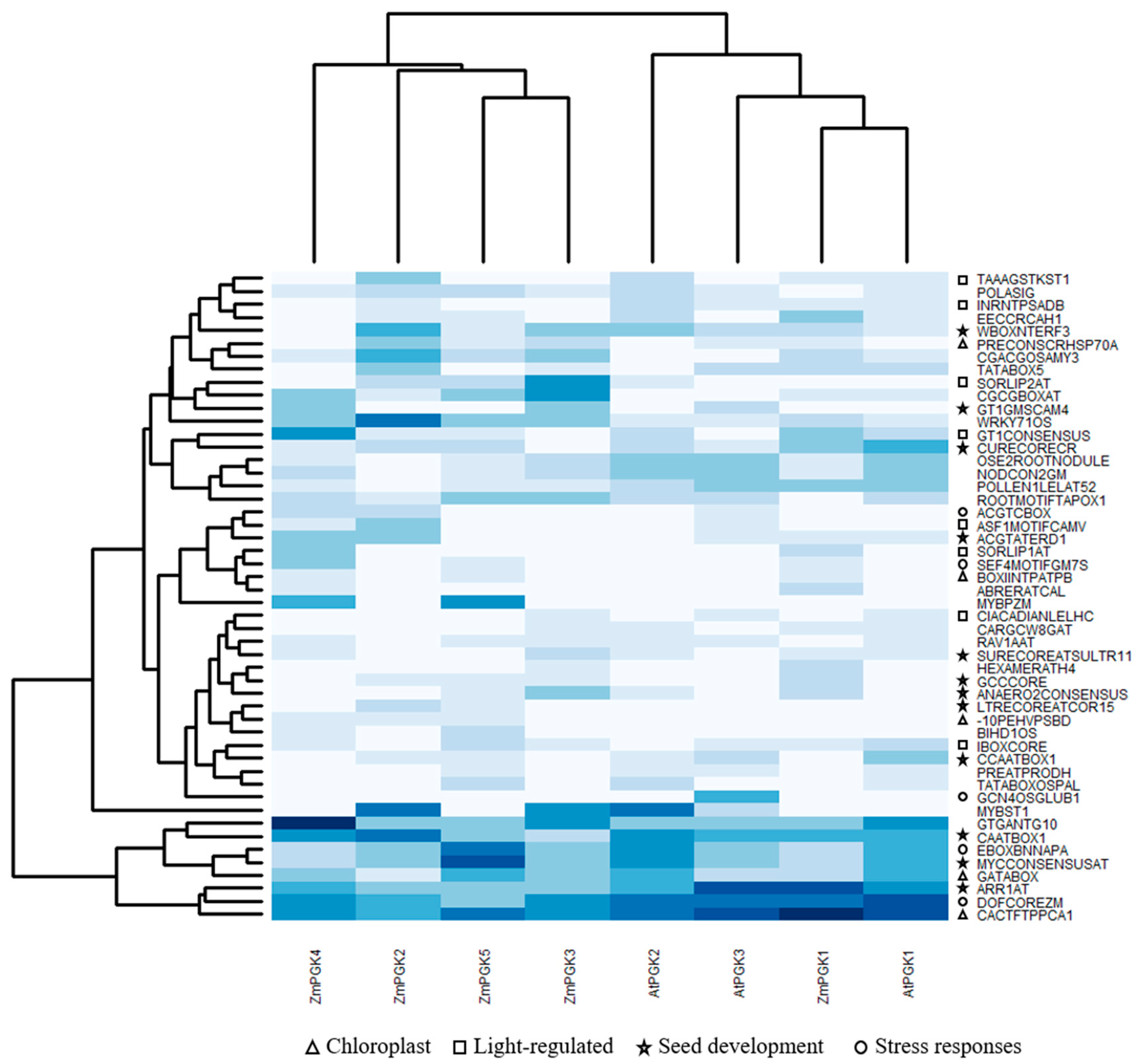
| PGK-Encoding Genes | Protein Identity (% Amino Acid) | ||||
|---|---|---|---|---|---|
| ZmPGK2 | ZmPGK3 | ZmPGK4 | ZmPGK5 | ||
| Zm00001d038579_T001.1 | ZmPGK1 | 95.4 | 80.9 | 66.3 | 21.8 |
| Zm00001d010672_T001.1 | ZmPGK2 | 100 | 81.1 | 70.4 | 18.5 |
| Zm00001d015376_T002.1 | ZmPGK3 | 100 | 59.6 | 20.2 | |
| Zm00001d043194_T001.1 | ZmPGK4 | 100 | 15.4 | ||
| Zm00001d032867_T002.1 | ZmPGK5 | 100 | |||
| Protein Name | Molecular Weight kDa; Amino Acids (With, Without Signal Peptide) | Theoretical pI ProtParam EMBL SMS | Prediction of Location: Predotar ChloroP TargetP Wolf PSORT | Predicted N-Glycosylation Sites NetNGlyc 1.0 (Position Amino Acid) |
|---|---|---|---|---|
| ZmPGK1 | 49.8, 42.8; (480aa, 409aa) | 6.29, 5.07 6.28, 4.88 6.69, 4.81 | Plastid Chloroplast (71aa) Chloroplast (71aa) Chloroplast | 109 NITD |
| ZmPGK2 | 49.6, 42.9; (477aa, 409aa) | 6.99, 5.13 7.36, 4.93 7.62, 4.88 | Plastid Chloroplast (68aa) Chloroplast (68aa) Chloroplast | 106 NITD |
| ZmPGK3 | 42.4; (402aa) | 5.65 5.52 5.58 | None None None Cytoplasm | No sites predicted |
| ZmPGK4 | 38.3, 32.9; (371aa, 318aa) | 7.02, 5.80 7.26, 5.73 7.55, 6.05 | Plastid Chloroplast (53aa) Chloroplast (53aa) Chloroplast | 22 NSTG 117 NITD |
| ZmPGK5 | 56.4; (509aa) | 6.19 6.15 6.50 | None None None Nucleus | 39 NFTG 63 NDSF 202 NSTG 389 NATS |
| Tissue | Km PGA (mM) | Km ATP (mM) |
|---|---|---|
| Leaf | 0.84 ± 0.11 | 0.06 ± 0.01 |
| Husk | 0.47 ± 0.06 | 0.24 ± 0.07 |
| Stem | 0.59 ± 0.10 | 0.08 ± 0.02 |
| Cob | 0.45 ± 0.06 | 0.24 ± 0.07 |
| Root | 0.50 ± 0.07 | 0.08 ± 0.02 |
| Localized | Monocots | Dicots | ||||
|---|---|---|---|---|---|---|
| Z. mays (C4) | S. bicolor (C4) | O. sativa (C3) | A. thaliana (C3) | V. vinifera (C3) | P. trichocarpa (C3) | |
| Plastid/Photosynthetic | ZmPGK1 Zm00001d038579 ZmPGK2 Zm00001d010672 ZmPGK4 Zm00001d043194 | SbPGKpla1 SORBI_3009G183700 | OsPGKpla1 AAT07576.1 OsPGKpla2 EEE55536.1 OsPGKpla3 NP_001172606.1 | AtPGK1 AT3G12780 AtPGK2 AT1G56190 | VvPGKpla1 XP_003634814.1 VvPGKpla2 XP_002263796.1 | PtPGKpla1 POPTR_010G171500v3 PtPGKpla2 POPTR_008G084500v3 |
| Cytoplasmic/Glycolytic | ZmPGK3 Zm00001d015376 | SbPGKcyt1 SORBI_3004G055200 SbPGKcyt2 SORBI_3010G221800 | OsPGKcyt1 ABL74575.1 OsPGK7cyt2 NP_001058317.1 | AtPGK3 AT1G79550 | VvPGKcyt1 XP_002263950.1 | PtPGKcyt1 POPTR_008G084400v3 PtPGKcyt2 POPTR_010G171600v3 |
| Nuclear | ZmPGK5 Zm00001d032867 | SbPGKnuc SORBI_3001G236800 | OsPGK5nuc1 ABB47707.1 OsPGK6nuc2 EEE51029.1 | - | VvPGKnuc CBI32769.3 | PtPGKnuc POPTR_016G091800v3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Massange-Sánchez, J.A.; Casados-Vázquez, L.E.; Juarez-Colunga, S.; Sawers, R.J.H.; Tiessen, A. The Phosphoglycerate Kinase (PGK) Gene Family of Maize (Zea mays var. B73). Plants 2020, 9, 1639. https://doi.org/10.3390/plants9121639
Massange-Sánchez JA, Casados-Vázquez LE, Juarez-Colunga S, Sawers RJH, Tiessen A. The Phosphoglycerate Kinase (PGK) Gene Family of Maize (Zea mays var. B73). Plants. 2020; 9(12):1639. https://doi.org/10.3390/plants9121639
Chicago/Turabian StyleMassange-Sánchez, Julio A., Luz E. Casados-Vázquez, Sheila Juarez-Colunga, Ruairidh J. H. Sawers, and Axel Tiessen. 2020. "The Phosphoglycerate Kinase (PGK) Gene Family of Maize (Zea mays var. B73)" Plants 9, no. 12: 1639. https://doi.org/10.3390/plants9121639





