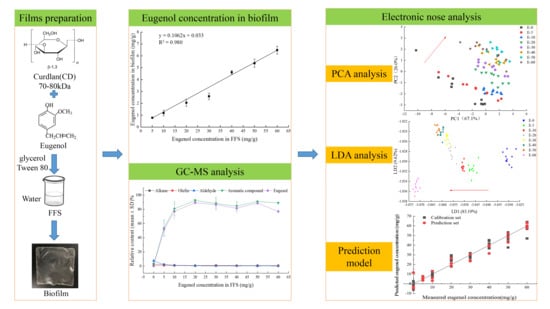
Rapid Non-Destructive Quantification of Eugenol in Curdlan Biofilms by Electronic Nose Combined with Gas Chromatography-Mass Spectrometry
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Biofilms Preparation
2.3. Quantitative Determination of Eugenol in the Biofilms
2.4. E-Nose Analysis
2.5. GC-MS Analysis
2.6. Statistical Analysis and Modeling
3. Results and Discussion
3.1. Quantitative Determination of Eugenol in the Biofilms
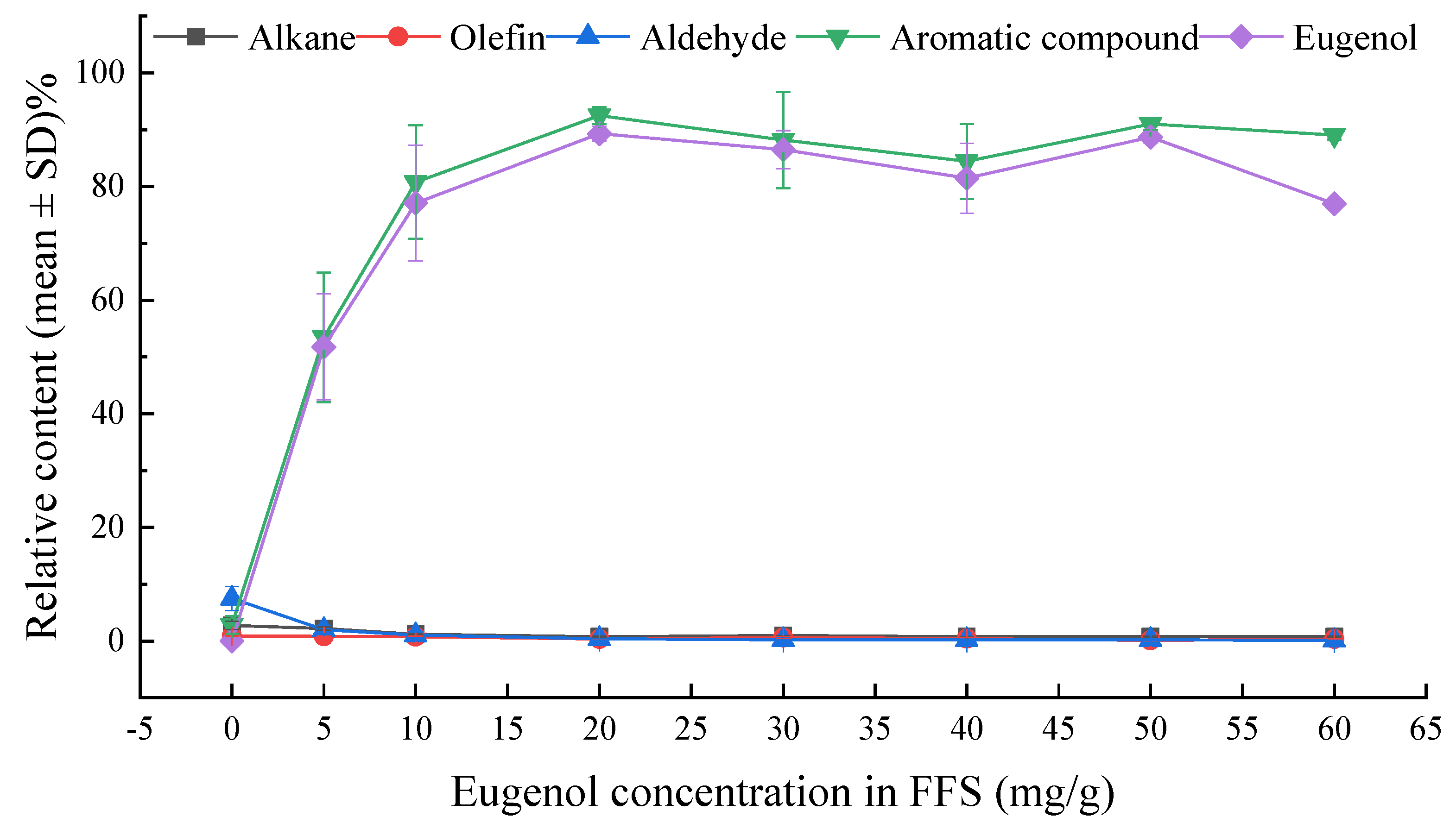
3.2. GC-MS Analysis
3.3. E-nose Analysis
3.3.1. Radar Graph
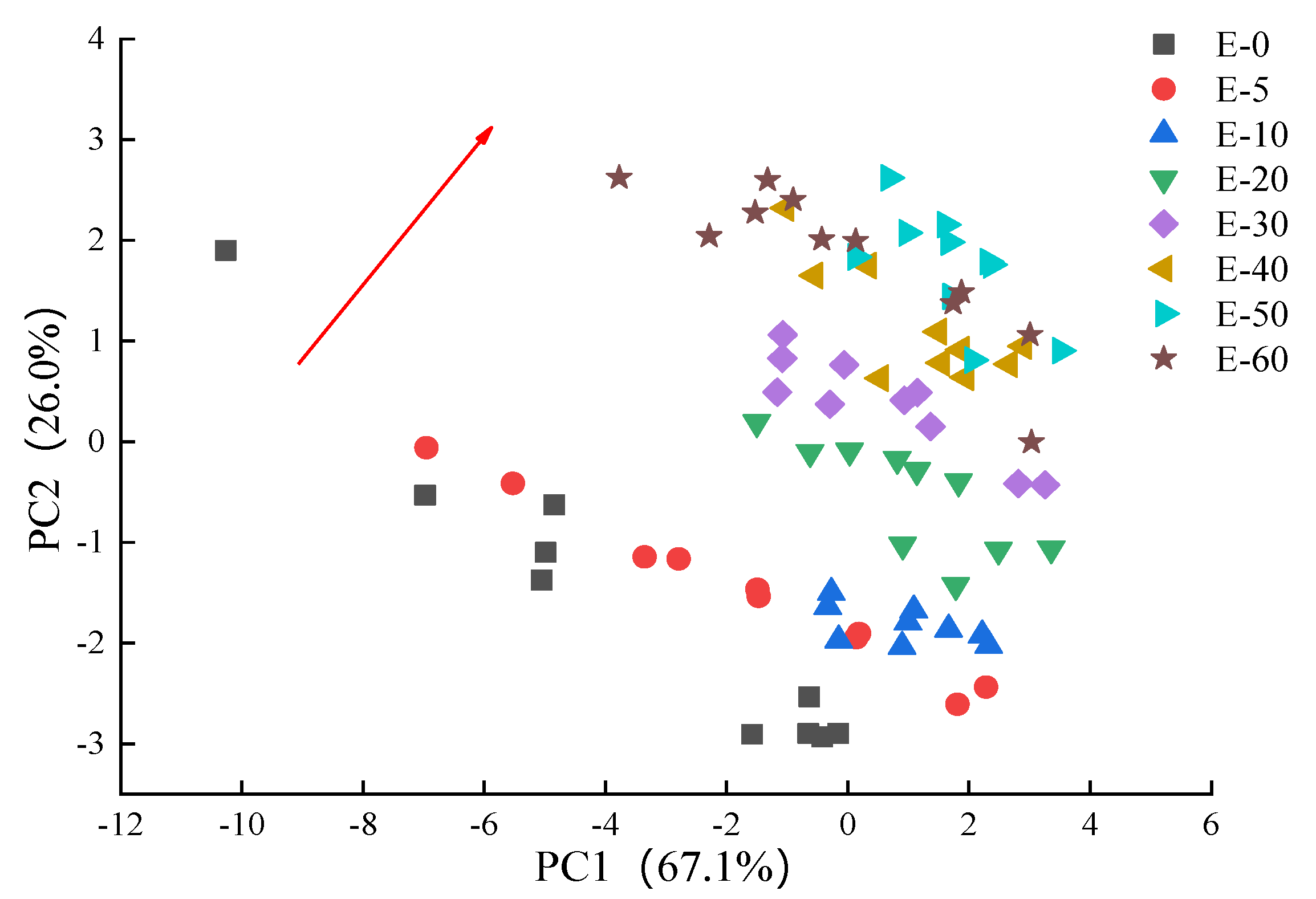
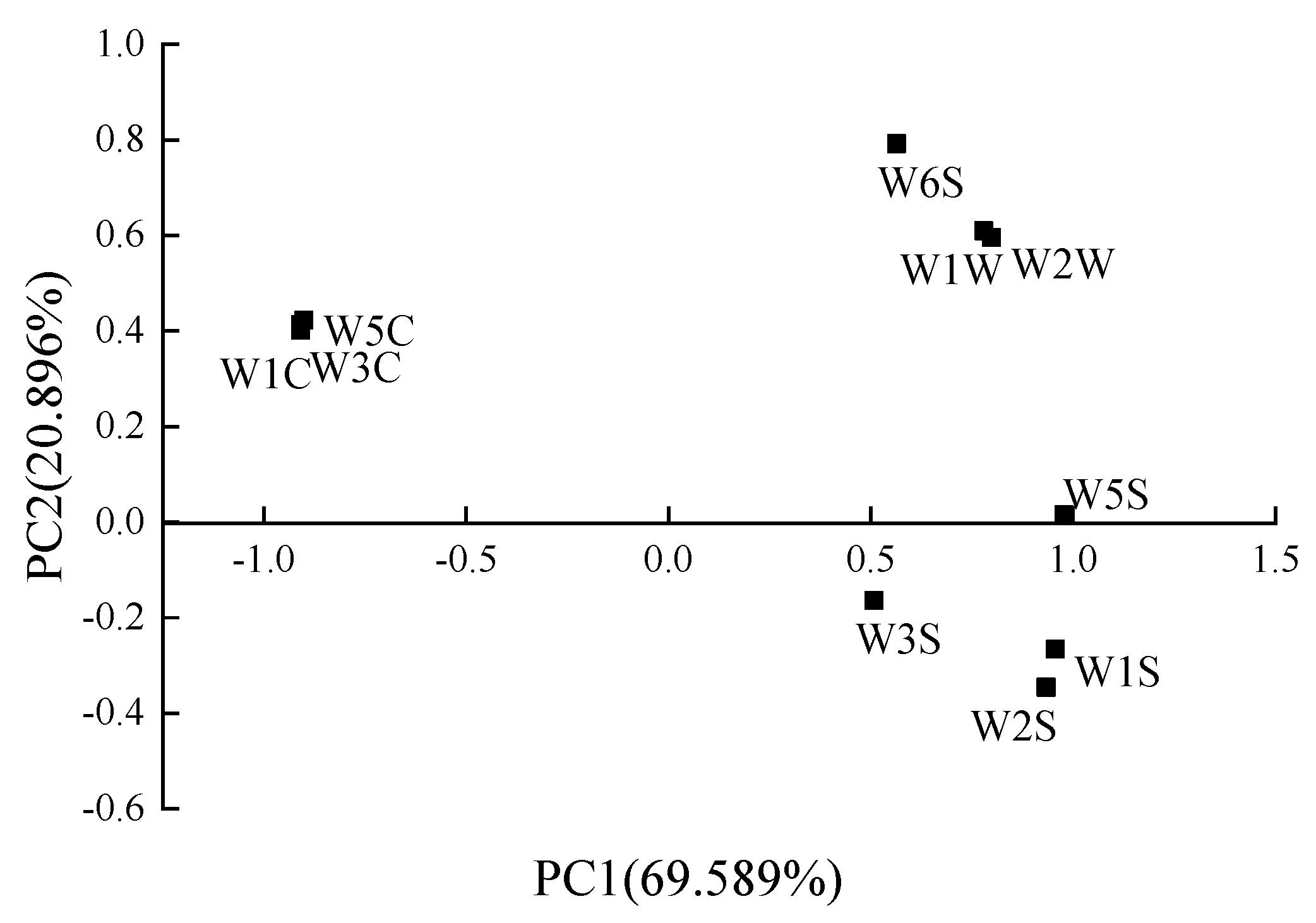
3.3.2. PCA and LDA Analysis
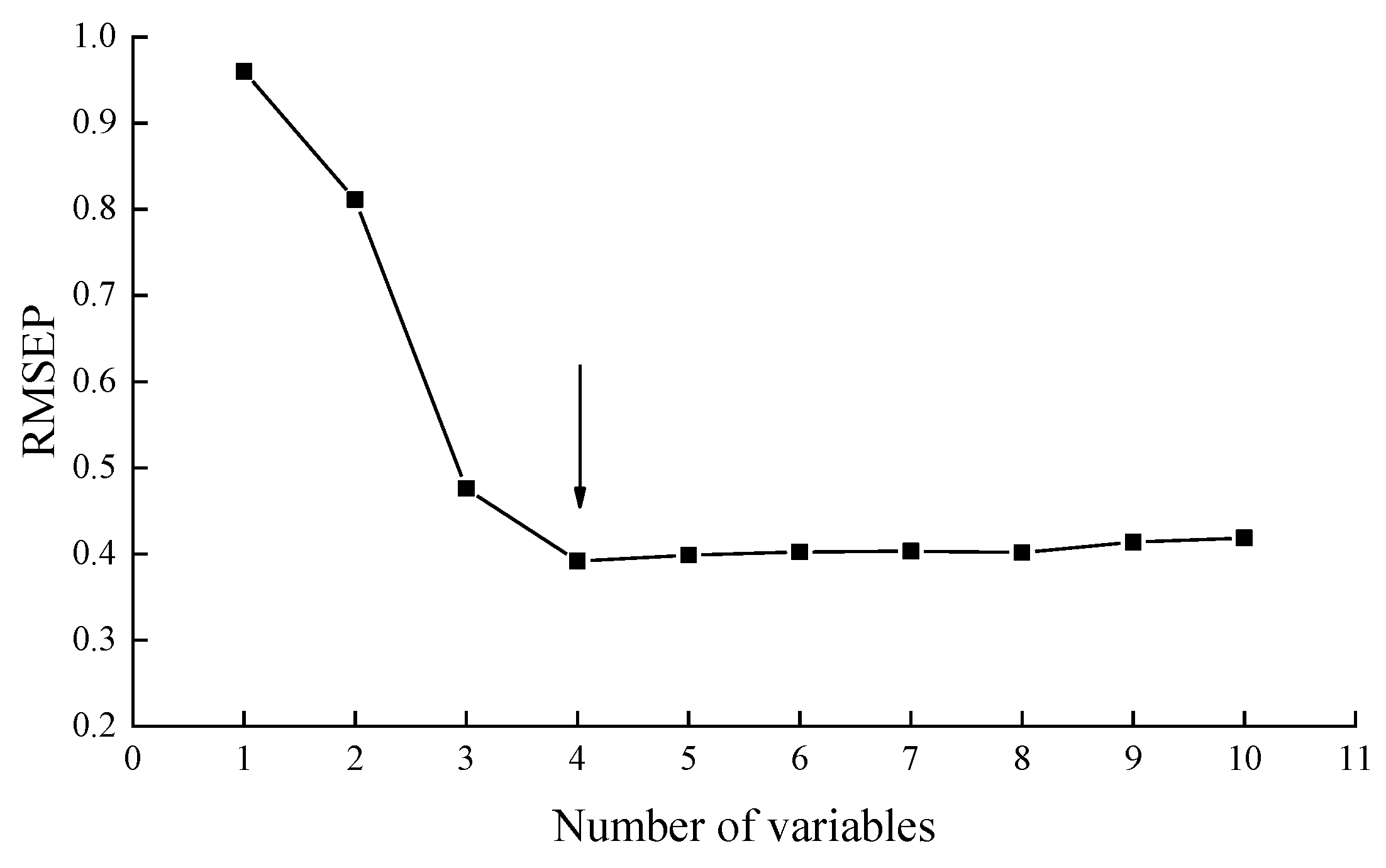
3.3.3. Featured Sensor Screening
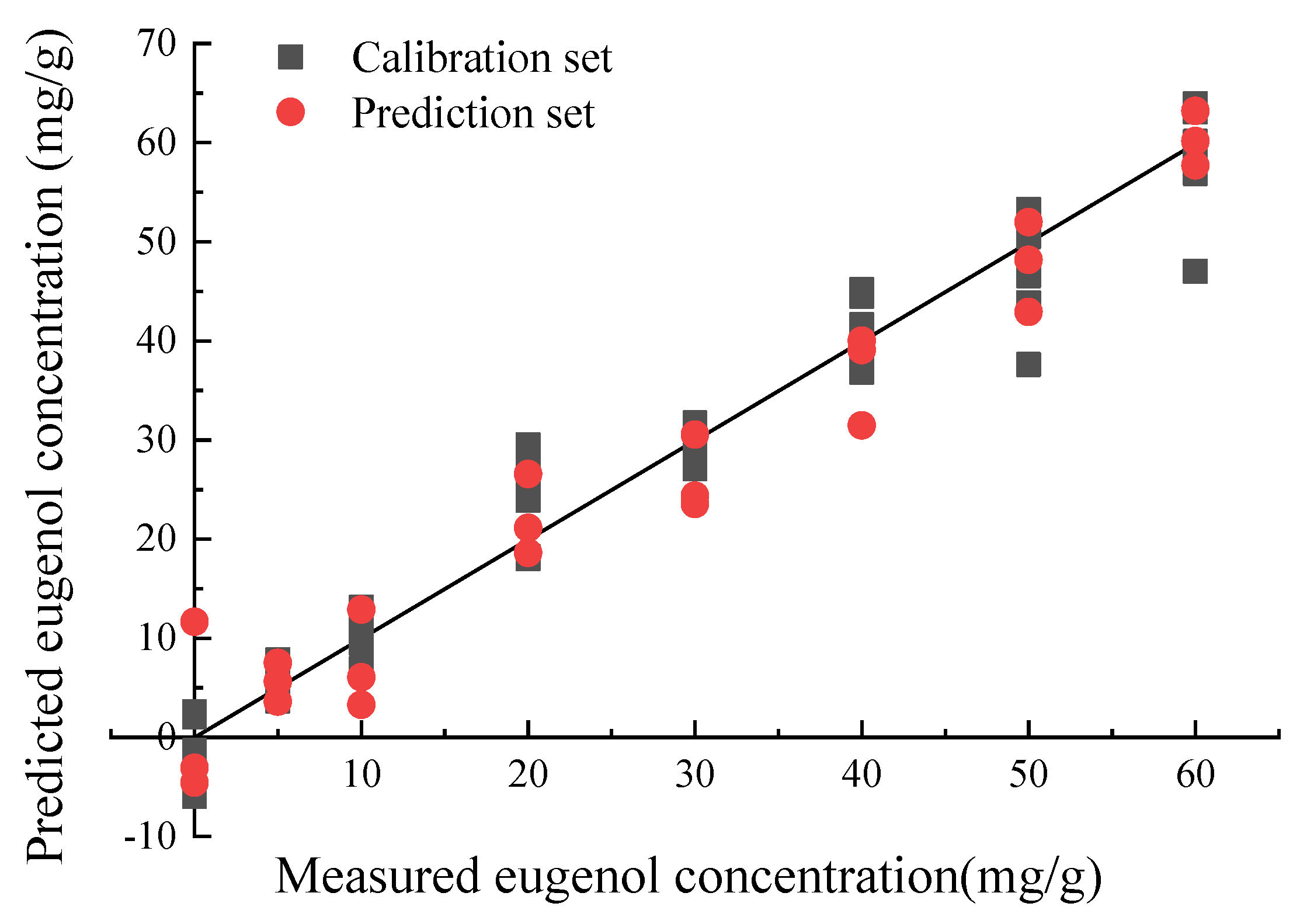
3.3.4. Quantitative Models
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhou, Y.; Xu, T.; Zhang, Y.; Zhang, C.; Lu, Z.; Lu, F.; Zhao, H. Effect of tea polyphenols on curdlan/chitosan blending film properties and its application to chilled meat preservation. Coatings 2019, 9, 262. [Google Scholar] [CrossRef]
- Alizadeh-Sani, M.; Ehsani, A.; Moghaddas Kia, E.; Khezerlou, A. Microbial gums: Introducing a novel functional component of edible coatings and packaging. Appl. Microbiol. Biotechnol. 2019, 103, 6853–6866. [Google Scholar] [CrossRef]
- Ahmad, M.; Nirmal, N.P.; Chuprom, J. Blend film based on fish gelatine/curdlan for packaging applications: Spectral, microstructural and thermal characteristics. RSC Adv. 2015, 5, 99044–99057. [Google Scholar] [CrossRef]
- Wang, K.; Du, L.; Zhang, C.; Lu, Z.; Lu, F.; Zhao, H. Preparation of chitosan/curdlan/carboxymethyl cellulose blended film and its characterization. J. Food Sci. Technol. 2019, 56, 5396–5404. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liu, Y.; Li, Y.; Lv, M.; Li, P.; Xu, H.; Wang, L. Preparation and characterization of novel curdlan/chitosan blending membranes for antibacterial applications. Carbohydr. Polym. 2011, 84, 952–959. [Google Scholar] [CrossRef]
- Wu, C.; Peng, S.; Wen, C.; Wang, X.; Fan, L.; Deng, R.; Pang, J. Structural characterization and properties of konjac glucomannan/curdlan blend films. Carbohydr. Polym. 2012, 89, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Dai, Q.; Guo, Y.; Wei, Y.; Wu, M.; Zhang, H. Glycyrrhiza polysaccharide-mediated synthesis of silver nanoparticles and their use for the preparation of nanocomposite curdlan antibacterial film. Int. J. Biol. Macromol. 2019, 141, 422–430. [Google Scholar] [CrossRef]
- Talón, E.; Vargas, M.; Chiralt, A.; González-Martínez, C. Antioxidant starch-based films with encapsulated eugenol. Application to sunflower oil preservation. LWT 2019, 113, 108290. [Google Scholar] [CrossRef]
- Talón, E.; Vargas, M.; Chiralt, A.; González-Martínez, C. Eugenol incorporation into thermoprocessed starch films using different encapsulating materials. Food Packag. Shelf Life 2019, 21, 100326. [Google Scholar] [CrossRef]
- Bonilla, J.; Poloni, T.; Lourenço, R.V.; Sobral, P.J.A. Antioxidant potential of eugenol and ginger essential oils with gelatin/chitosan films. Food Biosci. 2018, 23, 107–114. [Google Scholar] [CrossRef]
- Fernandes Nassar, S.; Dombre, C.; Gastaldi, E.; Touchaleaume, F.; Chalier, P. Soy protein isolate nanocomposite film enriched with eugenol, an antimicrobial agent: Interactions and properties. J. Appl. Polym. Sci. 2018, 135, 45941. [Google Scholar] [CrossRef]
- Cheng, J.; Wang, H.; Kang, S.; Xia, L.; Jiang, S.; Chen, M.; Jiang, S. An active packaging film based on yam starch with eugenol and its application for pork preservation. Food Hydrocoll. 2019, 96, 546–554. [Google Scholar] [CrossRef]
- Requena, R.; Vargas, M.; Chiralt, A. Eugenol and carvacrol migration from PHBV films and antibacterial action in different food matrices. Food Chem. 2019, 277, 38–45. [Google Scholar] [CrossRef]
- Yang, L.; Zhao, F.; Zeng, B. Electrochemical determination of eugenol using a three-dimensional molecularly imprinted poly (p-aminothiophenol-co-p-aminobenzoic acids) film modified electrode. Electrochim. Acta 2016, 210, 293–300. [Google Scholar] [CrossRef]
- Janes, S.E.; Price, C.S.; Thomas, D. Essential oil poisoning: N-acetylcysteine for eugenol-induced hepatic failure and analysis of a national database. Eur. J. Pediatr. 2005, 164, 520–522. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Feng, T.-T.; Wu, J.-H.; Du, M.; Qin, L.; Wang, Z.-Y.; Xu, X.-B. Vortex-assisted liquid-liquid micro-extraction followed by head space solid phase micro-extraction for the determination of eugenol in fish using GC-MS. Food Anal. Methods 2017, 11, 790–796. [Google Scholar] [CrossRef]
- Li, J.; Zhang, J.; Liu, Y. Optimization of solid-phase-extraction cleanup and validation of quantitative determination of eugenol in fish samples by gas chromatography-tandem mass spectrometry. Anal. Bioanal. Chem. 2015, 407, 6563–6568. [Google Scholar] [CrossRef]
- Shishov, A.; Gagarionova, S.; Bulatov, A. Deep eutectic mixture membrane-based microextraction: HPLC-FLD determination of phenols in smoked food samples. Food Chem. 2020, 314, 126097. [Google Scholar] [CrossRef]
- Saglam, O.; Dilgin, D.G.; Ertek, B.; Dilgin, Y. Differential pulse voltammetric determination of eugenol at a pencil graphite electrode. Mater. Sci. Eng. C Mater. Biol. Appl. 2016, 60, 156–162. [Google Scholar] [CrossRef]
- Afzali, D.; Zarei, S.; Fathirad, F.; Mostafavi, A. Gold nanoparticles modified carbon paste electrode for differential pulse voltammetric determination of eugenol. Mater. Sci. Eng. C Mater. Biol. Appl. 2014, 43, 97–101. [Google Scholar] [CrossRef]
- Voss, H.G.J.; Mendes Junior, J.J.A.; Farinelli, M.E.; Stevan, S.L., Jr. A prototype to detect the alcohol content of beers based on an electronic nose. Sensors (Basel) 2019, 19, 2646. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Wang, J.; Hong, X.; Wang, C. Fast determination of lycopene content and soluble solid content of cherry tomatoes using metal oxide sensors based electronic nose. Acta Aliment. 2016, 45, 182–189. [Google Scholar] [CrossRef]
- Suárez, J.I.; Arroyo, P.; Lozano, J.; Herrero, J.L.; Padilla, M. Bluetooth gas sensing module combined with smartphones for air quality monitoring. Chemosphere 2018, 205, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tian, F.; Nie, H.; Dang, L.; Li, G.; Ye, Q.; Kadri, C. Classification of multiple indoor air contaminants by an electronic nose and a hybrid support vector machine. Sens. Actuators B Chem. 2012, 174, 114–125. [Google Scholar] [CrossRef]
- Cui, S.; Wang, J.; Yang, L.; Wu, J.; Wang, X. Qualitative and quantitative analysis on aroma characteristics of ginseng at different ages using E-nose and GC-MS combined with chemometrics. J. Pharm. Biomed. Anal. 2015, 102, 64–77. [Google Scholar] [CrossRef]
- Zhang, W.; Pan, L.; Zhao, X.; Tu, K. A study on soluble solids content assessment using electronic nose: Persimmon fruit picked on different dates. Int. J. Food Prop. 2014, 19, 53–62. [Google Scholar] [CrossRef]
- Li, D.; Lei, T.; Zhang, S.; Shao, X.; Xie, C. A novel headspace integrated E-nose and its application in discrimination of Chinese medical herbs. Sens. Actuators B Chem. 2015, 221, 556–563. [Google Scholar] [CrossRef]
- Gu, D.-C.; Liu, W.; Yan, Y.; Wei, W.; Gan, J.-h.; Lu, Y.; Jiang, Z.-L.; Wang, X.-C.; Xu, C.-H. A novel method for rapid quantitative evaluating formaldehyde in squid based on electronic nose. LWT 2019, 101, 382–388. [Google Scholar] [CrossRef]
- Requena, R.; Vargas, M.; Chiralt, A. Release kinetics of carvacrol and eugenol from poly(hydroxybutyrate-co-hydroxyvalerate) (PHBV) films for food packaging applications. Eur. Polym. J. 2017, 92, 185–193. [Google Scholar] [CrossRef]
- Garrido-Miranda, K.A.; Rivas, B.L.; Pérez -Rivera, M.A.; Sanfuentes, E.A.; Peña-Farfal, C. Antioxidant and antifungal effects of eugenol incorporated in bionanocomposites of poly(3-hydroxybutyrate)-thermoplastic starch. LWT 2018, 98, 260–267. [Google Scholar] [CrossRef]
- Fan, J.; Zhang, W.; Zhou, T.; Zhang, D.; Zhang, D.; Zhang, L.; Wang, G.; Cao, F. Discrimination of malus taxa with different scent intensities using electronic nose and gas chromatography(-)mass spectrometry. Sensors (Basel) 2018, 18, 3429. [Google Scholar] [CrossRef] [PubMed]
- Long, Q.; Li, Z.; Han, B.; Gholam Hosseini, H.; Zhou, H.; Wang, S.; Luo, D. Discrimination of two cultivars of alpinia officinarum hance using an electronic nose and gas chromatography-mass spectrometry coupled with chemometrics. Sensors (Basel) 2019, 19, 572. [Google Scholar] [CrossRef]
- Huang, M.; Wang, Q.; Zhang, M.; Zhu, Q. Prediction of color and moisture content for vegetable soybean during drying using hyperspectral imaging technology. J. Food Eng. 2014, 128, 24–30. [Google Scholar] [CrossRef]
- Yang, Q.; Sun, D.-W.; Cheng, W. Development of simplified models for nondestructive hyperspectral imaging monitoring of TVB-N contents in cured meat during drying process. J. Food Eng. 2017, 192, 53–60. [Google Scholar] [CrossRef]
- Liu, C.; Liu, W.; Lu, X.; Chen, W.; Yang, J.; Zheng, L. Potential of multispectral imaging for real-time determination of colour change and moisture distribution in carrot slices during hot air dehydration. Food Chem. 2016, 195, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.-H.; Sun, D.-W.; Cheng, J.-H.; Pu, H. Mapping moisture contents in grass carp (Ctenopharyngodon idella) slices under different freeze drying periods by Vis-NIR hyperspectral imaging. LWT 2017, 75, 529–536. [Google Scholar] [CrossRef]
- Zhang, H.; Shao, W.; Qiu, S.; Wang, J.; Wei, Z. Collaborative analysis on the marked ages of rice wines by electronic tongue and nose based on different feature data sets. Sensors (Basel) 2020, 20, 1065. [Google Scholar] [CrossRef]
- Li, P.; Ren, Z.; Shao, K.; Tan, H.; Niu, Z. Research on distinguishing fish meal quality using different characteristic parameters based on electronic nose technology. Sensors (Basel) 2019, 19, 2146. [Google Scholar] [CrossRef]
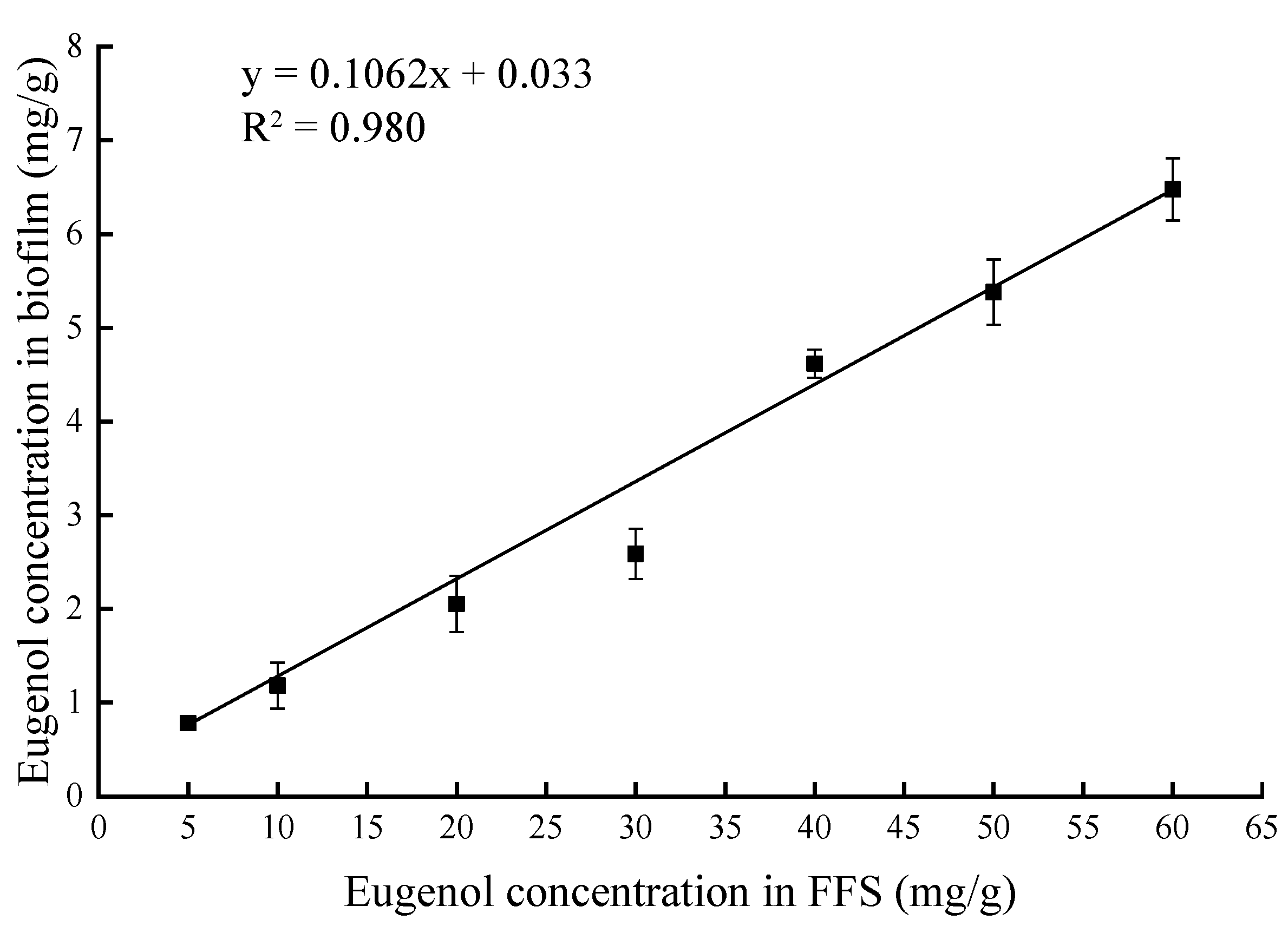



| Samples | Eugenol Concentration in FFS (mg/g) | Eugenol Concentration in Biofilm (mg/g) |
|---|---|---|
| E-5 | 5 | 0.782 ± 0.056 g |
| E-10 | 10 | 1.181 ± 0.245 f |
| E-20 | 20 | 2.053 ± 0.300 e |
| E-30 | 30 | 2.586 ± 0.268 d |
| E-40 | 40 | 4.617 ± 0.150 c |
| E-50 | 50 | 5.381 ± 0.349 b |
| E-60 | 60 | 6.476 ± 0.334 a |
| Peak No. | Volatile Compounds | Relative Concentration (Mean ± SD) % | |||||||
|---|---|---|---|---|---|---|---|---|---|
| E-0 | E-5 | E-10 | E-20 | E-30 | E-40 | E-50 | E-60 | ||
| Alkane | |||||||||
| 1 | Pentadecane, 2,6,10,14-tetramethyl- | 0.334 ± 0.029 b | 0.466 ± 0.045 a | 0.217 ± 0.031 c | 0.144 ± 0.007 d | 0.241 ± 0.010 c | 0.189 ± 0.003 cd | 0.231 ± 0.023 c | 0.184 ± 0.017 cd |
| 2 | Tetradecane | 0.543 ± 0.105 a | 0.207 ± 0.012 b | 0.191 ± 0.057 b | 0.107 ± 0.008 b | 0.130 ± 0.027 b | 0.134 ± 0.013 b | 0.095 ± 0.014 b | 0.131 ± 0.013 b |
| 3 | Hexadecane | 0.453 ± 0.189 a | 0.485 ± 0.212 a | 0.239 ± 0.016 ab | 0.279 ± 0.132 ab | 0.267 ± 0.045 ab | 0.187 ± 0.033 ab | 0.227 ± 0.006 ab | 0.150 ± 0.015 b |
| 4 | Dodecane | 0.494 ± 0.060 a | 0.165 ± 0.106 b | 0.119 ± 0.010bc | 0.031 ± 0.004 c | 0.045 ± 0.038 c | 0.027 ± 0.006 c | 0.018 ± 0.002 c | 0.025 ± 0.005 c |
| 5 | Eicosane | 0.476 ± 0.375 | 0.100 ± 0.034 | 0.139 ± 0.013 | N.D. | 0.040 ± 0.14 | 0.015 ± 0.004 | N.D. | 0.047 ± 0.025 |
| 6 | Heptadecane | 0.106 ± 0.006 b | 0.264 ± 0.067 a | 0.111 ± 0.018 b | 0.119 ± 0.025 b | 0.160 ± 0.001 b | 0.094 ± 0.002 b | 0.142 ± 0.044 b | 0.137 ± 0.014 b |
| 7 | Tridecane | 0.819 ± 0.004 a | 0.245 ± 0.048 b | 0.185 ± 0.023 c | 0.065 ± 0.007 d | 0.061 ± 0.032 d | 0.048 ± 0.001 d | 0.034 ± 0.007 d | 0.031 ± 0.002 d |
| 8 | Octadecane | 0.132 ± 0.010 a | 0.085 ± 0.031 b | 0.054 ± 0.011 bc | 0.042 ± 0.004 c | 0.058 ± 0.005 bc | 0.048 ± 0.017 c | 0.037 ± 0.012 c | 0.044 ± 0.005 c |
| 9 | Cyclopentadecane | 0.336 ± 0.014 | 0.181 ± 0.032 | N.D. | 0.046 ± 0.034 | N.D. | 0.018 ± 0.001 | 0.037 ± 0.003 | 0.026 ± 0.008 |
| ∑ | 2.796 ± 0.762 a | 2.198 ± 0.254 a | 1.185 ± 0.119 b | 0.834 ± 0.191 b | 0.982 ± 0.010 b | 0.744 ± 0.053 b | 0.802 ± 0.104 b | 0.762 ± 0.076 b | |
| Olefin | |||||||||
| 10 | 1-Octadecene | 0.318 ± 0.024 a | 0.215 ± 0.123 ab | 0.067 ± 0.005 b | 0.179 ± 0.025 ab | 0.263 ± 0.118 a | 0.072 ± 0.031 b | 0.070 ± 0.054 b | 0.083 ± 0.006 b |
| 11 | Naphthalene, 1,2,3,4-tetrahydro-1,6-dimethyl-4-(1-methylethyl)-, (1S-cis)- | 0.173 ± 0.068 ab | 0.195 ± 0.003 a | 0.129 ± 0.026 abc | 0.088 ± 0.026 bc | 0.123 ± 0.014 abc | 0.135 ± 0.048 abc | 0.071 ± 0.003 c | 0.051 ± 0.004 c |
| 12 | D-Limonene | 0.412 ± 0.072 | 0.186 ± 0.011 | 0.067 ± 0.005 | N.D. | N.D. | N.D. | N.D. | N.D. |
| 13 | Caryophyllene | N.D. | 0.050 ± 0.036 | 0.085 ± 0.020 | 0.022 ± 0.004 | 0.079 ± 0.026 | 0.063 ± 0.020 | 0.028 ± 0.002 | 0.134 ± 0.023 |
| 14 | 2-Tetradecene, (E)- | N.D. | 0.081 ± 0.019 | 0.059 ± 0.003 | N.D. | N.D. | N.D. | N.D. | N.D. |
| 15 | 1-Hexadecene | N.D. | 0.081 ± 0.005 | 0.130 ± 0.078 | 0.020 ± 0.003 | N.D. | N.D. | N.D. | N.D. |
| 16 | alpha-caryophyllene | N.D. | 0.205 ± 0.019 | 0.203 ± 0.008 | N.D. | N.D. | 0.224 ± 0.021 | N.D. | N.D. |
| 17 | Caryophyllene oxide | N.D. | N.D. | N.D. | 0.065 ± 0.012 | 0.149 ± 0.077 | 0.065 ± 0.016 | N.D. | 0.091 ± 0.002 |
| ∑ | 0.903 ± 0.060 a | 0.817 ± 0.083 ab | 0.742 ± 0.073 ab | 0.364 ± 0.053 de | 0.613 ± 0.155 bc | 0.445 ± 0.179 cd | 0.154 ± 0.038 e | 0.359 ± 0.036 de | |
| Aldehyde | |||||||||
| 18 | Heptanal | 1.192 ± 0.418 | 0.141 ± 0.176 | 0.064 ± 0.064 | N.D. | N.D. | N.D. | N.D. | N.D. |
| 19 | 2-Octenal, (E)- | 1.976 ± 0.521 a | 0.448 ± 0.210 b | 0.214 ± 0.028 b | 0.066 ± 0.014 b | 0.046 ± 0.012b | 0.048 ± 0.003b | 0.054 ± 0.013b | 0.022 ± 0.008b |
| 20 | Decanal | 0.488 ± 0.246 | 0.223 ± 0.065 | 0.160 ± 0.020 | 0.062 ± 0.007 | 0.035 ± 0.006 | N.D. | N.D. | 0.017 ± 0.002 |
| 21 | Nonanal | 1.379 ± 0.456 a | 0.566 ± 0.070 b | 0.470 ± 0.012 bc | 0.097 ± 0.024 bc | 0.052 ± 0.004 bc | 0.101 ± 0.015 bc | 0.086 ± 0.033 bc | 0.024 ± 0.003 c |
| 22 | 2-Nonenal, (E)- | 2.462 ± 0.657 a | 0.614 ± 0.212 b | 0.385 ± 0.078 b | 0.160 ± 0.039 b | 0.126 ± 0.055 b | 0.089 ± 0.014 b | 0.092 ± 0.023 b | 0.050 ± 0.002 b |
| ∑ | 7.497 ± 2.109 a | 1.994 ± 0.733 b | 1.293 ± 0.179 b | 0.385 ± 0.006 b | 0.259 ± 0.045 b | 0.237 ± 0.032 b | 0.233 ± 0.068 b | 0.095 ± 0.013 b | |
| Aromatic compound | |||||||||
| 23 | Eugenol | N.D. | 51.770 ± 9.310 | 77.086 ± 10.167 | 89.330 ± 1.268 | 86.498 ± 3.363 | 81.465 ± 6.145 | 88.700 ± 1.010 | 86.952 ± 0.909 |
| 24 | Benzene, 1,2-dimethoxy-4-(2-propenyl)- | 0.192 ± 0.056 b | 0.136 ± 0.014 b | 0.209 ± 0.001 b | 0.242 ± 0.037 b | 0.419 ± 0.087 a | 0.423 ± 0.025 a | 0.164 ± 0.026 b | 0.221 ± 0.063 b |
| 25 | Phenol, 2-methoxy-4-(1-propenyl)-, (E)- | 2.586 ± 1.683 a | 2.966 ± 0.140 a | 3.461 ± 0.216 a | 2.873 ± 0.230 a | 2.456 ± 0.015 a | 2.469 ± 0.409 a | 2.076 ± 0.212 a | 1.876 ± 0.007 a |
| 26 | Naphthalene | 0.137 ± 0.004 a | 0.070 ± 0.024 b | 0.035 ± 0.010 c | 0.032 ± 0.003 c | 0.039 ± 0.023 c | 0.021 ± 0.003 c | 0.019 ± 0.003 c | 0.027 ± 0.013 c |
| 27 | 1,1’-Biphenyl, 2,2’,5,5’-tetramethyl- | N.D. | N.D. | N.D. | N.D. | N.D. | 0.048 ± 0.002 | 0.088 ± 0.004 | N.D. |
| ∑ | 2.824 ± 1.568 c | 53.459 ± 11.417 b | 80.773 ± 9.974 a | 92.477 ± 1.463 a | 88.184 ± 8.461 a | 84.425 ± 6.584 a | 91.005 ± 1.132 a | 89.076 ± 0.841 a | |
| Modeling Method | Number of Variables | R2c | R2p | Root Mean Square Error of Prediction (RMSEP)/(mg/g) | Residual Predictive Deviation (RPD) |
|---|---|---|---|---|---|
| PLS | 10 | 0.962 | 0.952 | 4.612 | 4.530 |
| 4 | 0.955 | 0.948 | 4.706 | 4.440 | |
| SVM | 10 | 0.998 | 0.897 | 6.613 | 3.159 |
| 4 | 0.990 | 0.905 | 6.327 | 3.302 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, L.; Zhu, J.; Fan, X.; Zhang, C.; Tu, K.; Peng, J.; Wang, J.; Pan, L. Rapid Non-Destructive Quantification of Eugenol in Curdlan Biofilms by Electronic Nose Combined with Gas Chromatography-Mass Spectrometry. Sensors 2020, 20, 4441. https://doi.org/10.3390/s20164441
Han L, Zhu J, Fan X, Zhang C, Tu K, Peng J, Wang J, Pan L. Rapid Non-Destructive Quantification of Eugenol in Curdlan Biofilms by Electronic Nose Combined with Gas Chromatography-Mass Spectrometry. Sensors. 2020; 20(16):4441. https://doi.org/10.3390/s20164441
Chicago/Turabian StyleHan, Lu, Jingyi Zhu, Xia Fan, Chong Zhang, Kang Tu, Jing Peng, Jiahong Wang, and Leiqing Pan. 2020. "Rapid Non-Destructive Quantification of Eugenol in Curdlan Biofilms by Electronic Nose Combined with Gas Chromatography-Mass Spectrometry" Sensors 20, no. 16: 4441. https://doi.org/10.3390/s20164441
APA StyleHan, L., Zhu, J., Fan, X., Zhang, C., Tu, K., Peng, J., Wang, J., & Pan, L. (2020). Rapid Non-Destructive Quantification of Eugenol in Curdlan Biofilms by Electronic Nose Combined with Gas Chromatography-Mass Spectrometry. Sensors, 20(16), 4441. https://doi.org/10.3390/s20164441