Efficient Oxidation of Methyl Glycolate to Methyl Glyoxylate Using a Fusion Enzyme of Glycolate Oxidase, Catalase and Hemoglobin
Abstract
1. Introduction
2. Results and Discussion
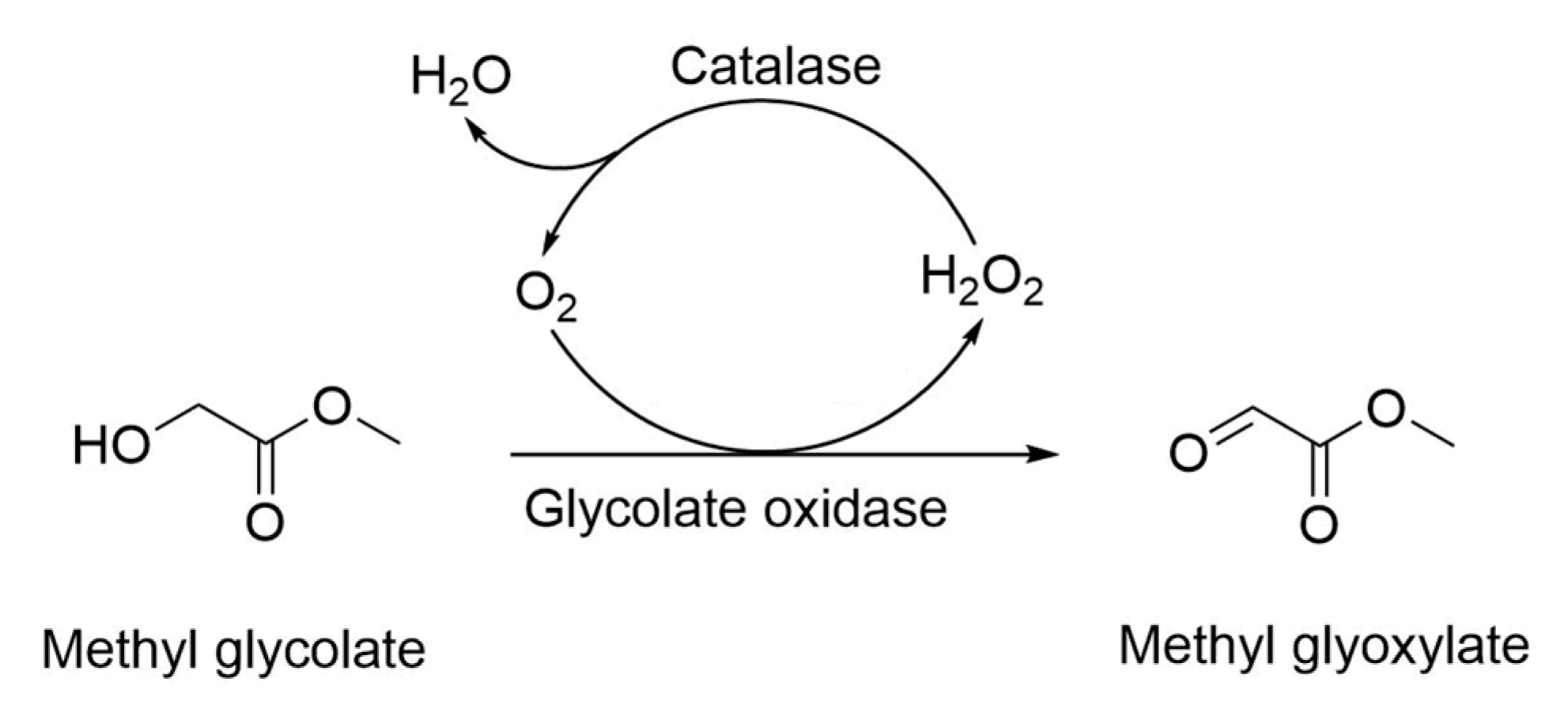
2.1. Cascade Catalysis of Glycolate Oxidase, Catalase and Hemoglobin
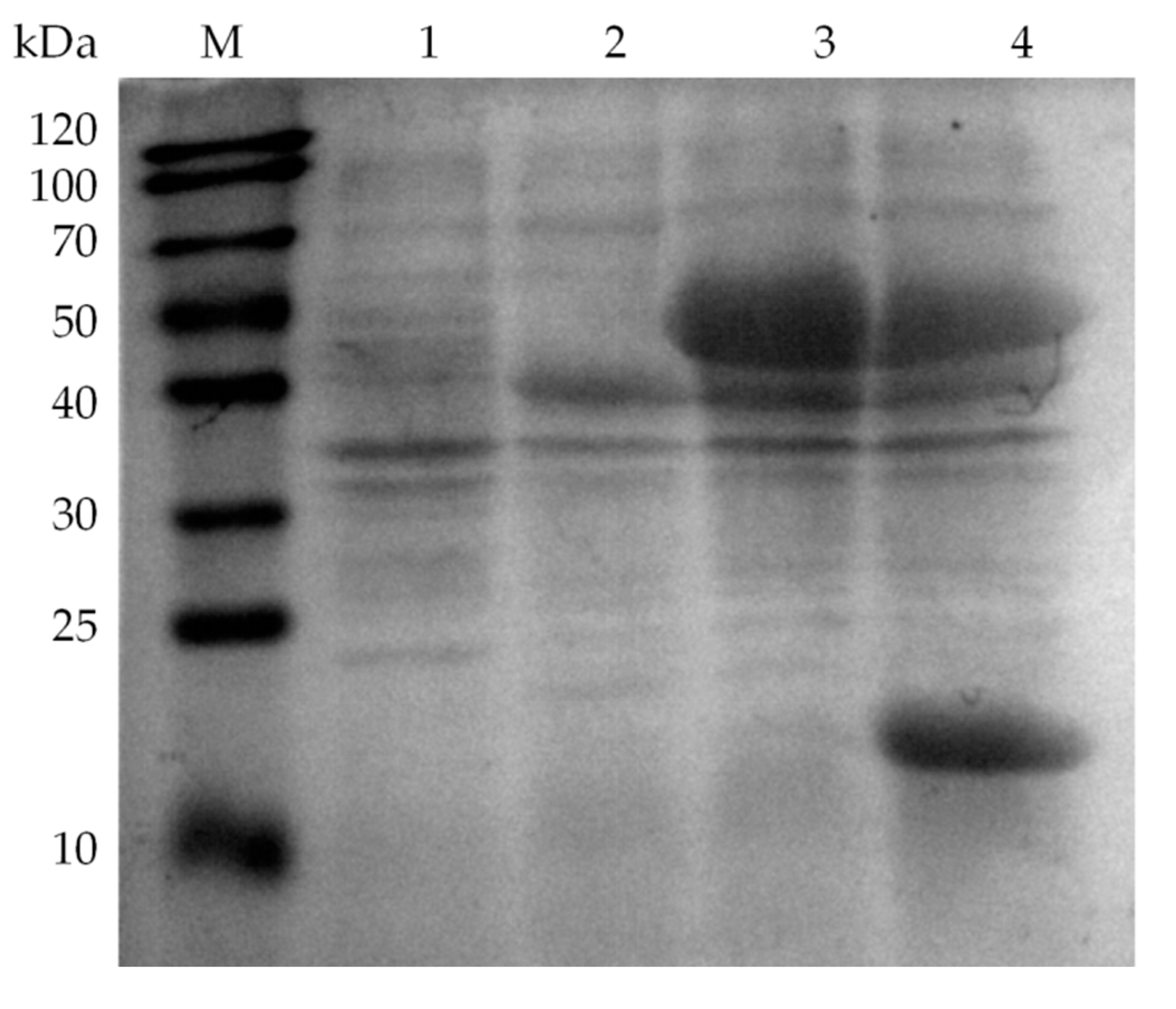
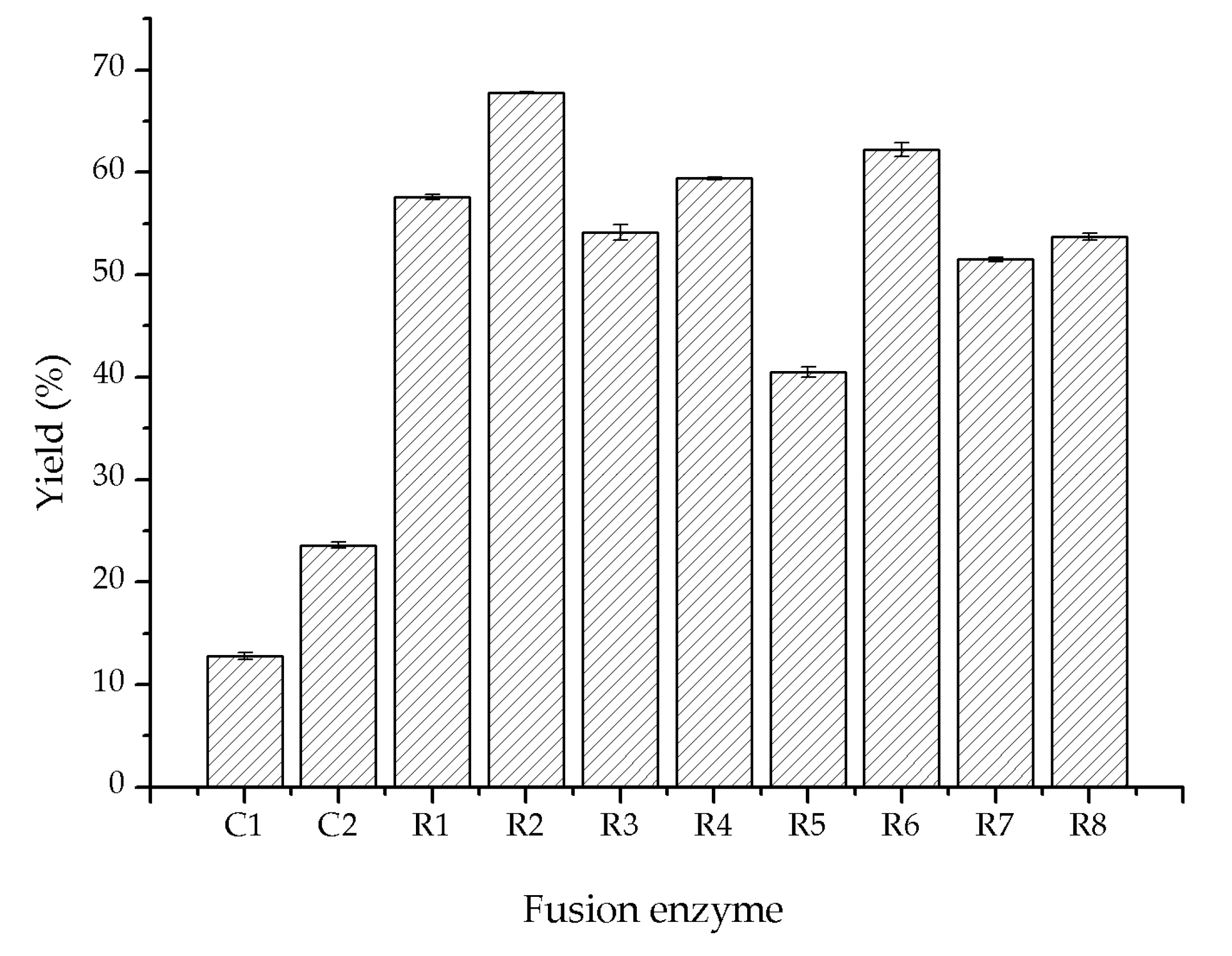
2.2. Fusion Expression of Glycolate Oxidase, Catalase and Hemoglobin
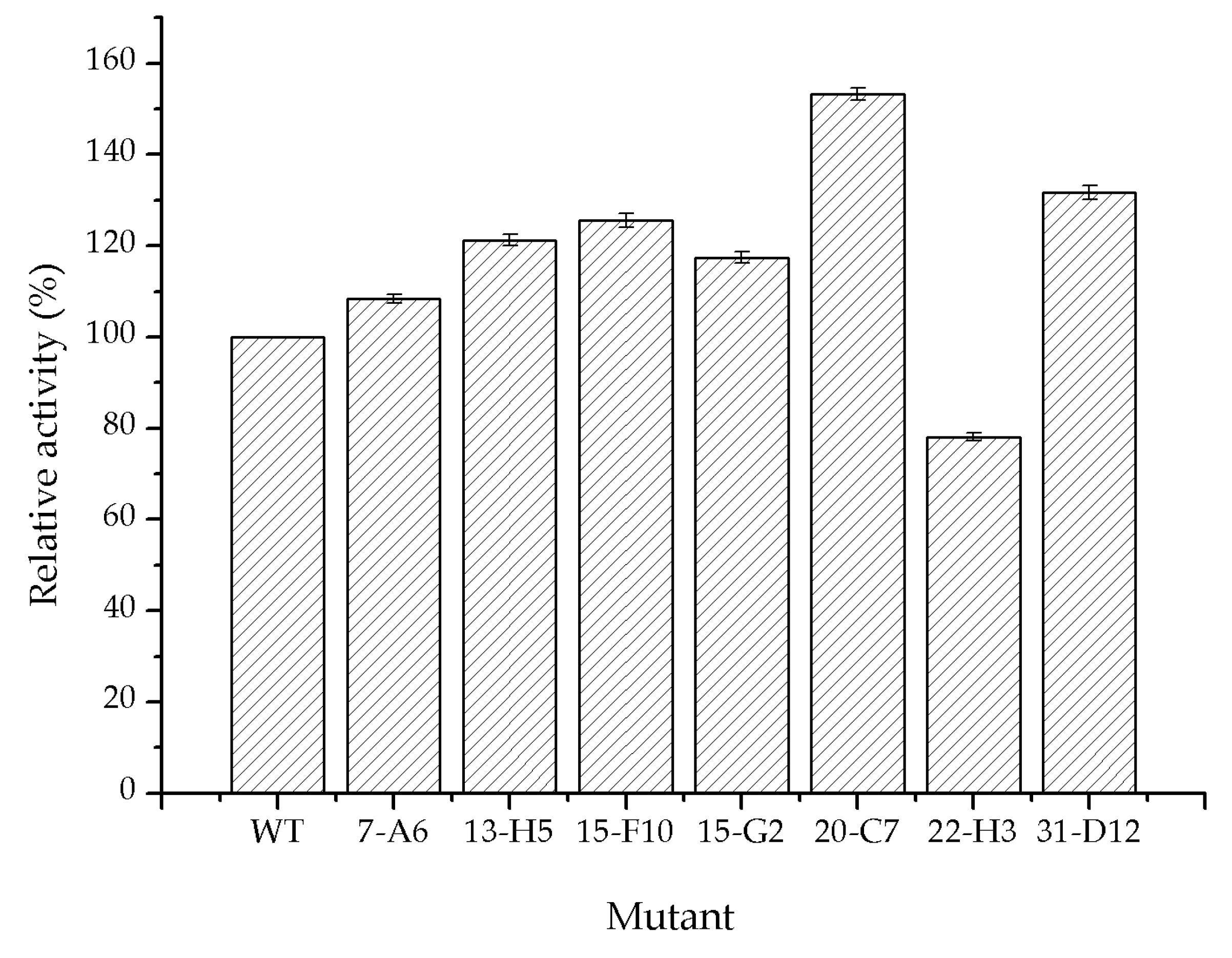
2.3. Directed Evolution of Glycolate Oxidase
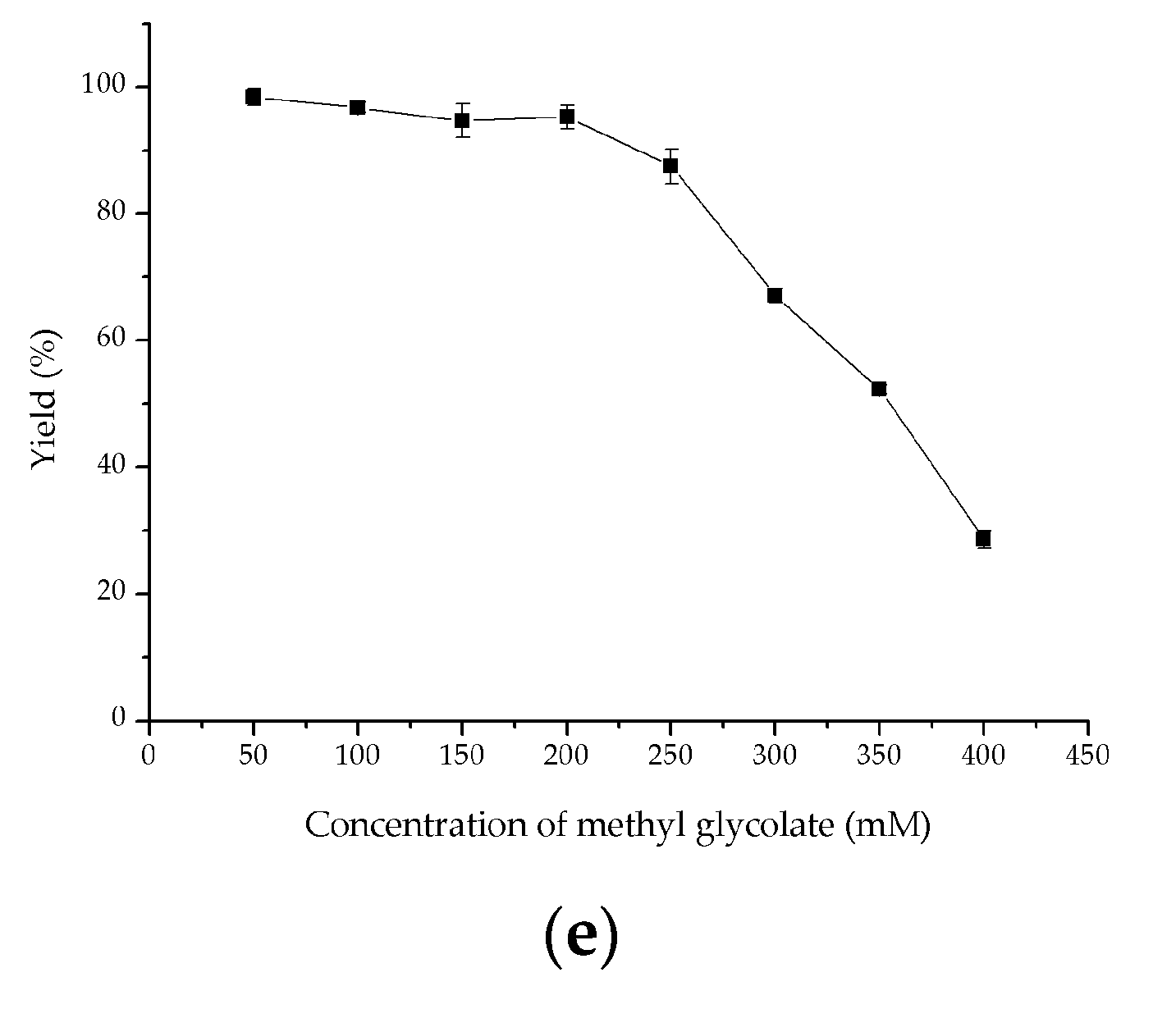
2.4. Investigation of Key Factors on Oxidation of Methyl Glycolate to Methyl Glyoxylate
3. Materials and Methods
3.1. Genes, Organisms and Chemicals
3.2. Over-Expression of Glycolate Oxidase, Catalase and Hemoglobin
3.3. Fusion Expression of Glycolate Oxidase, Catalase and Hemoglobin
3.4. Directed Evolution of Glycolate Oxidase SoGOX and Screening of the Evolved Mutants
3.5. HPLC Analyses of Methyl Glycolate and Methyl Glyoxylate
3.6. Site-Saturation Mutagenesis for the Sites M267 and S362 of SoGOX
3.7. Key Factors on Oxidation of Methyl Glycolate Using Crude Fusion Enzyme VsHGB-GSG-SoGOXmut-GGGGS-HpCAT
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hook, J.M. A simple and efficient synthesis of ethyl and methyl glyoxylate. Synth. Commun. 1984, 14, 83–87. [Google Scholar] [CrossRef]
- Bailey, P.D.; Brown, G.R.; Korber, F.; Reed, A.; Wilson, R.D. Asymmetric synthesis of pipecolic acid derivatives using the aza-Diels-Alder reaction. Tetrahedron Asymmetry 1991, 2, 1263–1282. [Google Scholar] [CrossRef]
- Ying, X.; Yu, S.; Huang, M.; Wei, R.; Meng, S.; Cheng, F.; Yu, M.; Ying, M.; Zhao, M.; Wang, Z. Engineering the enantioselectivity of yeast old yellow enzyme OYE2y in asymmetric reduction of (E/Z)-citral to (R)-citronellal. Molecules 2019, 24, 1057. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Chen, Z.; Wang, Y. Bioaldehydes and beyond: Expanding the realm of bioderived chemicals using biogenic aldehydes as platforms. Curr. Opin. Chem. Biol. 2020, 59, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Heidlindemann, M.; Hammel, M.; Schefflfflffler, U.; Mahrwald, R.; Hummel, W.; Berkessel, A.; Gröger, H. Chemoenzymatic synthesis of vitamin B5-intermediate (R)-pantolactone via combined asymmetric organo- and biocatalysis. J. Org. Chem. 2015, 80, 3387–3396. [Google Scholar] [CrossRef]
- Monti, D.; Ottolina, G.; Carrea, G.; Riva, S. Redox reactions catalyzed by isolated enzymes. Chem. Rev. 2011, 111, 4111–4140. [Google Scholar] [CrossRef]
- Kroutil, W.; Mang, H.; Edegger, K.; Faber, K. Biocatalytic oxidation of primary and secondary alcohols. Adv. Synth. Catal. 2004, 346, 125–142. [Google Scholar] [CrossRef]
- Liu, J.; Wu, S.; Li, Z. Recent advances in enzymatic oxidation of alcohols. Curr. Opin. Chem. Biol. 2018, 43, 77–86. [Google Scholar] [CrossRef]
- Jin, J.; Tan, T.; Wang, H.; Su, G. The Expression of spinach glycolate oxidase (GO) in E. coli and the application of GO in the production of glyoxylic acid. Mol. Biotechnol. 2003, 25, 207–214. [Google Scholar] [CrossRef]
- Isobe, K.; Watabe, S.; Yamada, M. Characterization and application of a glycolate dehydrogenase from Trichoderma harzianum AIU 353. J. Mol. Catal. B Enzym. 2012, 83, 94–99. [Google Scholar] [CrossRef]
- Nölke, G.; Houdelet, M.; Kreuzaler, F.; Peterhänsel, C.; Schillberg, S. The expression of a recombinant glycolate dehydrogenase polyprotein in potato (Solanum tuberosum) plastids strongly enhances photosynthesis and tuber yield. Plant Biotechnol. J. 2014, 12, 734–742. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Higashiyama, T.; Kishino, S.; Kataoka, M.; Ogawa, J.; Shimizu, S.; Isobe, K. Novel alcohol oxidase with glycolate oxidase activity from Ochrobactrum sp. AIU 033. J. Mol. Catal. B Enzym. 2014, 105, 41–48. [Google Scholar] [CrossRef]
- Dellero, Y.; Mauve, C.; Boex-Fontvieille, E.; Flesch, V.; Jossier, M.; Tcherkez, G.; Hodges, M. Experimental evidence for a hydride transfer mechanism in plant glycolate oxidase catalysis. J. Biol. Chem. 2015, 290, 1689–1698. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, T.P.; van Schie, M.M.C.H.; Ma, A.; Tieves, F.; Younes, S.H.H.; Fernändez-Fueyo, E.; Arends, I.W.C.E.; Riul, A., Jr.; Hollmanna, F. Efficient aerobic oxidation of trans-2-hexen-1-ol using the aryl alcohol oxidase from Pleurotus eryngii. Adv. Synth. Catal. 2019, 361, 2668–2672. [Google Scholar] [CrossRef]
- Gavagan, J.E.; Fager, S.K.; Seip, J.E.; Payne, M.S.; Anton, D.L.; DiCosimo, R. Glyoxylic acid production using microbial transformant catalysts. J. Org. Chem. 1995, 60, 3957–3963. [Google Scholar] [CrossRef]
- Ricca, E.; Brucher, B.; Schrittwieser, J.H. Multi-enzymatic cascade reactions: Overview and perspectives. Adv. Synth. Catal. 2011, 353, 2239–2262. [Google Scholar] [CrossRef]
- Li, G.; Lian, J.; Xue, H.; Jiang, Y.; Wu, M.; Lin, J.; Yang, L. Enzymatic preparation of pyruvate by a whole-cell biocatalyst coexpressing L-lactate oxidase and catalase. Process Biochem. 2020, 96, 113–121. [Google Scholar] [CrossRef]
- Luo, H.; Ji, Q.; Chang, Y.; Du, C. Expression and characterization of fusion protein of Vitreoscilla hemoglobin with spinach glycolate oxidase. In Proceedings of the 2011 International Conference on Remote Sensing, Environment and Transportation Engineering, Nanjing, China, 24 June 2011; pp. 7580–7583. [Google Scholar]
- Aalbers, F.S.; Fraaije, M.W. Enzyme fusions in biocatalysis: Coupling reactions by pairing enzymes. ChemBioChem 2019, 20, 20–28. [Google Scholar] [CrossRef]
- Kataoka, M.; Miyakawa, T.; Shimizu, S.; Tanokura, M. Enzymes useful for chiral compound synthesis: Structural biology, directed evolution, and protein engineering for industrial use. Appl. Microbiol. Biotechnol. 2016, 100, 5747–5757. [Google Scholar] [CrossRef]
- Markel, U.; Essani, K.D.; Besirlioglu, V.; Schiffffels, J.; Streit, W.R.; Schwaneberg, U. Advances in ultrahigh-throughput screening for directed enzyme evolution. Chem. Soc. Rev. 2020, 49, 233–262. [Google Scholar] [CrossRef]
- Hazell, S.L.; Evans, D.J., Jr.; Graha, D.Y. Helicobacter pylori catalase. J. Gen. Microbiol. 1991, 137, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Khosla, C.; Bailey, J.E. Characterization of the oxygen-dependent promoter of the Vitreoscilla hemoglobin gene in Escherichia coli. J. Bacteriol. 1989, 171, 5995–6004. [Google Scholar] [CrossRef]
- Volokita, M.; Somervillet, R.C. The primary structure of spinach glycolate oxidase from the DNA sequence of a cDNA clone. J. Biol. Chem. 1987, 262, 15825–15828. [Google Scholar]
- Macheroux, P.; Mulrooney, S.B.; Williams, C.H., Jr.; Massey, V. Direct expression of active spinach glycolate oxidase in Escherichia coli. Biochim. Biophys. Acta 1992, 1132, 11–16. [Google Scholar] [CrossRef]
- Macheroux, P.; Massey, V.; Thiele, D.J. Expression of spinach glycolate oxidase in Saccharomyces cerevisiae: Purification and characterization. Biochemistry 1991, 30, 4612–4619. [Google Scholar] [CrossRef] [PubMed]
- Stenberg, K.; Lindqvist, Y. High-level expression, purification, and crystallization of recombinant spinach glycolate oxidase in Escherichia coli. Protein Expr. Purif. 1996, 8, 295–298. [Google Scholar] [CrossRef]
- Guo, H.; Yang, Y.; Xue, F.; Zhang, H.; Huang, T.; Liu, W.; Liu, H.; Zhang, F.; Yang, M.; Liu, C.; et al. Effect of flexible linker length on the activity of fusion protein 4-coumaroyl-CoA ligase: Stilbene synthase. Mol. BioSyst. 2017, 13, 598–606. [Google Scholar] [CrossRef]
- Aalbers, F.S.; Fraaije, M.W. Design of artificial alcohol oxidases: Alcohol dehydrogenase-NADPH oxidase fusions for continuous oxidations. ChemBioChem 2019, 20, 51–56. [Google Scholar] [CrossRef]
- Ge, M.; Chi, X.; Zhang, A.; Luo, G.; Sun, G.; Xie, H.; Hei, Z. Intestinal NF-E2-related factor-2 expression and antioxidant activity changes in rats undergoing orthotopic liver autotransplantation. Oncol. Lett. 2013, 6, 1307–1312. [Google Scholar] [CrossRef]
- Hiraka, K.; Kojima, K.; Tsugawa, W.; Asano, R.; Ikebukuro, K.; Sode, K. Rational engineering of Aerococcus viridans L-lactate oxidase for the mediator modification to achieve quasi-direct electron transfer type lactate sensor. Biosens. Bioelectron. 2020, 151, 111974. [Google Scholar] [CrossRef]
- Dick, M.; Hartmann, R.; Weiergraber, O.H.; Bisterfeld, C.; Classen, T.; Schwarten, M.; Neudecker, P.; Willboldbd, D.; Pietruszka, J. Mechanism-based inhibition of an aldolase at high concentrations of its natural substrate acetaldehyde: Structural insights and protective strategies. Chem. Sci. 2016, 7, 4492–4502. [Google Scholar] [CrossRef] [PubMed]
- Kadkhodaei, S.; Memari, H.R.; Abbasiliasi, S.; Rezaei, M.A.; Movahedi, A.; Shung, T.J.; Ariff, A.B. Multiple overlap extension PCR (MOE-PCR): An effective technical shortcut to high throughput synthetic biology. RSC Adv. 2016, 6, 66682. [Google Scholar] [CrossRef]

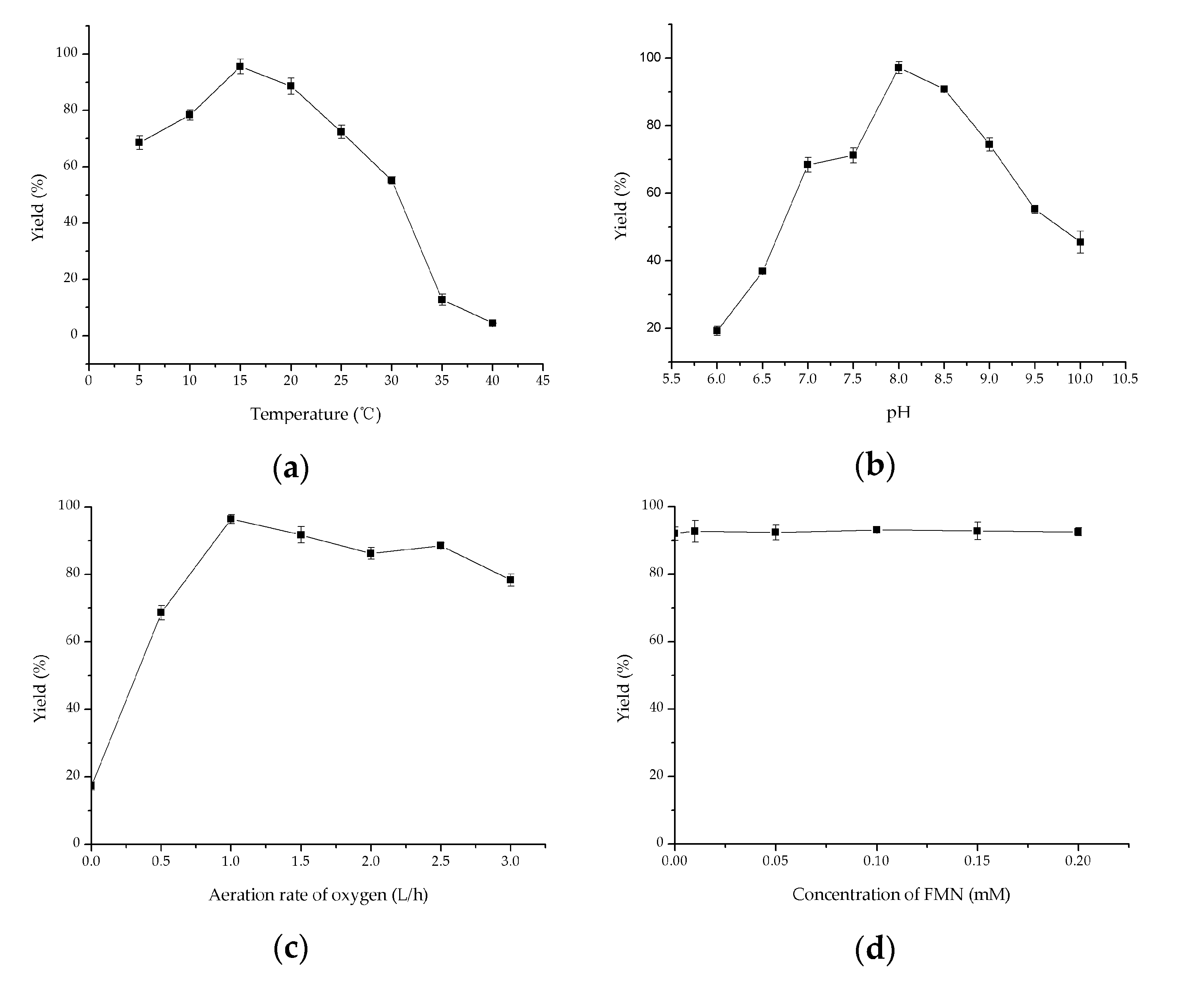
| The Protein(s) Expressed in E. coli Cells | Yield (%) |
|---|---|
| SoGOX | 21.1 ± 0.5 |
| SoGOX and HpCAT | 58.3 ± 1.3 |
| SoGOX, HpCAT and VsHGB | 65.2 ± 2.1 |
| SoGOX, HpCAT and VsHGB 2 | 88.7 ± 2.5 |
| Mutant | Amino Acid Substitution | H2O2 (µM) |
|---|---|---|
| WT | 45.5 | |
| 7-A6 | I73V | 61.2 |
| 13-H5 | R134H/K169R | 71.2 |
| 15-F10 | A138L/Y261F | 67.9 |
| 15-G2 | K190S | 66.9 |
| 20-C7 | M267S/S362G | 80.6 |
| 22-H3 | S314T | 59.1 |
| 31-D12 | K274S | 69.4 |
| Combinatorial Mutant | H2O2 (µM) 1 | Yield (%) 2 |
|---|---|---|
| WT | 45.5 | 26.3 |
| M267S/S362G | 80.6 | 45.3 |
| M267S/S362A | 54.5 | 33.7 |
| M267T/S362G | 96.7 | 50.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ying, X.; Wang, C.; Shao, S.; Wang, Q.; Zhou, X.; Bai, Y.; Chen, L.; Lu, C.; Zhao, M.; Wang, Z. Efficient Oxidation of Methyl Glycolate to Methyl Glyoxylate Using a Fusion Enzyme of Glycolate Oxidase, Catalase and Hemoglobin. Catalysts 2020, 10, 943. https://doi.org/10.3390/catal10080943
Ying X, Wang C, Shao S, Wang Q, Zhou X, Bai Y, Chen L, Lu C, Zhao M, Wang Z. Efficient Oxidation of Methyl Glycolate to Methyl Glyoxylate Using a Fusion Enzyme of Glycolate Oxidase, Catalase and Hemoglobin. Catalysts. 2020; 10(8):943. https://doi.org/10.3390/catal10080943
Chicago/Turabian StyleYing, Xiangxian, Can Wang, Shuai Shao, Qizhou Wang, Xueting Zhou, Yanbing Bai, Liang Chen, Chenze Lu, Man Zhao, and Zhao Wang. 2020. "Efficient Oxidation of Methyl Glycolate to Methyl Glyoxylate Using a Fusion Enzyme of Glycolate Oxidase, Catalase and Hemoglobin" Catalysts 10, no. 8: 943. https://doi.org/10.3390/catal10080943
APA StyleYing, X., Wang, C., Shao, S., Wang, Q., Zhou, X., Bai, Y., Chen, L., Lu, C., Zhao, M., & Wang, Z. (2020). Efficient Oxidation of Methyl Glycolate to Methyl Glyoxylate Using a Fusion Enzyme of Glycolate Oxidase, Catalase and Hemoglobin. Catalysts, 10(8), 943. https://doi.org/10.3390/catal10080943





