Alpha-Glucosidase- and Lipase-Inhibitory Phenalenones from a New Species of Pseudolophiostoma Originating from Thailand
Abstract
:1. Introduction
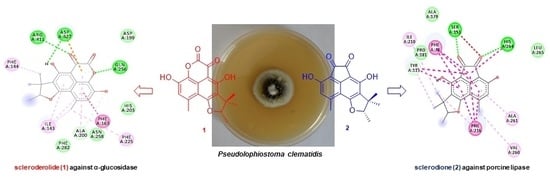
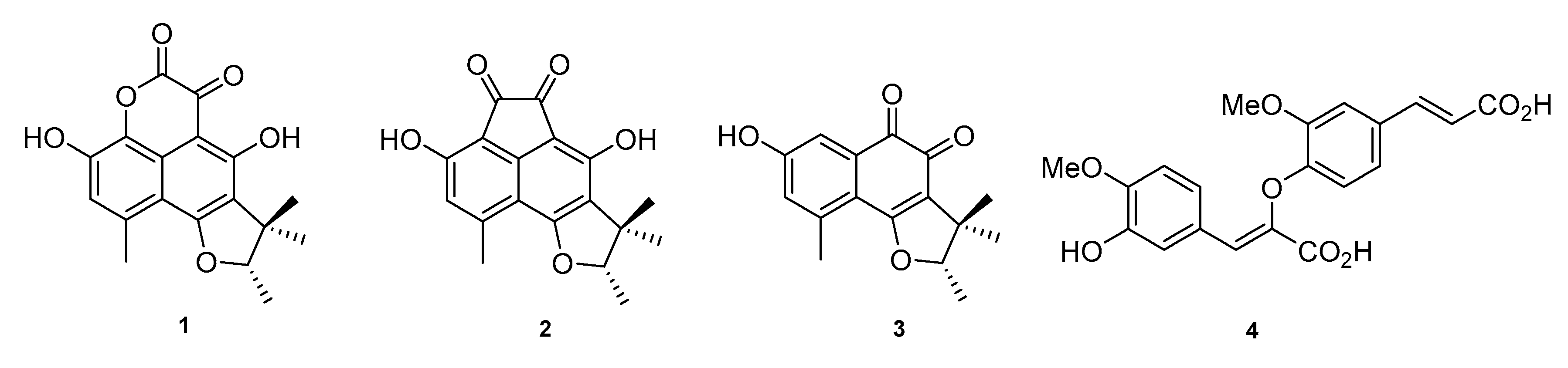
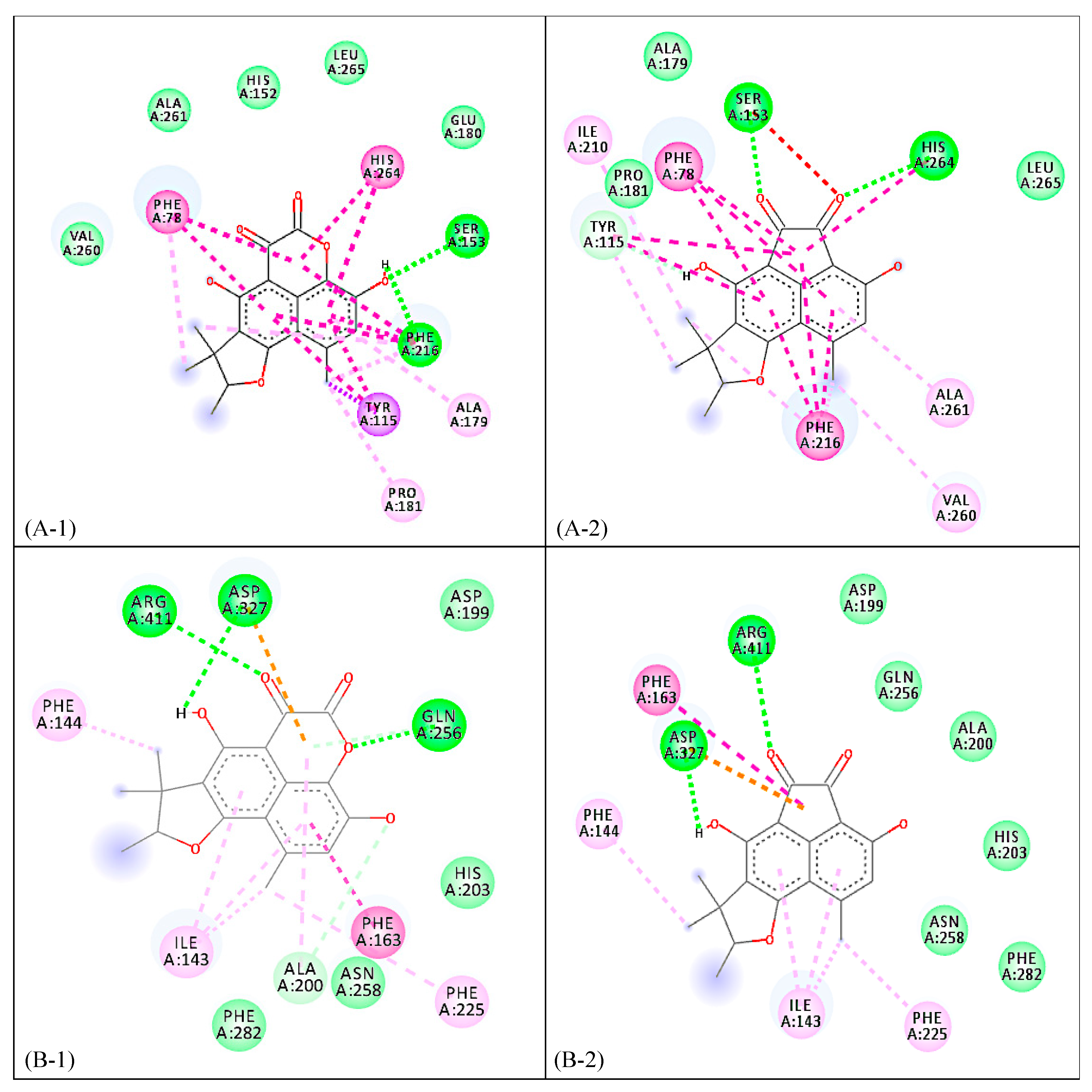
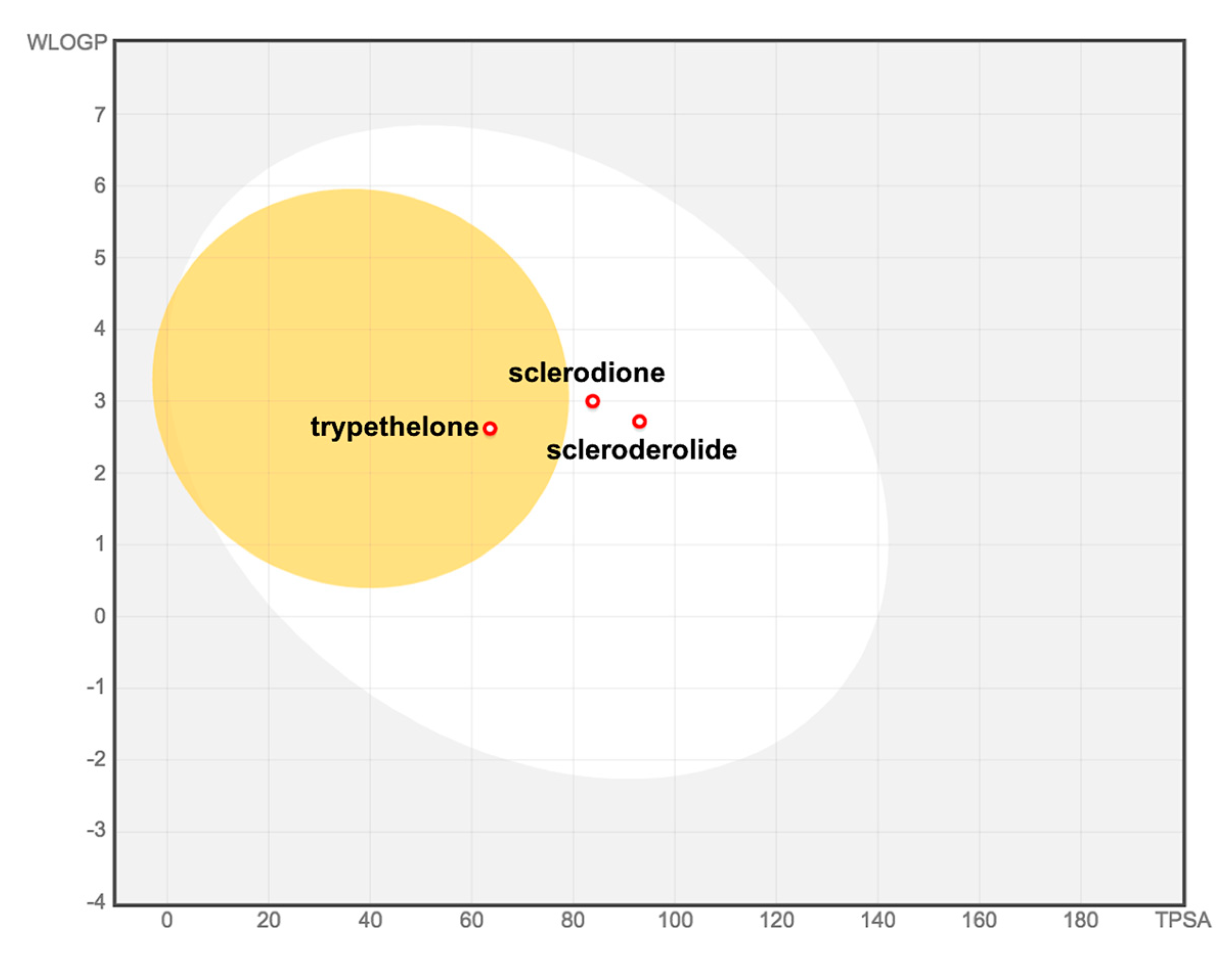
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Fermentation and Extraction
3.4. Isolation of 1–4
3.5. Biological Activity Assays
3.6. Computational Calculations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Al-Goblan, A.S.; Al-Alfi, M.A.; Khan, M.Z. Mechanism linking diabetes mellitus and obesity. Diabetes Metab. Syndr. Obes. Targets Ther. 2014, 7, 587–591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Silva, D.D.; Rapior, S.; Hyde, K.D.; Bahkali, A.H. Medicinal mushrooms in prevention and control of diabetes mellitus. Fungal Divers. 2012, 56, 1–29. [Google Scholar] [CrossRef]
- Hyde, K.D.; Xu, J.; Rapior, S.; Jeewon, R.; Lumyong, S.; Niego, A.G.T.; Abeywickrama, P.D.; Aluthmuhandiram, J.V.S.; Brahamanage, R.S.; Brooks, S.; et al. The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Divers. 2019, 97, 1–136. [Google Scholar] [CrossRef] [Green Version]
- Hyde, K.D.; Jones, E.B.G.; Liu, J.K.; Ariyawansa, H.; Boehm, E.; Boonmee, S.; Braun, U.; Chomnunti, P.; Crous, P.W.; Dai, D.Q.; et al. Families of Dothideomycetes. Fungal Divers. 2013, 63, 1–313. [Google Scholar] [CrossRef]
- Chooi, Y.H.; Muria-Gonzalez, M.J.; Solomon, P.S. A genome-wide survey of the secondary metabolite biosynthesis genes in the wheat pathogen Parastagonospora nodorum. Mycology 2014, 5, 192–206. [Google Scholar] [CrossRef] [PubMed]
- Phukhamsakda, C.; Macabeo, A.P.G.; Yuyama, K.T.; Hyde, K.D.; Stadler, M. Biofilm inhibitory abscisic acid derivatives from the plant-associated Dothideomycete fungus, Roussoella sp. Molecules 2018, 23, 2190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phukhamsakda, C.; Macabeo, A.P.G.; Huch, V.; Cheng, T.; Hyde, K.D.; Stadler, M. Sparticolins A–G, biologically active oxidized spirodioxynaphthalene derivatives from the Ascomycete Sparticola junci. J. Nat. Prod. 2019, 82, 2878–2885. [Google Scholar] [CrossRef]
- Thambugala, K.M.; Hyde, K.D.; Tanaka, K.; Tian, Q.; Wanasinghe, D.N.; Ariyawansa, H.A.; Jayasiri, S.C.; Boonmee, S.; Camporesi, E.; Hashimoto, A.; et al. Towards a natural classification and backbone tree for Lophiostomataceae, Floricolaceae, and Amorosiaceae fam. nov. Fungal Divers. 2015, 74, 199–266. [Google Scholar] [CrossRef]
- Hashimoto, A.; Hirayama, K.; Takahashi, H.; Matsumura, M.; Okada, G.; Chen, C.Y.; Huang, J.W.; Kakishima, M.; Ono, T.; Tanaka, K. Resolving the Lophiostoma bipolare complex: Generic delimitations within Lophiostomataceae. Stud. Mycol. 2018, 90, 161–189. [Google Scholar] [CrossRef]
- Ayer, W.A.; Hoyano, Y.; Pedras, M.S.; Clardy, J.; Arnold, E. Metabolites produced by the Scleroderris canker fungus, Gremmeniella abietina. Part 2. The structure of scleroderolide. Can. J. Chem. 1987, 65, 748–753. [Google Scholar] [CrossRef]
- Elsebai, M.F.; Natesan, L.; Kehraus, S.; Mohamed, I.E.; Schnakenburg, G.; Sasse, F.; Shaaban, S.; Gütschow, M.; König, G.M. HLE-inhibitory alkaloids with a polyketide skeleton from the marine-derived fungus Coniothyrium cereale. J. Nat. Prod. 2011, 74, 2282–2285. [Google Scholar] [CrossRef] [PubMed]
- Intaraudom, C.; Nitthithanasilp, S.; Rachtawee, P.; Boonruangprapa, T.; Prabpai, S.; Kongsaeree, P.; Pittayakhajonwut, P. Phenalenone derivatives and the unusual tricyclic sesterterpene acid from the marine fungus Lophiostoma bipolare BCC25910. Phytochemistry 2015, 120, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Ayer, W.A.; Kamada, M.; Ma, Y.-T. Sclerodin and related compounds from a plant disease causing fungus. Scleroderris yellow. Can. J. Chem. 1989, 67, 2089–2094. [Google Scholar] [CrossRef]
- Basnet, B.B.; Liu, L.; Zhao, W.; Liu, R.; Ma, K.; Bao, L.; Ren, J.; Wei, X.; Yu, H.; Wei, J.; et al. New 1,2-naphthoquinone-derived pigments from the mycobiont of lichen Trypethelium eluteriae Sprengel. Nat. Prod. Res. 2019, 33, 2044–2050. [Google Scholar] [CrossRef] [PubMed]
- Mathey, A.; Steffan, B.; Steglich, W. 1,2-Naphthochinon-Derivate aus Kulturen des Mycosymbionten der Flechte Trypethelium eluteriae (Trypetheliaceae). Liebigs Ann. der Chemie 1980, 779–785. [Google Scholar] [CrossRef]
- Ayer, W.A.; Hoyano, Y.; Soledade Pedras, M.; Van Altena, I. Metabolites produced by the Scleroderris canker fungus, Gremmeniella abietina. Part 1. Can. J. Chem. 1986, 64, 1585–1589. [Google Scholar] [CrossRef]
- Garcia-Conesa, M.T.; Plumb, G.W.; Waldron, K.W.; Ralph, J.; Williamson, G. Ferulic acid dehydrodimers from wheat bran: isolation, purification and antioxidant properties of 8-O-4-diferulic acid. Redox Rep. 1997, 3, 319–323. [Google Scholar] [CrossRef] [Green Version]
- Ralph, J.; Quideau, S.; Grabber, J.H.; Hatfield, R.D. Identification and synthesis of new ferulic acid dehydrodimers present in grass cell walls. J. Chem. Soc. Perkin Trans. 1 1994, 3485–3498. [Google Scholar] [CrossRef]
- Kokotos, G. Inhibition of digestive lipases by 2-oxo amide triacylglycerol analogues. J. Mol. Catal. B Enzym. 2003, 22, 255–269. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [Green Version]
- Benet, L.Z.; Hosey, C.M.; Ursu, O.; Oprea, T.I. BDDCS, the Rule of 5 and drugability. Adv. Drug Deliv. Rev. 2016, 101, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Zoete, V. A BOILED-Egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandargo, B.; Michehl, M.; Praditya, D.; Steinmann, E.; Stadler, M.; Surup, F. Antiviral meroterpenoid rhodatin and sesquiterpenoids rhodocoranes A-E from the wrinkled peach mushroom, Rhodotus palmatus. Org. Lett. 2019, 21, 3286–3289. [Google Scholar] [CrossRef] [PubMed]
- Macabeo, A.P.G.; Rubio, P.J.M.; Alejandro, G.J.D.; Knorn, M. An α-glucosidase inhibitor from Drepananthus philippinensis. Procedia Chem. 2015, 14, 36–39. [Google Scholar]
- Dechakhamphu, A.; Wongchum, N. Screening for anti-pancreatic lipase properties of 28 traditional Thai medicinal herbs. J. Trop. Biomed. 2015, 5, 1042–1045. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006, 25, 247–260. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. Software news and update AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar]
Sample Availability: Samples of the compounds are available from the authors. |
| Sample | Vs. α-glucosidase IC50 (µM) | Vs. lipase IC50 (µM) |
|---|---|---|
| 1 | 48.7 | 3.4 |
| 2 | 120.0 | 1.0 |
| 3 | >100 | >100 |
| 4 | nt | nt |
| N-deoxynojirimycin | 130.5 | - |
| Orlistat | - | 9.4 |
| Compound | Lipinski’s Rule of Five | Drug-Likeness | ||||
| Molecular Weight (g/mol) | Lipophilicity (MLogP) | H-bond Donors | H-bond Acceptors | Rule Violations | ||
| <500 | <5 | <5 | <10 | <2 | ||
| 1 | 328.32 | 1.40 | 2 | 6 | 0 | yes |
| 2 | 312.32 | 1.13 | 2 | 5 | 0 | yes |
| 3 | 272.30 | 1.06 | 1 | 4 | 0 | yes |
| 1 | 2 | 3 | |
|---|---|---|---|
| Mutagenicity | High risk | Medium risk | None |
| Tumorigenicity | High risk | High risk | None |
| Irritant Effect | None | None | None |
| Reproductive Toxicity | None | None | High risk |
| cLogP | 2.62 | 2.88 | 2.44 |
| Water Solubility | −4.91 (moderately soluble) | −5.29 (moderately soluble) | −3.34 (soluble) |
| Compound | IC50 vs. L929 (μg/mL) | IC50 vs. KB3.1 (μg/mL) |
|---|---|---|
| 1 | 15 | 21 |
| 2 | 27 | 18 |
| 3 | 2.7 | 4.2 |
| Epothilone B | 9 × 10−4 | 4 × 10−5 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Macabeo, A.P.G.; Pilapil, L.A.E.; Garcia, K.Y.M.; Quimque, M.T.J.; Phukhamsakda, C.; Cruz, A.J.C.; Hyde, K.D.; Stadler, M. Alpha-Glucosidase- and Lipase-Inhibitory Phenalenones from a New Species of Pseudolophiostoma Originating from Thailand. Molecules 2020, 25, 965. https://doi.org/10.3390/molecules25040965
Macabeo APG, Pilapil LAE, Garcia KYM, Quimque MTJ, Phukhamsakda C, Cruz AJC, Hyde KD, Stadler M. Alpha-Glucosidase- and Lipase-Inhibitory Phenalenones from a New Species of Pseudolophiostoma Originating from Thailand. Molecules. 2020; 25(4):965. https://doi.org/10.3390/molecules25040965
Chicago/Turabian StyleMacabeo, Allan Patrick G., Luis Agustin E. Pilapil, Katherine Yasmin M. Garcia, Mark Tristan J. Quimque, Chayanard Phukhamsakda, Allaine Jean C. Cruz, Kevin D. Hyde, and Marc Stadler. 2020. "Alpha-Glucosidase- and Lipase-Inhibitory Phenalenones from a New Species of Pseudolophiostoma Originating from Thailand" Molecules 25, no. 4: 965. https://doi.org/10.3390/molecules25040965