NAD+ Metabolism and Regulation: Lessons From Yeast
Abstract
1. Introduction
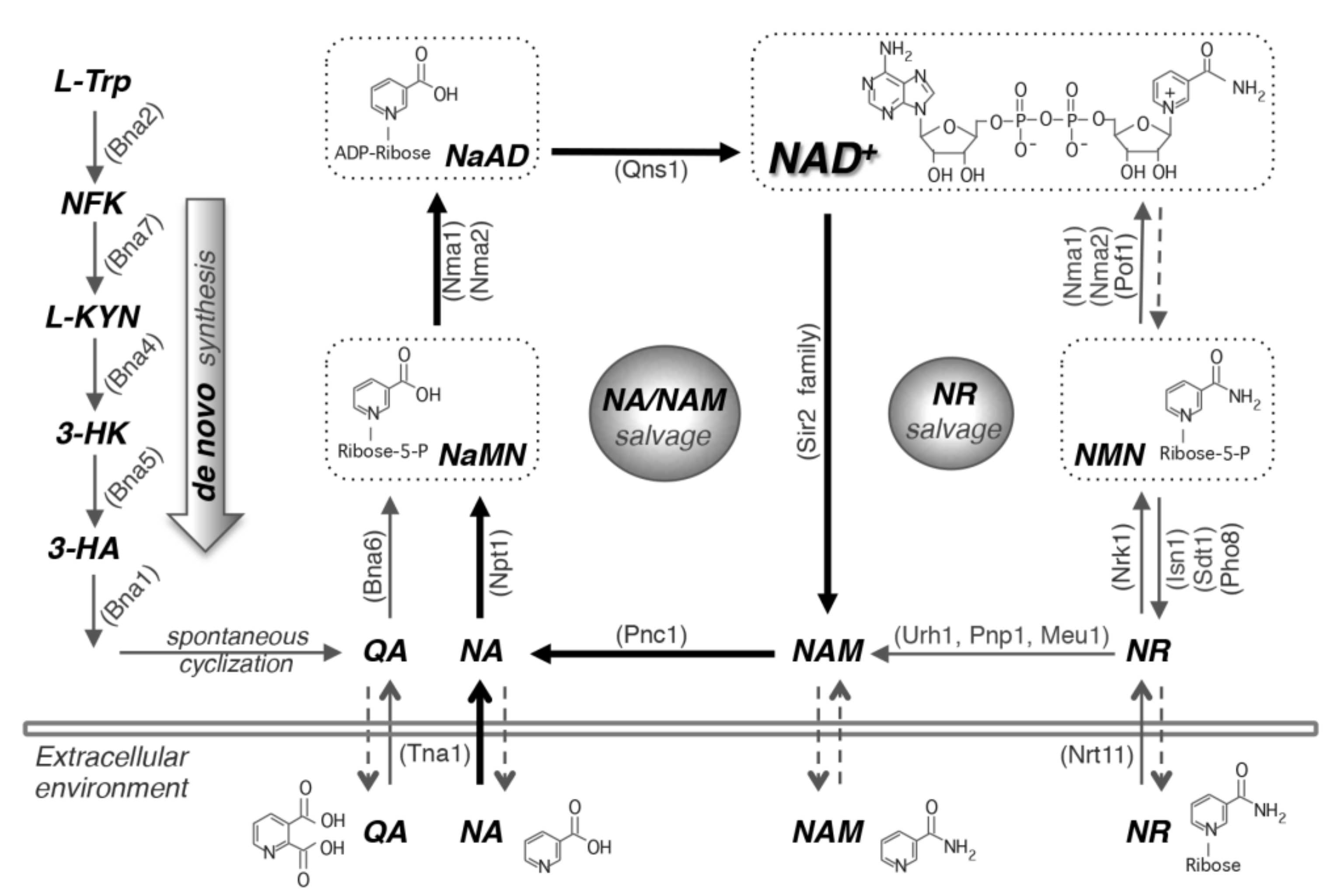
2. NAD+ Biosynthesis Pathways
3. NAD+ and Its Derivatives in Redox Reactions
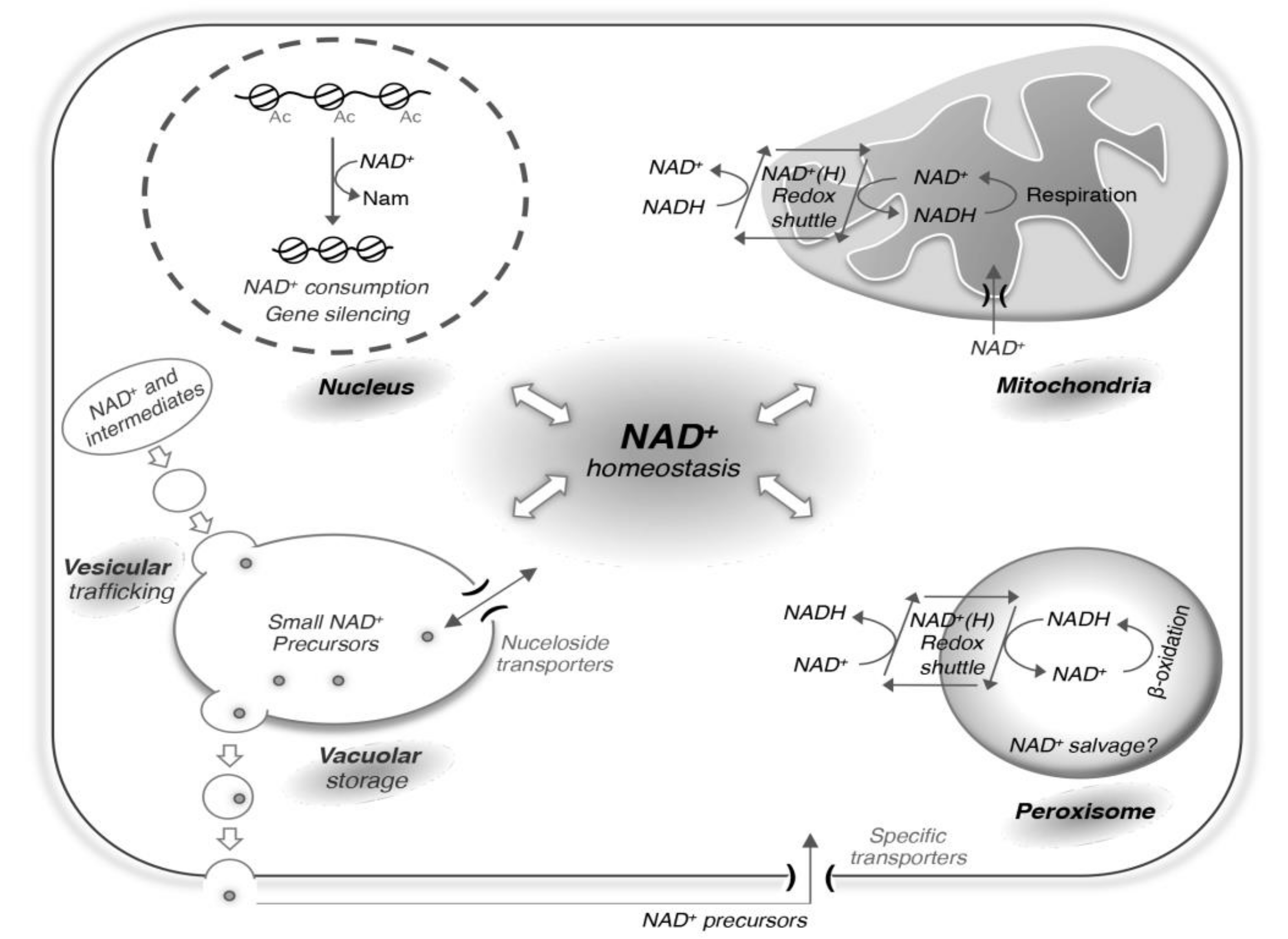
4. Balancing the Redox State of NAD+ and NADH
5. Cellular Processes that Consume NAD+
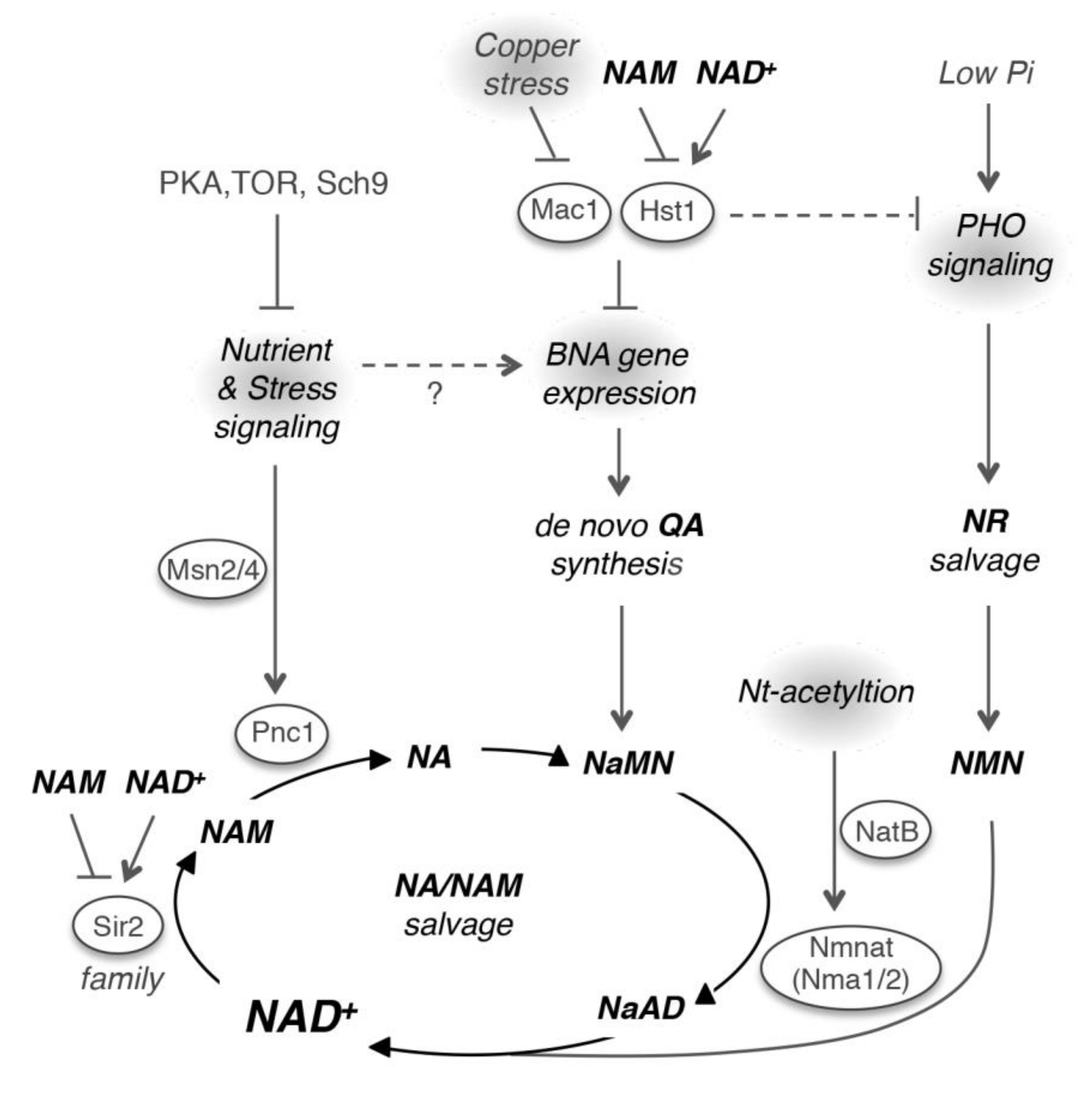
6. Regulation of NAD+ Homeostasis
7. NAD+ and Diseases
8. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kato, M.; Lin, S.J. Regulation of NAD+ metabolism, signaling and compartmentalization in the yeast Saccharomyces cerevisiae. DNA Repair 2014, 23, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Nikiforov, A.; Kulikova, V.; Ziegler, M. The human NAD metabolome: Functions, metabolism and compartmentalization. Crit. Rev. Biochem. Mol. Biol. 2015, 50, 284–297. [Google Scholar] [CrossRef] [PubMed]
- Chini, C.C.; Tarrago, M.G.; Chini, E.N. NAD and the aging process: Role in life, death and everything in between. Mol. Cell. Endocrinol. 2017, 455, 62–74. [Google Scholar] [CrossRef] [PubMed]
- Imai, S.I.; Guarente, L. NAD and sirtuins in aging and disease. Trends Cell Biol. 2014, 24, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, J.; Baur, J.A.; Imai, S.I. NAD(+) Intermediates: The Biology and Therapeutic Potential of NMN and NR. Cell Metab. 2018, 27, 513–528. [Google Scholar] [CrossRef]
- Garten, A.; Schuster, S.; Penke, M.; Gorski, T.; de Giorgis, T.; Kiess, W. Physiological and pathophysiological roles of NAMPT and NAD metabolism. Nat. Rev. Endocrinol. 2015, 11, 535–546. [Google Scholar] [CrossRef]
- Verdin, E. NAD(+) in aging, metabolism, and neurodegeneration. Science 2015, 350, 1208–1213. [Google Scholar] [CrossRef]
- Canto, C.; Menzies, K.J.; Auwerx, J. NAD(+) Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell Metab. 2015, 22, 31–53. [Google Scholar] [CrossRef]
- Yang, Y.; Sauve, A.A. NAD+ metabolism: Bioenergetics, signaling and manipulation for therapy. Biochim. Biophys. Acta 2016, 1864, 1787–1800. [Google Scholar] [CrossRef]
- Liu, H.W.; Smith, C.B.; Schmidt, M.S.; Cambronne, X.A.; Cohen, M.S.; Migaud, M.E.; Brenner, C.; Goodman, R.H. Pharmacological bypass of NAD(+) salvage pathway protects neurons from chemotherapy-induced degeneration. Proc. Natl. Acad. Sci. USA 2018, 115, 10654–10659. [Google Scholar] [CrossRef] [PubMed]
- Poyan Mehr, A.; Tran, M.T.; Ralto, K.M.; Leaf, D.E.; Washco, V.; Messmer, J.; Lerner, A.; Kher, A.; Kim, S.H.; Khoury, C.C.; et al. De novo NAD(+) biosynthetic impairment in acute kidney injury in humans. Nat. Med. 2018, 24, 1351–1359. [Google Scholar] [CrossRef] [PubMed]
- Schwarcz, R.; Bruno, J.P.; Muchowski, P.J.; Wu, H.Q. Kynurenines in the mammalian brain: When physiology meets pathology. Nat. Rev. Neurosci. 2012, 13, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.D.; Maqsood, S.; Huang, J.Y.; Pan, Y.; Harkcom, W.; Li, W.; Sauve, A.; Verdin, E.; Jaffrey, S.R. Activation of SIRT3 by the NAD(+) precursor nicotinamide riboside protects from noise-induced hearing loss. Cell Metab. 2014, 20, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.A.; Harder, J.M.; Foxworth, N.E.; Cochran, K.E.; Philip, V.M.; Porciatti, V.; Smithies, O.; John, S.W. Vitamin B3 modulates mitochondrial vulnerability and prevents glaucoma in aged mice. Science 2017, 355, 756–760. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.B.; Kubota, S.; Ban, N.; Yoshida, M.; Santeford, A.; Sene, A.; Nakamura, R.; Zapata, N.; Kubota, M.; Tsubota, K.; et al. NAMPT-Mediated NAD(+) Biosynthesis Is Essential for Vision In Mice. Cell Rep. 2016, 17, 69–85. [Google Scholar] [CrossRef] [PubMed]
- Belenky, P.; Racette, F.G.; Bogan, K.L.; McClure, J.M.; Smith, J.S.; Brenner, C. Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+. Cell 2007, 129, 473–484. [Google Scholar] [CrossRef]
- Elmore, J.G.; Feinstein, A.R. Joseph Goldberger: An Unsung Hero of American Clinical Epidemiology. Ann. Intern. Med. 1994, 121, 372–375. [Google Scholar] [CrossRef]
- Rajakumar, K. Pellagra in the United States: A historical perspective. South. Med. J. 2000, 93, 272–277. [Google Scholar] [CrossRef]
- Elvehjem, C.A.; Madden, R.J.; Strong, F.M.; Woolley, D.W. The isolation and identification of the anti-black tongue factor. J. Biol. Chem. 1938, 123, 137–149. [Google Scholar] [CrossRef]
- Axelrod, A.; Madden, R.J.; Elvehjem, C. The effect of a nicotinic acid deficiency upon the coenzyme I content of animal tissues. J. Biol. Chem. 1939, 131, 85–93. [Google Scholar]
- Axelrod, A.; Spies, T.D.; Elvehjem, C. The effect of a nicotinic acid deficiency upon the coenzyme I content of the human erythrocyte and muscle. J. Biol. Chem. 1941, 138, 667–676. [Google Scholar]
- Harden, A.; Young, W.J. The alcoholic ferment of yeast-juice. Proc. R Soc. Lond. Ser. B Contain. Pap. Biol. Character 1906, 77, 405–420. [Google Scholar]
- Harden, A.; Young, W.J. The alcoholic ferment of yeast-juice. Part II.—The coferment of yeast-juice. Proc. R Soc. Lond. Ser. B Contain. Pap. A Biol. Character 1906, 78, 369–375. [Google Scholar]
- Barnett, J.A. A history of research on yeasts 5: The fermentation pathway. Yeast 2003, 20, 509–543. [Google Scholar] [CrossRef] [PubMed]
- Von Euler, H.; Myraback, K. Garungs-co-Enzym der Hefe. I. Z. Physiol. Chem. 1923, 131, 179–203. [Google Scholar] [CrossRef]
- Warburg, O.; Christian, W.; Griese, A. Wasserstoffübertragendes Co-Ferment, seine Zusammensetzung und Wirkungsweise. Biochem. Z 1935, 282, 157–205. [Google Scholar]
- Jackson, M.D.; Schmidt, M.T.; Oppenheimer, N.J.; Denu, J.M. Mechanism of nicotinamide inhibition and transglycosidation by Sir2 histone/protein deacetylases. J. Biol. Chem. 2003, 278, 50985–50998. [Google Scholar] [CrossRef]
- Bitterman, K.J.; Anderson, R.M.; Cohen, H.Y.; Latorre-Esteves, M.; Sinclair, D.A. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 2002, 277, 45099–45107. [Google Scholar] [CrossRef]
- Klaidman, L.; Morales, M.; Kem, S.; Yang, J.; Chang, M.L.; Adams, J.D., Jr. Nicotinamide offers multiple protective mechanisms in stroke as a precursor for NAD+, as a PARP inhibitor and by partial restoration of mitochondrial function. Pharmacology 2003, 69, 150–157. [Google Scholar] [CrossRef]
- Chong, Z.Z.; Lin, S.H.; Maiese, K. Nicotinamide modulates mitochondrial membrane potential and cysteine protease activity during cerebral vascular endothelial cell injury. J. Vasc. Res. 2002, 39, 131–147. [Google Scholar] [CrossRef]
- Croft, T.; Raj, C.J.T.; Salemi, M.; Phinney, B.S.; Lin, S.-J. A functional link between NAD+ homeostasis and N-terminal protein acetylation in Saccharomyces cerevisiae. J. Biol. Chem. 2018, 293, 2927–2938. [Google Scholar] [CrossRef]
- Lu, S.P.; Kato, M.; Lin, S.J. Assimilation of endogenous nicotinamide riboside is essential for calorie restriction-mediated life span extension in Saccharomyces cerevisiae. J. Biol. Chem. 2009, 284, 17110–17119. [Google Scholar] [CrossRef]
- Ohashi, K.; Kawai, S.; Murata, K. Secretion of quinolinic acid, an intermediate in the kynurenine pathway, for utilization in NAD+ biosynthesis in the yeast Saccharomyces cerevisiae. Eukaryot. Cell 2013, 12, 648–653. [Google Scholar] [CrossRef]
- James Theoga Raj, C.; Croft, T.; Venkatakrishnan, P.; Groth, B.; Dhugga, G.; Cater, T.; Lin, S.J. The copper-sensing transcription factor Mac1, the histone deacetylase Hst1, and nicotinic acid regulate de novo NAD(+) biosynthesis in budding yeast. J. Biol. Chem. 2019, 294, 5562–5575. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.P.; Lin, S.J. Phosphate-responsive signaling pathway is a novel component of NAD+ metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 2011, 286, 14271–14281. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Lin, S.J. YCL047C/POF1 Is a Novel Nicotinamide Mononucleotide Adenylyltransferase (NMNAT) in Saccharomyces cerevisiae. J. Biol. Chem. 2014, 289, 15577–15587. [Google Scholar] [CrossRef]
- Bedalov, A.; Hirao, M.; Posakony, J.; Nelson, M.; Simon, J.A. NAD+-dependent deacetylase Hst1p controls biosynthesis and cellular NAD+ levels in Saccharomyces cerevisiae. Mol. Cell. Biol. 2003, 23, 7044–7054. [Google Scholar] [CrossRef]
- Medvedik, O.; Lamming, D.W.; Kim, K.D.; Sinclair, D.A. MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol. 2007, 5, e261. [Google Scholar] [CrossRef]
- Anderson, R.M.; Bitterman, K.J.; Wood, J.G.; Medvedik, O.; Sinclair, D.A. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature 2003, 423, 181–185. [Google Scholar] [CrossRef]
- Gallo, C.M.; Smith, D.L., Jr.; Smith, J.S. Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol. Cell. Biol. 2004, 24, 1301–1312. [Google Scholar] [CrossRef]
- Bieganowski, P.; Seidle, H.F.; Wojcik, M.; Brenner, C. Synthetic lethal and biochemical analyses of NAD and NADH kinases in Saccharomyces cerevisiae establish separation of cellular functions. J. Biol. Chem. 2006, 281, 22439–22445. [Google Scholar] [CrossRef] [PubMed]
- Pinson, B.; Ceschin, J.; Saint-Marc, C.; Daignan-Fornier, B. Dual control of NAD+ synthesis by purine metabolites in yeast. eLife 2019, 8, e43808. [Google Scholar] [CrossRef] [PubMed]
- Krehl, W.A.; Teply, L.J.; Sarma, P.S.; Elvehjem, C.A. Growth-Retarding Effect of Corn in Nicotinic Acid-Low Rations and Its Counteraction by Tryptophane. Science 1945, 101, 489–490. [Google Scholar] [CrossRef] [PubMed]
- Panozzo, C.; Nawara, M.; Suski, C.; Kucharczyka, R.; Skoneczny, M.; Becam, A.M.; Rytka, J.; Herbert, C.J. Aerobic and anaerobic NAD+ metabolism in Saccharomyces cerevisiae. FEBS Lett. 2002, 517, 97–102. [Google Scholar] [CrossRef]
- Emanuelli, M.; Amici, A.; Carnevali, F.; Pierella, F.; Raffaelli, N.; Magni, G. Identification and characterization of a second NMN adenylyltransferase gene in Saccharomyces cerevisiae. Protein Expr. Purif. 2003, 27, 357–364. [Google Scholar] [CrossRef]
- Emanuelli, M.; Carnevali, F.; Lorenzi, M.; Raffaelli, N.; Amici, A.; Ruggieri, S.; Magni, G. Identification and characterization of YLR328W, the Saccharomyces cerevisiae structural gene encoding NMN adenylyltransferase. Expression and characterization of the recombinant enzyme. FEBS Lett. 1999, 455, 13–17. [Google Scholar] [CrossRef]
- Bieganowski, P.; Pace, H.C.; Brenner, C. Eukaryotic NAD+ synthetase Qns1 contains an essential, obligate intramolecular thiol glutamine amidotransferase domain related to nitrilase. J. Biol. Chem. 2003, 278, 33049–33055. [Google Scholar] [CrossRef]
- Preiss, J.; Handler, P. Biosynthesis of diphosphopyridine nucleotide. I. Identification of intermediates. J. Biol. Chem. 1958, 233, 488–492. [Google Scholar]
- Preiss, J.; Handler, P. Biosynthesis of diphosphopyridine nucleotide. II. Enzymatic aspects. J. Biol. Chem. 1958, 233, 493–500. [Google Scholar]
- Sporty, J.; Lin, S.J.; Kato, M.; Ognibene, T.; Stewart, B.; Turteltaub, K.; Bench, G. Quantitation of NAD+ biosynthesis from the salvage pathway in Saccharomyces cerevisiae. Yeast 2009, 26, 363–369. [Google Scholar] [CrossRef]
- Preiss, J.; Handler, P. Enzymatic synthesis of nicotinamide mononucleotide. J. Biol. Chem. 1957, 225, 759–770. [Google Scholar] [PubMed]
- Avalos, J.L.; Bever, K.M.; Wolberger, C. Mechanism of Sirtuin Inhibition by Nicotinamide: Altering the NAD+ Cosubstrate Specificity of a Sir2 Enzyme. Mol. Cell 2005, 17, 855–868. [Google Scholar] [CrossRef] [PubMed]
- Rankin, P.W.; Jacobson, E.L.; Benjamin, R.C.; Moss, J.; Jacobson, M.K. Quantitative studies of inhibitors of ADP-ribosylation in vitro and in vivo. J. Biol. Chem. 1989, 264, 4312–4317. [Google Scholar] [PubMed]
- Sethi, J.K.; Empson, R.M.; Galione, A. Nicotinamide inhibits cyclic ADP-ribose-mediated calcium signalling in sea urchin eggs. Biochem. J. 1996, 319, 613–617. [Google Scholar] [CrossRef] [PubMed]
- Ghislain, M.; Talla, E.; Francois, J.M. Identification and functional analysis of the Saccharomyces cerevisiae nicotinamidase gene, PNC1. Yeast 2002, 19, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Rowen, J.W.; Kornberg, A. The phosphorolysis of nicotinamide riboside. J. Biol. Chem. 1951, 193, 497–507. [Google Scholar] [PubMed]
- Nishizuka, Y.; Hayaishi, O. Mammalian pyridine ribonucleoside phosphokinase. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1971; Volume 18, pp. 141–144. [Google Scholar]
- Sasiak, K.; Saunders, P.P. Purification and properties of a human nicotinamide ribonucleoside kinase. Arch. Biochem. Biophys. 1996, 333, 414–418. [Google Scholar] [CrossRef]
- Saunders, P.P.; Spindler, C.D.; Tan, M.-T.; Alvarez, E.; Robins, R.K. Tiazofurin is phosphorylated by three enzymes from Chinese hamster ovary cells. Cancer Res. 1990, 50, 5269–5274. [Google Scholar]
- Bieganowski, P.; Brenner, C. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell 2004, 117, 495–502. [Google Scholar] [CrossRef]
- Tempel, W.; Rabeh, W.M.; Bogan, K.L.; Belenky, P.; Wojcik, M.; Seidle, H.F.; Nedyalkova, L.; Yang, T.; Sauve, A.A.; Park, H.W.; et al. Nicotinamide riboside kinase structures reveal new pathways to NAD+. PLoS Biol. 2007, 5, e263. [Google Scholar] [CrossRef]
- Bogan, K.L.; Evans, C.; Belenky, P.; Song, P.; Burant, C.F.; Kennedy, R.; Brenner, C. Identification of Isn1 and Sdt1 as glucose- and vitamin-regulated nicotinamide mononucleotide and nicotinic acid mononucleotide [corrected] 5′-nucleotidases responsible for production of nicotinamide riboside and nicotinic acid riboside. J. Biol. Chem. 2009, 284, 34861–34869. [Google Scholar] [CrossRef] [PubMed]
- Llorente, B.; Dujon, B. Transcriptional regulation of the Saccharomyces cerevisiae DAL5 gene family and identification of the high affinity nicotinic acid permease TNA1 (YGR260w). FEBS Lett. 2000, 475, 237–241. [Google Scholar] [CrossRef]
- Belenky, P.A.; Moga, T.G.; Brenner, C. Saccharomyces cerevisiae YOR071C encodes the high affinity nicotinamide riboside transporter Nrt1. J. Biol. Chem. 2008, 283, 8075–8079. [Google Scholar] [CrossRef] [PubMed]
- Pullman, M.E.; San Pietro, A.; Colowick, S.P. On the structure of reduced diphosphopyridine nucleotide. J. Biol. Chem. 1954, 206, 129–141. [Google Scholar] [PubMed]
- Kornberg, A.; Pricer, W. On the structure of triphosphopyridine nucleotide. J. Biol. Chem. 1950, 186, 557–567. [Google Scholar] [PubMed]
- Friedkin, M.; Lehninger, A.L. Phosphorylation coupled to electron transport between dihydrodiphosphopyridine nucleotide and oxygen. J. Biol. Chem. 1948, 174, 757–758. [Google Scholar]
- Friedkin, M.; Lehninger, A.L. Esterification of inorganic phosphate coupled to electron transport between dihydrodiphosphopyridine nucleotide and oxygen. I. J. Biol. Chem. 1949, 178, 611–623. [Google Scholar]
- Kornberg, A. Enzymatic synthesis of triphosphopyridine nucleotide. J. Biol. Chem. 1950, 182, 805–813. [Google Scholar]
- Shi, F.; Kawai, S.; Mori, S.; Kono, E.; Murata, K. Identification of ATP-NADH kinase isozymes and their contribution to supply of NADP (H) in Saccharomyces cerevisiae. FEBS J. 2005, 272, 3337–3349. [Google Scholar] [CrossRef]
- Strand, M.K.; Stuart, G.R.; Longley, M.J.; Graziewicz, M.A.; Dominick, O.C.; Copeland, W.C. POS5 gene of Saccharomyces cerevisiae encodes a mitochondrial NADH kinase required for stability of mitochondrial DNA. Eukaryot. Cell 2003, 2, 809–820. [Google Scholar] [CrossRef]
- Agledal, L.; Niere, M.; Ziegler, M. The phosphate makes a difference: Cellular functions of NADP. Redox Rep. 2010, 15, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Minard, K.I.; McAlister-Henn, L. Antioxidant function of cytosolic sources of NADPH in yeast. Free Radic. Biol. Med. 2001, 31, 832–843. [Google Scholar] [CrossRef]
- Jamieson, D.J. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 1998, 14, 1511–1527. [Google Scholar] [CrossRef]
- Inoue, Y.; Matsuda, T.; Sugiyama, K.-i.; Izawa, S.; Kimura, A. Genetic analysis of glutathione peroxidase in oxidative stress response of Saccharomyces cerevisiae. J. Biol. Chem. 1999, 274, 27002–27009. [Google Scholar] [CrossRef]
- Todisco, S.; Agrimi, G.; Castegna, A.; Palmieri, F. Identification of the mitochondrial NAD+ transporter in Saccharomyces cerevisiae. J. Biol. Chem. 2006, 281, 1524–1531. [Google Scholar] [CrossRef]
- Alano, C.C.; Tran, A.; Tao, R.; Ying, W.; Karliner, J.S.; Swanson, R.A. Differences among cell types in NAD+ compartmentalization: A comparison of neurons, astrocytes, and cardiac myocytes. J. Neurosci. Res. 2007, 85, 3378–3385. [Google Scholar] [CrossRef]
- Cronin, V.; Maras, B.; Barra, D.; Doonan, S. The amino acid sequence of the aspartate aminotransferase from baker’s yeast (Saccharomyces cerevisiae). Biochem. J. 1991, 277, 335–340. [Google Scholar] [CrossRef]
- Morin, P.J.; Subramanian, G.S.; Gilmore, T.D. AAT1, a gene encoding a mitochondrial aspartate aminotransferase in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1992, 1171, 211–214. [Google Scholar] [CrossRef]
- Thompson, L.M.; Sutherland, P.; Steffan, J.S.; McAlister-Henn, L. Gene sequence and primary structure of mitochondrial malate dehydrogenase from Saccharomyces cerevisiae. Biochemistry 1988, 27, 8393–8400. [Google Scholar] [CrossRef]
- Minard, K.I.; McAlister-Henn, L. Isolation, nucleotide sequence analysis, and disruption of the MDH2 gene from Saccharomyces cerevisiae: Evidence for three isozymes of yeast malate dehydrogenase. Mol. Cell. Biol. 1991, 11, 370–380. [Google Scholar] [CrossRef]
- Young, E.T.; Pilgrim, D. Isolation and DNA sequence of ADH3, a nuclear gene encoding the mitochondrial isozyme of alcohol dehydrogenase in Saccharomyces cerevisiae. Mol. Cell. Biol. 1985, 5, 3024–3034. [Google Scholar] [CrossRef] [PubMed]
- Bakker, B.M.; Bro, C.; Kötter, P.; Luttik, M.A.; Van Dijken, J.P.; Pronk, J.T. The mitochondrial alcohol dehydrogenase Adh3p is involved in a redox shuttle in Saccharomyces cerevisiae. J. Bacteriol. 2000, 182, 4730–4737. [Google Scholar] [CrossRef] [PubMed]
- Thielen, J.; Ciriacy, M. Biochemical basis of mitochondrial acetaldehyde dismutation in Saccharomyces cerevisiae. J. Bacteriol. 1991, 173, 7012–7017. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Albertyn, J.; Hohmann, S.; Thevelein, J.M.; Prior, B.A. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 1994, 14, 4135–4144. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-T.; Rahaim, P.; Robbins, P.; Yocum, R.R. Cloning, sequence, and disruption of the Saccharomyces diastaticus DAR1 gene encoding a glycerol-3-phosphate dehydrogenase. J. Bacteriol. 1994, 176, 7091–7095. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, P.; André, L.; Ansell, R.; Blomberg, A.; Adler, L. Cloning and characterization of GPD2, a second gene encoding sn-glycerol 3-phosphate dehydrogenase (NAD+) in Saccharomyces cerevisiae, and its comparison with GPD1. Mol. Microbiol. 1995, 17, 95–107. [Google Scholar] [CrossRef]
- Rønnow, B.; Kielland-Brandt, M.C. GUT2, a gene for mitochondrial glycerol 3-phosphate dehydrogenase of Saccharomyces cerevisiae. Yeast 1993, 9, 1121–1130. [Google Scholar] [CrossRef]
- Easlon, E.; Tsang, F.; Skinner, C.; Wang, C.; Lin, S.J. The malate-aspartate NADH shuttle components are novel metabolic longevity regulators required for calorie restriction-mediated life span extension in yeast. Genes Dev. 2008, 22, 931–944. [Google Scholar] [CrossRef]
- Bakker, B.M.; Overkamp, K.M.; van Maris, A.J.; Kotter, P.; Luttik, M.A.; van Dijken, J.P.; Pronk, J.T. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2001, 25, 15–37. [Google Scholar] [CrossRef]
- AbdelRaheim, S.R.; Cartwright, J.L.; Gasmi, L.; McLennan, A.G. The NADH diphosphatase encoded by the Saccharomyces cerevisiae NPY1 nudix hydrolase gene is located in peroxisomes. Arch. Biochem. Biophys. 2001, 388, 18–24. [Google Scholar] [CrossRef]
- Small, W.C.; McAlister-Henn, L. Identification of a cytosolically directed NADH dehydrogenase in mitochondria of Saccharomyces cerevisiae. J. Bacteriol. 1998, 180, 4051–4055. [Google Scholar] [CrossRef] [PubMed]
- Luttik, M.A.; Overkamp, K.M.; Kötter, P.; de Vries, S.; van Dijken, J.P.; Pronk, J.T. The Saccharomyces cerevisiae NDE1 andNDE2 Genes Encode Separate Mitochondrial NADH Dehydrogenases Catalyzing the Oxidation of Cytosolic NADH. J. Biol. Chem. 1998, 273, 24529–24534. [Google Scholar] [CrossRef] [PubMed]
- Mann, P.J.G.; Quastel, J.H. Nicotinamide, Cozymase and Tissue Metabolism. Nature 1941, 147, 326–327. [Google Scholar] [CrossRef][Green Version]
- Handler, P.; Klein, J.R. The inactivation of pyridine nucleotides by animal tissues in vitro. J. Biol. Chem. 1942, 144, 453–454. [Google Scholar]
- Walters, R.W.; Matheny, T.; Mizoue, L.S.; Rao, B.S.; Muhlrad, D.; Parker, R. Identification of NAD+ capped mRNAs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2017, 114, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Landry, J.; Sutton, A.; Tafrov, S.T.; Heller, R.C.; Stebbins, J.; Pillus, L.; Sternglanz, R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 2000, 97, 5807–5811. [Google Scholar] [CrossRef]
- Kraus, W.L. PARPs and ADP-Ribosylation: 50 Years … and Counting. Mol. Cell 2015, 58, 902–910. [Google Scholar] [CrossRef]
- Hecht, A.; Strahl-Bolsinger, S.; Grunstein, M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 1996, 383, 92–96. [Google Scholar] [CrossRef]
- Strahl-Bolsinger, S.; Hecht, A.; Luo, K.; Grunstein, M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997, 11, 83–93. [Google Scholar] [CrossRef]
- Smith, J.S.; Boeke, J.D. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997, 11, 241–254. [Google Scholar] [CrossRef]
- Moazed, D. Common themes in mechanisms of gene silencing. Mol. Cell 2001, 8, 489–498. [Google Scholar] [CrossRef]
- Lin, Y.-y.; Lu, J.-y.; Zhang, J.; Walter, W.; Dang, W.; Wan, J.; Tao, S.-C.; Qian, J.; Zhao, Y.; Boeke, J.D.; et al. Protein Acetylation Microarray Reveals NuA4 Controls Key Metabolic Target Regulating Gluconeogenesis. Cell 2009, 136, 1073–1084. [Google Scholar] [CrossRef] [PubMed]
- Downey, M.; Knight, B.; Vashisht Ajay, A.; Seller, C.A.; Wohlschlegel, J.A.; Shore, D.; Toczyski, D.P. Gcn5 and Sirtuins Regulate Acetylation of the Ribosomal Protein Transcription Factor Ifh1. Curr. Biol. 2013, 23, 1638–1648. [Google Scholar] [CrossRef] [PubMed]
- Finkel, T.; Deng, C.-X.; Mostoslavsky, R. Recent progress in the biology and physiology of sirtuins. Nature 2009, 460, 587–591. [Google Scholar] [CrossRef]
- Smith, J.S.; Brachmann, C.B.; Celic, I.; Kenna, M.A.; Muhammad, S.; Starai, V.J.; Avalos, J.L.; Escalante-Semerena, J.C.; Grubmeyer, C.; Wolberger, C.; et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 2000, 97, 6658–6663. [Google Scholar] [CrossRef]
- Lin, S.J.; Defossez, P.A.; Guarente, L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 2000, 289, 2126–2128. [Google Scholar] [CrossRef]
- Alston, T.A.; Abeles, R.H. Substrate specificity of nicotinamide methyltransferase isolated from porcine liver. Arch. Biochem. Biophys. 1988, 260, 601–608. [Google Scholar] [CrossRef]
- Aksoy, S.; Szumlanski, C.L.; Weinshilboum, R.M. Human liver nicotinamide N-methyltransferase. cDNA cloning, expression, and biochemical characterization. J. Biol. Chem. 1994, 269, 14835–14840. [Google Scholar]
- Lin, S.J.; Ford, E.; Haigis, M.; Liszt, G.; Guarente, L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004, 18, 12–16. [Google Scholar] [CrossRef]
- Chen, Y.G.; Kowtoniuk, W.E.; Agarwal, I.; Shen, Y.; Liu, D.R. LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat. Chem. Biol. 2009, 5, 879. [Google Scholar] [CrossRef]
- Zhang, H.; Zhong, H.; Zhang, S.; Shao, X.; Ni, M.; Cai, Z.; Chen, X.; Xia, Y. NAD tagSeq reveals that NAD+-capped RNAs are mostly produced from a large number of protein-coding genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 12072–12077. [Google Scholar] [CrossRef]
- Jiao, X.; Doamekpor, S.K.; Bird, J.G.; Nickels, B.E.; Tong, L.; Hart, R.P.; Kiledjian, M. 5′ end nicotinamide adenine dinucleotide cap in human cells promotes RNA decay through DXO-mediated deNADding. Cell 2017, 168, 1015–1027. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, Y.; Nakagawa, T.; Mao, X.; DiAntonio, A.; Milbrandt, J. NMNAT1 inhibits axon degeneration via blockade of SARM1-mediated NAD+ depletion. Elife 2016, 5, e19749. [Google Scholar] [CrossRef] [PubMed]
- Essuman, K.; Summers, D.W.; Sasaki, Y.; Mao, X.; DiAntonio, A.; Milbrandt, J. The SARM1 toll/interleukin-1 receptor domain possesses intrinsic NAD+ cleavage activity that promotes pathological axonal degeneration. Neuron 2017, 93, 1334–1343. [Google Scholar] [CrossRef] [PubMed]
- Camacho-Pereira, J.; Tarragó, M.G.; Chini, C.C.; Nin, V.; Escande, C.; Warner, G.M.; Puranik, A.S.; Schoon, R.A.; Reid, J.M.; Galina, A. CD38 dictates age-related NAD decline and mitochondrial dysfunction through an SIRT3-dependent mechanism. Cell Metab. 2016, 23, 1127–1139. [Google Scholar] [CrossRef] [PubMed]
- Aksnes, H.; Ree, R.; Arnesen, T. Co-translational, post-translational, and non-catalytic roles of N-terminal acetyltransferases. Mol. Cell 2019, 73, 1097–1114. [Google Scholar] [CrossRef]
- Priault, M.; Salin, B.; Schaeffer, J.; Vallette, F.d.; Di Rago, J.; Martinou, J. Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ. 2005, 12, 1613. [Google Scholar] [CrossRef]
- Reggiori, F.; Klionsky, D.J. Autophagic processes in yeast: Mechanism, machinery and regulation. Genetics 2013, 194, 341–361. [Google Scholar] [CrossRef]
- Delorme-Axford, E.; Guimaraes, R.S.; Reggiori, F.; Klionsky, D.J. The yeast Saccharomyces cerevisiae: An overview of methods to study autophagy progression. Methods 2015, 75, 3–12. [Google Scholar] [CrossRef]
- Jungmann, J.; Reins, H.A.; Lee, J.; Romeo, A.; Hassett, R.; Kosman, D.; Jentsch, S. MAC1, a nuclear regulatory protein related to Cu-dependent transcription factors is involved in Cu/Fe utilization and stress resistance in yeast. EMBO J. 1993, 12, 5051–5056. [Google Scholar] [CrossRef]
- Graden, J.A.; Winge, D.R. Copper-mediated repression of the activation domain in the yeast Mac1p transcription factor. Proc. Natl. Acad. Sci. USA 1997, 94, 5550–5555. [Google Scholar] [CrossRef] [PubMed]
- Gross, C.; Kelleher, M.; Iyer, V.R.; Brown, P.O.; Winge, D.R. Identification of the copper regulon in Saccharomyces cerevisiae by DNA microarrays. J. Biol. Chem. 2000, 275, 32310–32316. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.T.; Posewitz, M.C.; Srinivasan, C.; Winge, D.R. Mapping of the DNA binding domain of the copper-responsive transcription factor Mac1 from Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 23805–23811. [Google Scholar] [CrossRef] [PubMed]
- Serpe, M.; Joshi, A.; Kosman, D.J. Structure-function analysis of the protein-binding domains of Mac1p, a copper-dependent transcriptional activator of copper uptake in Saccharomyces cerevisiae. J. Biol. Chem. 1999, 274, 29211–29219. [Google Scholar] [CrossRef]
- Zhu, Z.; Labbe, S.; Pena, M.M.; Thiele, D.J. Copper differentially regulates the activity and degradation of yeast Mac1 transcription factor. J. Biol. Chem. 1998, 273, 1277–1280. [Google Scholar] [CrossRef]
- Anderson, R.M.; Bitterman, K.J.; Wood, J.G.; Medvedik, O.; Cohen, H.; Lin, S.S.; Manchester, J.K.; Gordon, J.I.; Sinclair, D.A. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 2002, 277, 18881–18890. [Google Scholar] [CrossRef]
- Tsang, F.; James, C.; Kato, M.; Myers, V.; Ilyas, I.; Tsang, M.; Lin, S.J. Reduced Ssy1-Ptr3-Ssy5 (SPS) Signaling Extends Replicative Life Span by Enhancing NAD+ Homeostasis in Saccharomyces cerevisiae. J. Biol. Chem. 2015, 290, 12753–12764. [Google Scholar] [CrossRef]
- Tsang, F.; Lin, S.J. Less is more: Nutrient limitation induces cross-talk of nutrient sensing pathways with NAD(+) homeostasis and contributes to longevity. Front. Biol. 2015, 10, 333–357. [Google Scholar] [CrossRef][Green Version]
- Carroll, A.S.; Bishop, A.C.; DeRisi, J.L.; Shokat, K.M.; O’Shea, E.K. Chemical inhibition of the Pho85 cyclin-dependent kinase reveals a role in the environmental stress response. Proc. Natl. Acad. Sci. USA 2001, 98, 12578–12583. [Google Scholar] [CrossRef]
- Swinnen, E.; Wanke, V.; Roosen, J.; Smets, B.; Dubouloz, F.; Pedruzzi, I.; Cameroni, E.; De Virgilio, C.; Winderickx, J. Rim15 and the crossroads of nutrient signalling pathways in Saccharomyces cerevisiae. Cell Div. 2006, 1, 3. [Google Scholar] [CrossRef]
- Wei, M.; Fabrizio, P.; Hu, J.; Ge, H.; Cheng, C.; Li, L.; Longo, V.D. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet. 2008, 4, e13. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.P.; Lin, S.J. Regulation of yeast sirtuins by NAD(+) metabolism and calorie restriction. Biochim. Biophys. Acta 2010, 1804, 1567–1575. [Google Scholar] [CrossRef] [PubMed]
- Okabe, K.; Yaku, K.; Tobe, K.; Nakagawa, T. Implications of altered NAD metabolism in metabolic disorders. J. Biomed. Sci. 2019, 26, 34. [Google Scholar] [CrossRef] [PubMed]
- Strømland, Ø.; Niere, M.; Nikiforov, A.A.; VanLinden, M.R.; Heiland, I.; Ziegler, M. Keeping the balance in NAD metabolism. Biochem. Soc. Trans. 2019, 47, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Demarest, T.G.; Babbar, M.; Okur, M.N.; Dan, X.; Croteau, D.L.; Fakouri, N.B.; Mattson, M.P.; Bohr, V.A. NAD+ metabolism in aging and cancer. Annu. Rev. Cancer Biol. 2019, 3, 105–130. [Google Scholar] [CrossRef]
- Pehar, M.; Harlan, B.A.; Killoy, K.M.; Vargas, M.R. Nicotinamide adenine dinucleotide metabolism and neurodegeneration. Antioxid. Redox Signal. 2018, 28, 1652–1668. [Google Scholar] [CrossRef] [PubMed]
- Yaku, K.; Okabe, K.; Hikosaka, K.; Nakagawa, T. NAD Metabolism in Cancer Therapeutics. Front. Oncol. 2018, 8, 622. [Google Scholar] [CrossRef]
- Kim, M.Y.; Zhang, T.; Kraus, W.L. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005, 19, 1951–1967. [Google Scholar] [CrossRef]
- Bajrami, I.; Kigozi, A.; Van Weverwijk, A.; Brough, R.; Frankum, J.; Lord, C.J.; Ashworth, A. Synthetic lethality of PARP and NAMPT inhibition in triple-negative breast cancer cells. EMBO Mol. Med. 2012, 4, 1087–1096. [Google Scholar] [CrossRef]
- Sampath, D.; Zabka, T.S.; Misner, D.L.; O’Brien, T.; Dragovich, P.S. Inhibition of nicotinamide phosphoribosyltransferase (NAMPT) as a therapeutic strategy in cancer. Pharmacol. Ther. 2015, 151, 16–31. [Google Scholar] [CrossRef]
- Heske, C.M. Beyond Energy Metabolism: Exploiting the Additional Roles of NAMPT for Cancer Therapy. Front. Oncol. 2019, 9, 1514. [Google Scholar] [CrossRef] [PubMed]
- Konen, J.M.; Fradette, J.J.; Gibbons, D.L. The Good, the Bad and the Unknown of CD38 in the Metabolic Microenvironment and Immune Cell Functionality of Solid Tumors. Cells 2019, 9, 52. [Google Scholar] [CrossRef] [PubMed]
- Hegyi, J.; Schwartz, R.A.; Hegyi, V. Pellagra: Dermatitis, dementia, and diarrhea. Int. J. Dermatol. 2004, 43, 1–5. [Google Scholar] [CrossRef]
- Lunn, E.R.; Perry, V.H.; Brown, M.C.; Rosen, H.; Gordon, S. Absence of Wallerian Degeneration does not Hinder Regeneration in Peripheral Nerve. Eur. J. Neurosci. 1989, 1, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Mack, T.G.; Reiner, M.; Beirowski, B.; Mi, W.; Emanuelli, M.; Wagner, D.; Thomson, D.; Gillingwater, T.; Court, F.; Conforti, L.; et al. Wallerian degeneration of injured axons and synapses is delayed by a Ube4b/Nmnat chimeric gene. Nat. Neurosci. 2001, 4, 1199–1206. [Google Scholar] [CrossRef]
- Conforti, L.; Wilbrey, A.; Morreale, G.; Janeckova, L.; Beirowski, B.; Adalbert, R.; Mazzola, F.; Di Stefano, M.; Hartley, R.; Babetto, E.; et al. WldS protein requires Nmnat activity and a short N-terminal sequence to protect axons in mice. J. Cell Biol. 2009, 184, 491–500. [Google Scholar] [CrossRef]
- Yamagishi, Y.; Tessier-Lavigne, M. An Atypical SCF-like Ubiquitin Ligase Complex Promotes Wallerian Degeneration through Regulation of Axonal Nmnat2. Cell Rep. 2016, 17, 774–782. [Google Scholar] [CrossRef]
- Rossi, F.; Geiszler, P.C.; Meng, W.; Barron, M.R.; Prior, M.; Herd-Smith, A.; Loreto, A.; Lopez, M.Y.; Faas, H.; Pardon, M.-C.; et al. NAD-biosynthetic enzyme NMNAT1 reduces early behavioral impairment in the htau mouse model of tauopathy. Behav. Brain Res. 2018, 339, 140–152. [Google Scholar] [CrossRef]
- Hasbani, D.M.; O’Malley, K.L. WldS mice are protected against the Parkinsonian mimetic MPTP. Exp. Neurol. 2006, 202, 93–99. [Google Scholar] [CrossRef]
- Zhai, R.G.; Zhang, F.; Hiesinger, P.R.; Cao, Y.; Haueter, C.M.; Bellen, H.J. NAD synthase NMNAT acts as a chaperone to protect against neurodegeneration. Nature 2008, 452, 887–891. [Google Scholar] [CrossRef]
- Chiang, P.W.; Wang, J.; Chen, Y.; Fu, Q.; Zhong, J.; Chen, Y.; Yi, X.; Wu, R.; Gan, H.; Shi, Y.; et al. Exome sequencing identifies NMNAT1 mutations as a cause of Leber congenital amaurosis. Nat. Genet. 2012, 44, 972–974. [Google Scholar] [CrossRef] [PubMed]
- Falk, M.J.; Zhang, Q.; Nakamaru-Ogiso, E.; Kannabiran, C.; Fonseca-Kelly, Z.; Chakarova, C.; Audo, I.; Mackay, D.S.; Zeitz, C.; Borman, A.D.; et al. NMNAT1 mutations cause Leber congenital amaurosis. Nat. Genet. 2012, 44, 1040–1045. [Google Scholar] [CrossRef] [PubMed]
- Koenekoop, R.K.; Wang, H.; Majewski, J.; Wang, X.; Lopez, I.; Ren, H.; Chen, Y.; Li, Y.; Fishman, G.A.; Genead, M.; et al. Mutations in NMNAT1 cause Leber congenital amaurosis and identify a new disease pathway for retinal degeneration. Nat. Genet. 2012, 44, 1035–1039. [Google Scholar] [CrossRef] [PubMed]
- Perrault, I.; Hanein, S.; Zanlonghi, X.; Serre, V.; Nicouleau, M.; Defoort-Delhemmes, S.; Delphin, N.; Fares-Taie, L.; Gerber, S.; Xerri, O.; et al. Mutations in NMNAT1 cause Leber congenital amaurosis with early-onset severe macular and optic atrophy. Nat. Genet. 2012, 44, 975–977. [Google Scholar] [CrossRef]
- Ali, Y.O.; Allen, H.M.; Yu, L.; Li-Kroeger, D.; Bakhshizadehmahmoudi, D.; Hatcher, A.; McCabe, C.; Xu, J.; Bjorklund, N.; Taglialatela, G. NMNAT2: HSP90 complex mediates proteostasis in proteinopathies. PLoS Biol. 2016, 14, e1002472. [Google Scholar] [CrossRef]
- Ocampo, A.; Liu, J.; Barrientos, A. NAD+ salvage pathway proteins suppress proteotoxicity in yeast models of neurodegeneration by promoting the clearance of misfolded/oligomerized proteins. Hum. Mol. Genet. 2013, 22, 1699–1708. [Google Scholar] [CrossRef]
- Gerdts, J.; Summers, D.W.; Sasaki, Y.; DiAntonio, A.; Milbrandt, J. Sarm1-mediated axon degeneration requires both SAM and TIR interactions. J. Neurosci. 2013, 33, 13569–13580. [Google Scholar] [CrossRef]
- Osterloh, J.M.; Yang, J.; Rooney, T.M.; Fox, A.N.; Adalbert, R.; Powell, E.H.; Sheehan, A.E.; Avery, M.A.; Hackett, R.; Logan, M.A. dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science 2012, 337, 481–484. [Google Scholar] [CrossRef]
- Gerdts, J.; Brace, E.; Sasaki, Y.; DiAntonio, A.; Milbrandt, J. SARM1 activation triggers axon degeneration locally via NAD+ destruction. Science 2015, 348, 453–457. [Google Scholar] [CrossRef]
- Mecocci, P.; MacGarvey, U.; Beal, M.F. Oxidative damage to mitochondrial DNA is increased in Alzheimer’s disease. Ann. Neurol. Off. J. Am. Neurol. Assoc. Child Neurol. Soc. 1994, 36, 747–751. [Google Scholar] [CrossRef]
- Giasson, B.I.; Duda, J.E.; Murray, I.V.; Chen, Q.; Souza, J.M.; Hurtig, H.I.; Ischiropoulos, H.; Trojanowski, J.Q.; Lee, V.M.-Y. Oxidative damage linked to neurodegeneration by selective α-synuclein nitration in synucleinopathy lesions. Science 2000, 290, 985–989. [Google Scholar] [CrossRef] [PubMed]
- Emerit, J.; Edeas, M.; Bricaire, F. Neurodegenerative diseases and oxidative stress. Biomed. Pharmacother. 2004, 58, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B. Oxidative stress and neurodegeneration: Where are we now? J. Neurochem. 2006, 97, 1634–1658. [Google Scholar] [CrossRef] [PubMed]
- Carrì, M.T.; Valle, C.; Bozzo, F.; Cozzolino, M. Oxidative stress and mitochondrial damage: Importance in non-SOD1 ALS. Front. Cell. Neurosci. 2015, 9, 41. [Google Scholar] [CrossRef] [PubMed]
- Beckman, J.S.; Carson, M.; Smith, C.D.; Koppenol, W.H. ALS, SOD and peroxynitrite. Nature 1993, 364, 584. [Google Scholar] [CrossRef]
- Deng, H.-X.; Hentati, A.; Tainer, J.A.; Iqbal, Z.; Cayabyab, A.; Hung, W.-Y.; Getzoff, E.D.; Hu, P.; Herzfeldt, B.; Roos, R.P. Amyotrophic lateral sclerosis and structural defects in Cu, Zn superoxide dismutase. Science 1993, 261, 1047–1051. [Google Scholar] [CrossRef]
- Bowling, A.C.; Schulz, J.B.; Brown Jr, R.H.; Beal, M.F. Superoxide dismutase activity, oxidative damage, and mitochondrial energy metabolism in familial and sporadic amyotrophic lateral sclerosis. J. Neurochem. 1993, 61, 2322–2325. [Google Scholar] [CrossRef]
- Lombard, D.B.; Alt, F.W.; Cheng, H.-L.; Bunkenborg, J.; Streeper, R.S.; Mostoslavsky, R.; Kim, J.; Yancopoulos, G.; Valenzuela, D.; Murphy, A. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell. Biol. 2007, 27, 8807–8814. [Google Scholar] [CrossRef]
- Ahn, B.-H.; Kim, H.-S.; Song, S.; Lee, I.H.; Liu, J.; Vassilopoulos, A.; Deng, C.-X.; Finkel, T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc. Natl. Acad. Sci. USA 2008, 105, 14447–14452. [Google Scholar] [CrossRef]
- Schlicker, C.; Gertz, M.; Papatheodorou, P.; Kachholz, B.; Becker, C.F.; Steegborn, C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J. Mol. Biol. 2008, 382, 790–801. [Google Scholar] [CrossRef]
- Shimazu, T.; Hirschey, M.D.; Hua, L.; Dittenhafer-Reed, K.E.; Schwer, B.; Lombard, D.B.; Li, Y.; Bunkenborg, J.; Alt, F.W.; Denu, J.M. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010, 12, 654–661. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Brown, K.; Hirschey, M.D.; Verdin, E.; Chen, D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010, 12, 662–667. [Google Scholar] [CrossRef] [PubMed]
- Someya, S.; Yu, W.; Hallows, W.C.; Xu, J.; Vann, J.M.; Leeuwenburgh, C.; Tanokura, M.; Denu, J.M.; Prolla, T.A. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell 2010, 143, 802–812. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.; Yang, Y.; Zhou, Y.; Maharana, C.; Lu, D.; Peng, W.; Liu, Y.; Wan, R.; Marosi, K.; Misiak, M. Mitochondrial SIRT3 mediates adaptive responses of neurons to exercise and metabolic and excitatory challenges. Cell Metab. 2016, 23, 128–142. [Google Scholar] [CrossRef]
- Buck, E.; Bayer, H.; Lindenberg, K.S.; Hanselmann, J.; Pasquarelli, N.; Ludolph, A.C.; Weydt, P.; Witting, A. Comparison of Sirtuin 3 Levels in ALS and Huntington’s Disease—Differential Effects in Human Tissue Samples vs. Transgenic Mouse Models. Front. Mol. Neurosci. 2017, 10, 156. [Google Scholar] [CrossRef]
- Tan, W.; Pasinelli, P.; Trotti, D. Role of mitochondria in mutant SOD1 linked amyotrophic lateral sclerosis. Biochim. Biophys. Acta 2014, 1842, 1295–1301. [Google Scholar] [CrossRef]
- Song, W.; Song, Y.; Kincaid, B.; Bossy, B.; Bossy-Wetzel, E. Mutant SOD1G93A triggers mitochondrial fragmentation in spinal cord motor neurons: Neuroprotection by SIRT3 and PGC-1α. Neurobiol. Dis. 2013, 51, 72–81. [Google Scholar] [CrossRef]
- Chandler, J.L.; Gholson, R.K. De novo biosynthesis of nicotinamide adenine dinucleotide in Escherichia coli: Excretion of quinolinic acid by mutants lacking quinolinate phosphoribosyl transferase. J. Bacteriol. 1972, 111, 98–102. [Google Scholar] [CrossRef]
- Grose, J.H.; Bergthorsson, U.; Roth, J.R. Regulation of NAD synthesis by the trifunctional NadR protein of Salmonella enterica. J. Bacteriol. 2005, 187, 2774–2782. [Google Scholar] [CrossRef]
- Frye, R.A. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 2000, 273, 793–798. [Google Scholar] [CrossRef]
- Brachmann, C.B.; Sherman, J.M.; Devine, S.E.; Cameron, E.E.; Pillus, L.; Boeke, J.D. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995, 9, 2888–2902. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ryu, D.; Wu, Y.; Gariani, K.; Wang, X.; Luan, P.; D’Amico, D.; Ropelle, E.R.; Lutolf, M.P.; Aebersold, R. NAD+ repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 2016, 352, 1436–1443. [Google Scholar] [CrossRef] [PubMed]
- Katsyuba, E.; Mottis, A.; Zietak, M.; De Franco, F.; van der Velpen, V.; Gariani, K.; Ryu, D.; Cialabrini, L.; Matilainen, O.; Liscio, P.; et al. De novo NAD(+) synthesis enhances mitochondrial function and improves health. Nature 2018, 563, 354–359. [Google Scholar] [CrossRef] [PubMed]
- Braidy, N.; Grant, R. Kynurenine pathway metabolism and neuroinflammatory disease. Neural Regen. Res. 2017, 12, 39–42. [Google Scholar] [CrossRef]
- Amaral, M.; Outeiro, T.F.; Scrutton, N.S.; Giorgini, F. The causative role and therapeutic potential of the kynurenine pathway in neurodegenerative disease. J. Mol. Med. 2013, 91, 705–713. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Croft, T.; Venkatakrishnan, P.; Lin, S.-J. NAD+ Metabolism and Regulation: Lessons From Yeast. Biomolecules 2020, 10, 330. https://doi.org/10.3390/biom10020330
Croft T, Venkatakrishnan P, Lin S-J. NAD+ Metabolism and Regulation: Lessons From Yeast. Biomolecules. 2020; 10(2):330. https://doi.org/10.3390/biom10020330
Chicago/Turabian StyleCroft, Trevor, Padmaja Venkatakrishnan, and Su-Ju Lin. 2020. "NAD+ Metabolism and Regulation: Lessons From Yeast" Biomolecules 10, no. 2: 330. https://doi.org/10.3390/biom10020330
APA StyleCroft, T., Venkatakrishnan, P., & Lin, S.-J. (2020). NAD+ Metabolism and Regulation: Lessons From Yeast. Biomolecules, 10(2), 330. https://doi.org/10.3390/biom10020330



