Ellagic Acid and Urolithins A and B Differentially Regulate Fat Accumulation and Inflammation in 3T3-L1 Adipocytes While Not Affecting Adipogenesis and Insulin Sensitivity
Abstract
1. Introduction
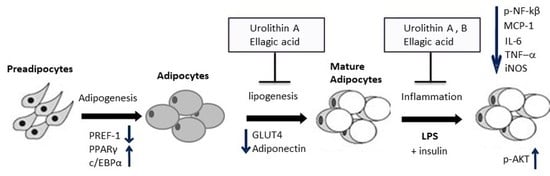
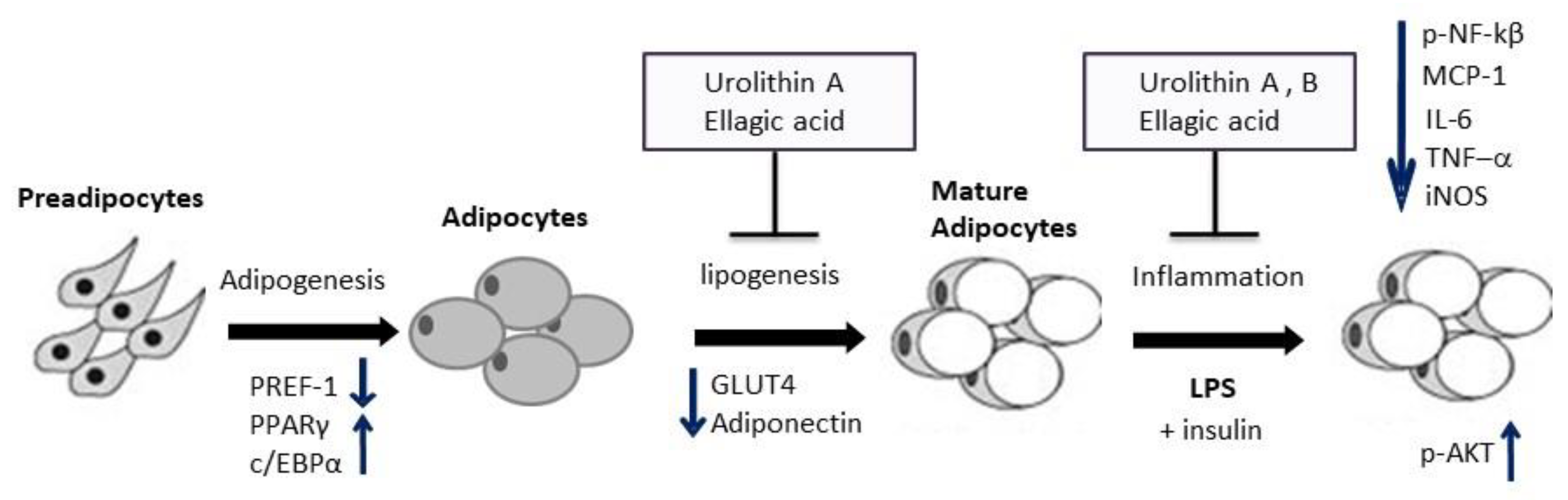
2. Results and Discussion
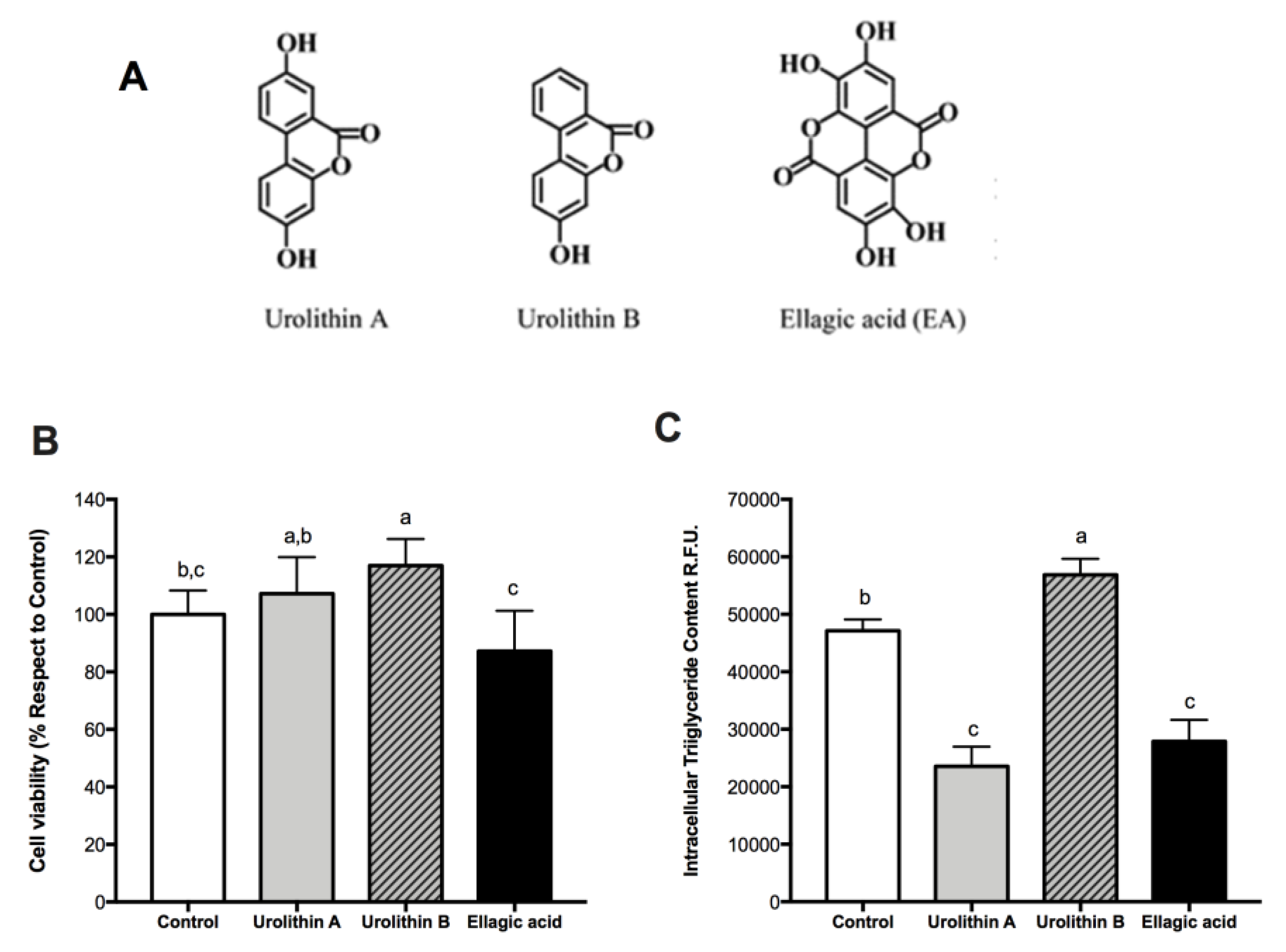
2.1. Effect of Ellagic Acid and Urolithins A and B on Cell Viability
2.2. Effect of Ellagic Acid and Urolithins A and B on Intracellular Triglyceride Content
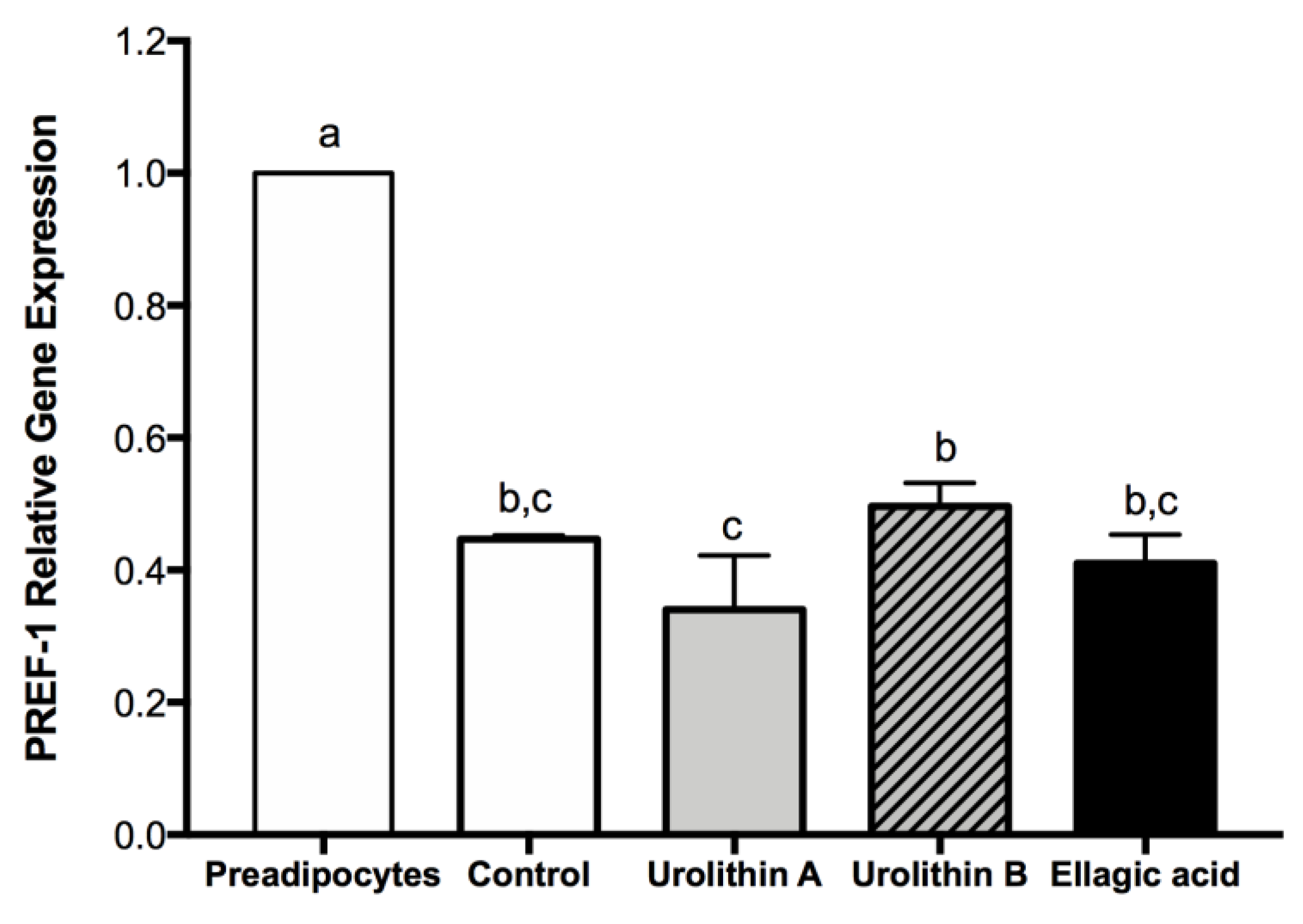
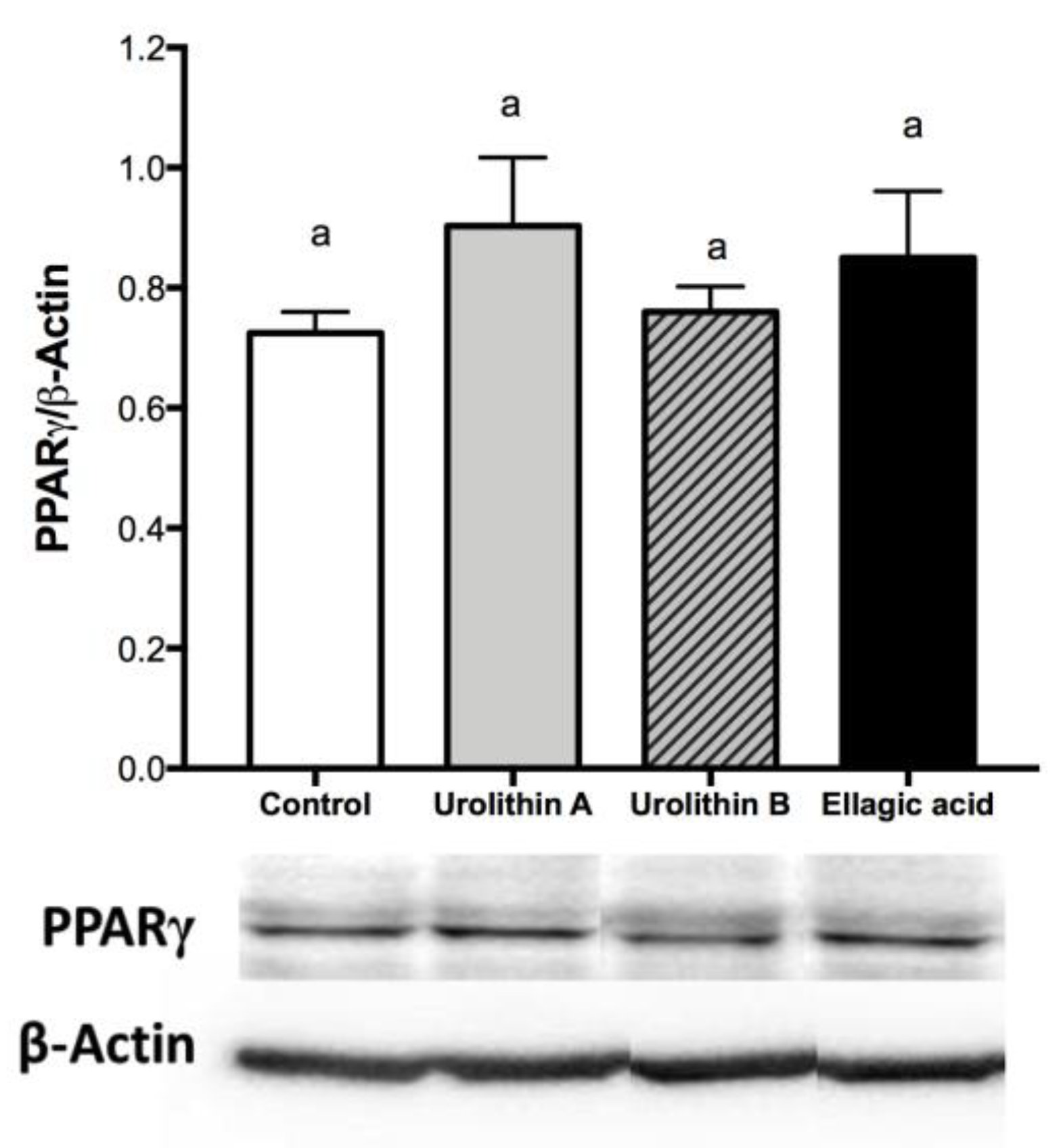
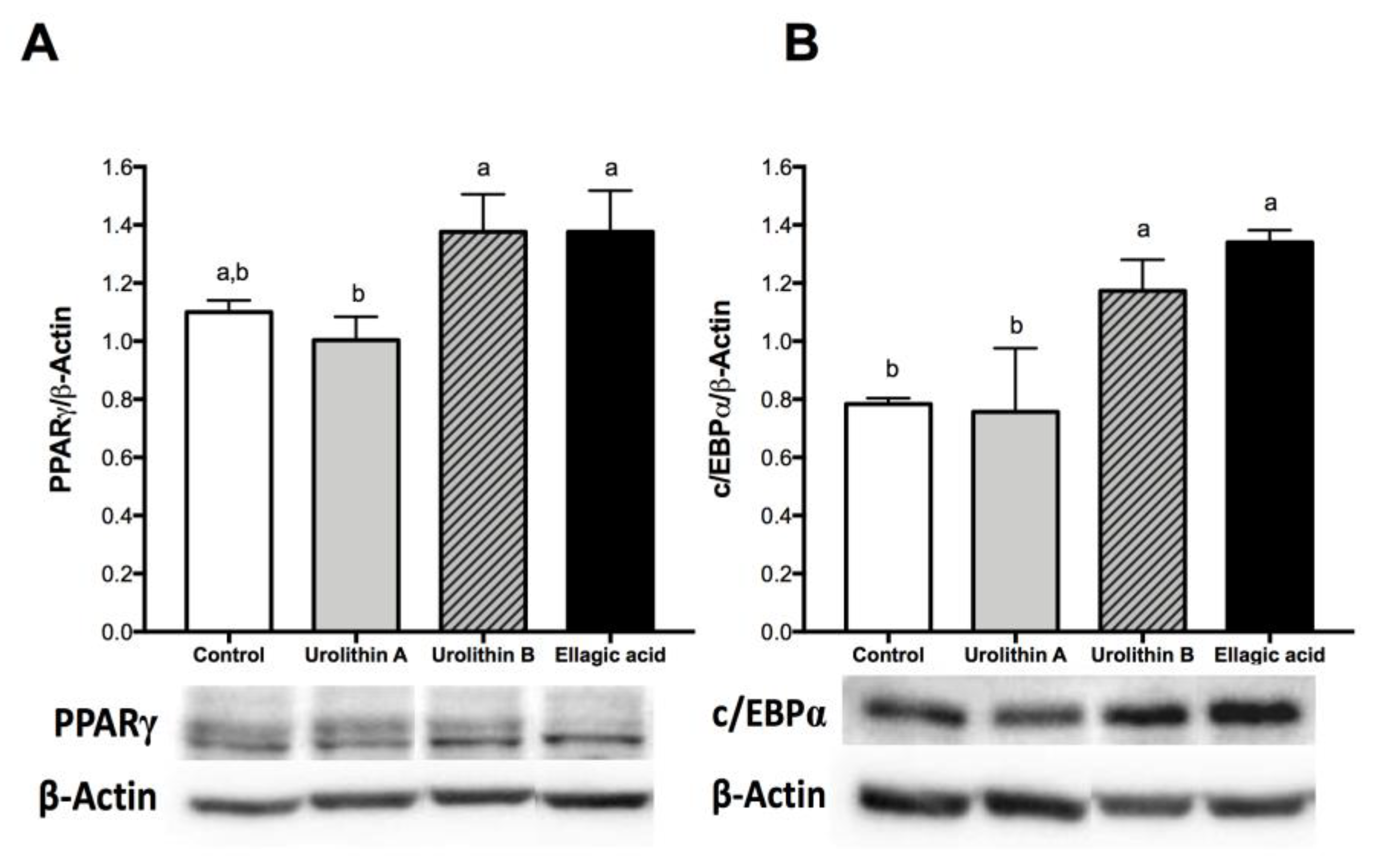
2.3. Effect of Ellagic Acid and Urolithins A and B on Transcription Factors Related to Adipocyte Differentiation
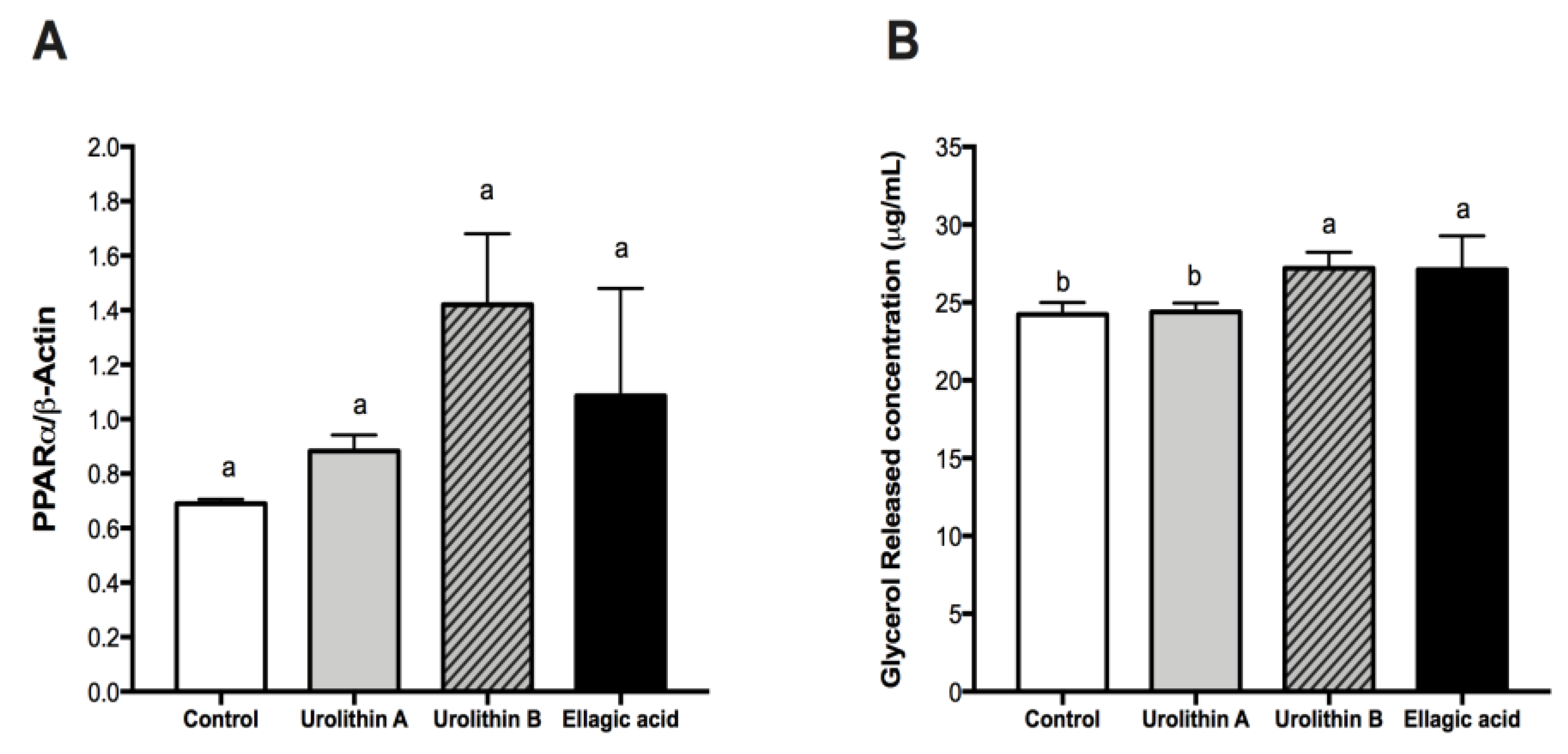
2.4. Effect of Ellagic Acid and Urolithins A and B on PPARα and Glycerol Release
2.5. Effect of Ellagic Acid and Urolithins A and B on Adipokines, Lipogenesis, and Lipolysis
2.6. Effect of Ellagic Acid and Urolithins A and B on LPS-Induced Inflammation and Insulin Sensitivity
3. Materials and Methods
3.1. Chemicals
3.2. Cell Culture
3.3. Cell Viability Assay
3.4. Cell Differentiation and Treatments for Adipogenesis Assays
3.5. Quantification of Lipid Content
3.6. Quantification of Glycerol Release
3.7. Ellagic Acid and Urolithins A and B on LPS-Induced Inflammation and Insulin Sensitivity
3.8. Western Blot Analysis
3.9. Real-Time Quantitative RT-PCR
3.10. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ACC1 | acetyl-CoA carboxylase 1 |
| ACC2 | acetyl-CoA carboxylase 2 |
| AKT | protein kinase B |
| AP1 | activator protein 1 |
| AP2 | adipocyte protein 2 |
| ATGL | adipose triglyceride lipase |
| c/EBPα | CCAAT-enhancer-binding protein alpha |
| COX-2 | cyclooxygenase 2 |
| DMSO | dimethyl sulfoxide |
| EA | ellagic acid |
| FASN | fatty acid synthase |
| GLUT4 | glucose transporter type 4 |
| HSL | hormone sensitive lipase |
| IL-6 | interleukin 6 |
| iNOS | inducible nitric oxide synthase |
| LPS | lipopolysaccharide |
| MCP-1 | monocyte chemoattractant protein-1 |
| NF-κB | nuclear factor κB |
| p-AKT | phosphorylated protein kinase B |
| p-NF-κB | phosphorylated nuclear factor κB |
| PPARα | peroxisome proliferator-activated receptor alpha |
| PPARγ | peroxisome proliferator-activated receptor gamma |
| SCD1 | stearoyl-CoA desaturase |
| TNFα | tumor necrosis factor alpha |
| Uro | urolithin |
References
- Marwan, A.G.; Nagel, C.W. Characterization of cranberry benzoates and their antimicrobial properties. J. Food Sci. 1986, 51, 1069–1070. [Google Scholar] [CrossRef]
- Daniel, E.M.; Krupnick, A.S.; Heur, Y.H.; Blinzler, A.J.; Nims, R.W.; Stoner, G.D. Extraction, stability and quantitation of ellagic acid in various fruits and nuts. J. Food Compos. Anal. 1989, 2, 338–349. [Google Scholar] [CrossRef]
- Chen, H.; Zuo, Y.; Deng, Y. Separation and determination of flavonoids and other phenolic compounds in cranberry juice by high-performance liquid chromatography. J. Chromatogr. A 2001, 913, 387–395. [Google Scholar] [CrossRef]
- Espín, J.C.; Larrosa, M.; García-Conesa, M.T.; Tomás-Barberán, F. Biological significance of urolithins, the gut microbial ellagic acid-derived metabolites: The evidence so far. Evid-Based Compl. Alt. 2013, 2013, 270418. [Google Scholar] [CrossRef] [PubMed]
- Cerdá, B.; Espín, J.C.; Parra, S.; Martínez, P.; Tomás-Barberán, F.A. The potent in vitro antioxidant ellagitannins from pomegranate juice are metabolized into bioavailable but poor antioxidant hydroxy-6H-dibenzopyran-6-one derivatives by the colonic microflora of healthy humans. Eur. J. Nutr. 2004, 43, 205–220. [Google Scholar]
- Pfundstein, B.; Haubner, R.; Würtele, G.; Gehres, N.; Cornelia, M.; Owen, R.W.U. Pilot walnut intervention study of urolithin bioavailability in human volunteers. J. Agric. Food Chem. 2014, 62, 10264–10273. [Google Scholar] [CrossRef]
- Tomás-Barberán, F.A.; González-Sarrías, A.; García-Villalba, R.; Núñez-Sánchez, M.A.; Selma, M.V.; García-Conesa, M.T.; Espín, J.C. Urolithins, the rescue of “old” metabolites to understand a “new” concept: Metabotypes as a nexus among phenolic metabolism, microbiota dysbiosis, and host health status. Mol. Nutr. Food Res. 2017, 61, 1500901. [Google Scholar] [CrossRef]
- Seeram, N.P.; Aronson, W.J.; Zhang, Y.; Henning, S.M.; Moro, A.; Lee, R.; Sartippour, M.; Harris, D.M.; Rettig, M.; Suchard, M.A.; et al. Pomegranate ellagitannin-derived metabolites inhibit prostate cancer growth and localize to the mouse prostate gland. J. Agric. Food Chem. 2007, 55, 7732–7737. [Google Scholar] [CrossRef]
- Larrosa, M.; Tomás-Barberán, F.A.; Espín, J.C. The dietary hydrolysable tannin punicalagin releases ellagic acid that induces apoptosis in human colon adenocarcinoma Caco-2 cells by using the mitochondrial pathway. J. Nutr. Biochem. 2006, 17, 611–625. [Google Scholar] [CrossRef]
- Larrosa, M.; González-Sarras, A.; García-Conesa, M.T.; Tomás-Barberán, F.A.; Espín, J.C. Urolithins, ellagic-acid derived metabolites produced by human colonic microflora, exhibit estrogenic and antiestrogenic activities. J. Agric. Food Chem. 2006, 54, 1611–1620. [Google Scholar] [CrossRef]
- Bialonska, D.; Kasimsetty, S.G.; Khan, S.I.; Ferreira, D. Urolithins, intestinal microbial metabolites of pomegranate ellagitannins, exhibit potent antioxidant activity in a cell-based assay. J. Agric. Food Chem. 2010, 57, 10181–10186. [Google Scholar] [CrossRef] [PubMed]
- González-Sarrías, A.; Larrosa, M.; Tomás-Barberán, F.A.; Dolara, P.; Espín, J.C. NF-κB dependent anti-inflammatory activity of urolithins, gut microbiota ellagic acid-derived metabolites, in human colonic fibroblasts. Brit. J. Nutr. 2010, 26, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Mele, L.; Mena, P.; Piemontese, A.; Marino, V.; López-Gutiérrez, N.; Bernini, F.; Brighenti, F.; Zanotti, I.; Del Rio, D. Antiatherogenic effects of ellagic acid and urolithins in vitro. Arch. Biochem. Biophys. 2016, 599, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.; Ma, H.; Liu, W.; Niesen, D.B.; Shah, N.; Crews, R.; Rose, K.N.; Vattem, D.A.; Seeram, N.P. Pomegranate’s neuroprotective effects against Alzheimer’s disease are mediated by urolithins, its ellagitannin-gut microbial derived metabolites. ACS Chem. Neurosci. 2016, 20, 26–33. [Google Scholar] [CrossRef]
- Hotamisligil, G.S. Inflammation and metabolic disorders. Nature 2006, 444, 860–867. [Google Scholar] [CrossRef]
- Green, H.; Kehinde, O. An established preadipose cell line and its differentiation in culture II. Factors affecting adipose conversion. Cell 1975, 5, 19–27. [Google Scholar] [CrossRef]
- Wong-a-nan, N.; Inthanon, K.; Saiai, A.; Inta, A.; Nimlamool, W.; Chomdej, S.; Kittakoop, P.; Wongkham, W. Lipogenesis inhibition and adipogenesis regulation via PPARγ pathway in 3T3-L1 cells by Zingiber cassumunar Roxb. rhizome extracts. EJBAS 2018, 5, 289–297. [Google Scholar] [CrossRef]
- Rosen, E.D.; Hsu, C.H.; Wang, X.; Sakai, S.; Freeman, M.W.; Gonzalez, F.J.; Spiegelman, B.M. C/EBPalpha induces adipogenesis through PPARgamma: A unified pathway. Genes Dev. 2002, 16, 22–26. [Google Scholar] [CrossRef]
- El-Jack, A.K.; Hamm, J.K.; Pilch, P.F.; Farmer, S.R. Reconstitution of insulin-sensitive glucose transport in fibroblasts requires expression of both PPARγ and C/EBPα. J. Biol. Chem. 1999, 274, 7946–7951. [Google Scholar] [CrossRef]
- Wu, Z.; Rosen, E.D.; Brun, R.; Hauser, S.; Adelmont, G.; Troy, A.E.; McKeon, C.; Darlington, G.J.; Spiegelman, B.M. Cross-regulation of C/EBPα and PPARγ controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol. Cell 1999, 3, 151–158. [Google Scholar] [CrossRef]
- Cabrero, A.; Alegret, M.; Sánchez, R.M.; Adzet, T.; Laguna, J.C.; Vázquez, M. Bezafibrate reduces mRNA levels of adipocyte markers and increases fatty acid oxidation in primary culture of adipocytes. Diabetes 2001, 50, 1883–1890. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, M.; Roglans, N.; Cabrero, A.; Rodríguez, C.; Adzet, T.; Alegret, M.; Sanchez, R.M.; Laguna, J.C. Bezafibrate induces Acyl-CoA oxidase mRNA levels and fatty acid peroxisomal beta-oxidation in rat white adipose tissue. Mol. Cell Biochem. 2001, 216, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Cisneros-Zevallos, L.; Bang, W.Y. Modulation of Adipogenesis and Inflammation in Adipocytes by Ellagic Acid and Metabolites Urolithins A and B; AGFD 93, American Chemical Society Annual Meeting Abstracts, Cornucopia; American Chemical Society: Washington, DC, USA, 2013; Volume 45, Available online: http://agfd.sites.acs.org/cornucopia13F.pdf (accessed on 10 March 2020).
- Kaestner, K.H.; Christy, R.J.; Lane, M.D. CCAAT/enhancer binding protein gene promoter: Binding of nuclear factors during differentiation of 3T3-L1 preadipocytes. Proc. Natl. Acad. Sci. USA 1991, 88, 2593–2597. [Google Scholar]
- Wu, Z.; Xie, Y.; Morrison, R.F.; Bucher, N.L.R.; Farmer, S.R. PPARγ induces the insulin-dependent glucose transporter GLUT4 in the absence of C/EBPα during the conversion of 3T3 fibroblasts into adipocytes. J. Clin. Invest. 1998, 101, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Armoni, M.; Kritz, N.; Harel, C.; Bar-Yoseph, F.; Chen, H.; Quon, M.J.; Karnieli, E. Peroxisome proliferator-activated receptor-γ represses GLUT4 promoter activity in primary adipocytes, and rosiglitazone alleviates this effect. J. Biol. Chem. 2003, 278, 30614–30623. [Google Scholar] [CrossRef] [PubMed]
- Madsen, M.S.; Siersbaek, R.; Boergesen, M.; Nielsen, R.; Mandrup, S. Peroxisome proliferator-activated receptor γ and C/EBPα synergistically activate key metabolic adipocyte genes by assisted loading. Mol. Cell Biol. 2014, 34, 939–954. [Google Scholar] [CrossRef]
- Park, S.K. Raspberry ketone, a naturally occurring phenolic compound, inhibits adipogenic and lipogenic gene expression in 3T3-L1 adipocytes. Pharm. Biol. 2015, 53, 870–875. [Google Scholar] [CrossRef]
- Goto, T.; Lee, J.Y.; Teraminami, A.; Kim, Y.; Hirai, S.; Uemura, T.; Inoue, H.; Takahashi, N.; Kawada, T. Activation of peroxisome proliferator-activated receptor-alpha stimulates both differentiation and fatty acid oxidation in adipocytes. J. Lipid Res. 2011, 52, 873–884. [Google Scholar] [CrossRef]
- Wang, W.; Lin, Q.; Lin, R.; Zhang, J.; Ren, F.; Zhang, J.; Ji, M.; Li, Y. PPARα agonist fenofibrate attenuates TNF-α-induced CD40 expression in 3T3-L1 adipocytes via the SIRT1-dependent signaling pathway. Expl. Cell Res. 2013, 319, 1523–1533. [Google Scholar] [CrossRef]
- Guerre-Millo, M.; Gervois, P.; Raspé, E.; Madsen, L.; Poulain, P.; Derudas, B.; Herbert, J.-M.; Winegar, D.A.; Willson, T.M.; Fruchart, J.-C.; et al. Peroxisome proliferator-activated receptor α activators improve insulin sensitivity and reduce adiposity. J. Biol. Chem. 2000, 275, 16638–16642. [Google Scholar] [CrossRef]
- Fu, Y.; Luo, N.; Klein, R.L.; Garvey, W.T. Adiponectin promotes adipocyte differentiation, insulin sensitivity, and lipid accumulation. J. Lipid Res. 2005, 46, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, A.; Yamauchi, T.; Takekawa, S.; Hada, Y.; Ito, Y.; Maki, T.; Kadowaki, T. Peroxisome proliferator–activated receptor (PPAR) α activation increases adiponectin receptors and reduces obesity-related inflammation in adipose tissue comparison of activation of PPAR α, PPAR γ, and their combination. Diabetes 2005, 54, 3358–3370. [Google Scholar] [CrossRef] [PubMed]
- Kang, I.; Kim, Y.; Tomás-Barberán, F.A.; Espín, J.C.; Chung, S. Urolithin A, C, and D, but not iso-urolithin A and urolithin B. attenuate triglyceride accumulation in human cultures of adipocytes and hepatocytes. Mol. Nutr. Food Res. 2016, 60, 1129–1138. [Google Scholar] [CrossRef] [PubMed]
- Cortés-Martín, A.; García-Villalba, R.; González-Sarrías, A.; Romo-Vaquero, M.; Loria-Kohen, V.; Ramírez-de-Molina, A.; Tomás-Barberán, F.A.; Selma, M.V.; Espín, J.C. The gut microbiota urolithin metabotypes revisited: The human metabolism of ellagic acid is mainly determined by aging. Food Funct. 2018, 9, 4100–4106. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, Y.; Nishii, S.; Zaima, N.; Moriyama, T.; Kawamura, Y. Ellagic acid improves hepatic steatosis and serum lipid composition through reduction of serum resistin levels and transcriptional activation of hepatic PPARα in obese, diabetic KK-A(y) mice. Biochem. Biophys. Res. Commun. 2013, 434, 486–491. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of the relative gene expression data using real-time quantitative PCR and the 2-DDCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ortiz-Quesda, A.G.; Lombardini, L.; Cisneros-Zevallos, L. Antioxidants in pecan nut cultivars [Carya illinoinensis (Wangenh.) K. Koch]. In Nuts and Seeds in Health and Disease Prevention; Preedy, V., Watson, R., Eds.; Academic Press: Cambridge, MA, USA, 2011; Chapter 104; pp. 881–889. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cisneros-Zevallos, L.; Bang, W.Y.; Delgadillo-Puga, C. Ellagic Acid and Urolithins A and B Differentially Regulate Fat Accumulation and Inflammation in 3T3-L1 Adipocytes While Not Affecting Adipogenesis and Insulin Sensitivity. Int. J. Mol. Sci. 2020, 21, 2086. https://doi.org/10.3390/ijms21062086
Cisneros-Zevallos L, Bang WY, Delgadillo-Puga C. Ellagic Acid and Urolithins A and B Differentially Regulate Fat Accumulation and Inflammation in 3T3-L1 Adipocytes While Not Affecting Adipogenesis and Insulin Sensitivity. International Journal of Molecular Sciences. 2020; 21(6):2086. https://doi.org/10.3390/ijms21062086
Chicago/Turabian StyleCisneros-Zevallos, Luis, Woo Young Bang, and Claudia Delgadillo-Puga. 2020. "Ellagic Acid and Urolithins A and B Differentially Regulate Fat Accumulation and Inflammation in 3T3-L1 Adipocytes While Not Affecting Adipogenesis and Insulin Sensitivity" International Journal of Molecular Sciences 21, no. 6: 2086. https://doi.org/10.3390/ijms21062086
APA StyleCisneros-Zevallos, L., Bang, W. Y., & Delgadillo-Puga, C. (2020). Ellagic Acid and Urolithins A and B Differentially Regulate Fat Accumulation and Inflammation in 3T3-L1 Adipocytes While Not Affecting Adipogenesis and Insulin Sensitivity. International Journal of Molecular Sciences, 21(6), 2086. https://doi.org/10.3390/ijms21062086