Ιn Vitro Screening of γ-Aminobutyric Acid and Autoinducer-2 Signalling in Lactic Acid Bacteria Exhibiting Probiotic Potential Isolated from Natural Black Conservolea Olives
Abstract
1. Introduction
2. Materials and Methods
2.1. Low pH Assay, Bile Salts Assay, and Bile Salt Hydrolase (BSH) Activity
2.2. Safety Assessment of the Selected Strains
2.3. Screening for AI-2 Activity
2.4. Detection of Extracellular GABA
2.5. Statistical Analysis
3. Results and Discussion
3.1. Low pH Assay, Bile Salts Assay, and BSH Activity
3.2. Safety Assessment of the Selected Strains
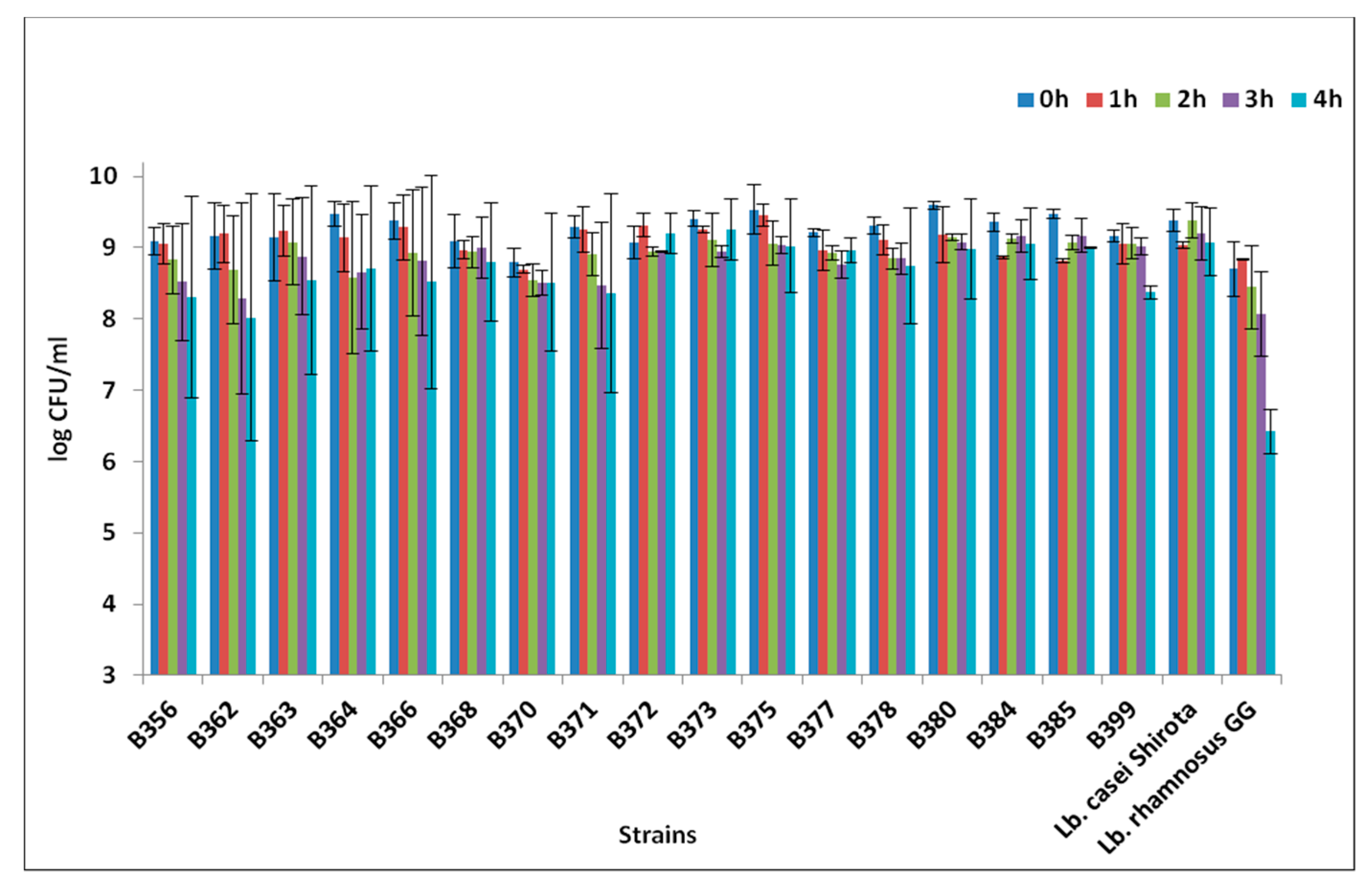
3.3. AI-2 Activity
3.4. Detection of Extracellular GABA
4. Conclusions
Author Contributions
Conflicts of Interest
References
- Choi, E.A.; Chang, H.C. Cholesterol-lowering effects of a putative probiotic strain Lactobacillus plantarum EM isolated from kimchi. LWT Food Sci. Technol. 2015, 62, 210–217. [Google Scholar] [CrossRef]
- He, T.; Priebe, M.G.; Zhong, Y.; Huang, C.; Harmsen, H.J.M. Effects of yogurt and bifidobacteria supplementation on the colonic microbiota in lactose intolerant subjects. J. Appl. Microbiol. 2007, 104, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.E.; Choi, S.C.; Park, K.S.; Park, M.I.; Shin, J.E. Change of Fecal Flora and Effectiveness of the Short-term VSL#3 Probiotic Treatment in Patients with Functional Constipation. J. Neurogastroenterol. Motil. 2015, 21, 111–120. [Google Scholar] [PubMed]
- McFarland, L.V. Probiotics for the primary and secondary prevention of C. difficile Infections: A meta-analysis and systematic review. Antibiotics 2015, 4, 160–178. [Google Scholar] [CrossRef] [PubMed]
- McFarland, L.V. Meta-analysis of probiotics for the prevention of traveler’s diarrhea. Travel Med. Infect. Dis. 2007, 5, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Maragkoudakis, P.A.; Zoumpopoulou, G.; Miaris, C.; Kalantzopoulos, G.; Pot, B.; Tsakalidou, E. Probiotic potential of Lactobacillus strains isolated from dairy products. Int. Dairy J. 2006, 16, 189–199. [Google Scholar] [CrossRef]
- Zago, M.; Fornasari, M.E.; Carminati, D.; Burns, P.; Suàrez, V.; Vinerola, G.; Reinheimer, G.; Giraffa, G. Characterisation and probiotic potential of Lactobacillus plantarum strains isolated from cheeses. Food Microbiol. 2011, 28, 1033–1040. [Google Scholar] [CrossRef]
- Pavli, F.G.; Argyri, A.A.; Papadopoulou, O.S.; Nychas, G.-J.E.; Chorianopoulos, N.G.; Tassou, C.C. Probiotic potential of lactic acid bacteria from traditional fermented dairy and meat products: Assessment by in vitro tests and molecular characterization. J. Probiotics Health 2016, 4, 157. [Google Scholar] [CrossRef]
- Botta, C.; Langerholc, T.; Cencič, A.; Cocolin, L. In vitro selection and characterization of new probiotic candidates from table olive microbiota. PLoS ONE 2014, 9, e94457. [Google Scholar] [CrossRef]
- Argyri, A.A.; Zoumpopoulou, G.; Karatzas, K.-A.G.; Tsakalidou, E.; Nychas, G.-J.E.; Panagou, E.Z.; Tassou, C.C. Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food Microbiol. 2013, 33, 282–291. [Google Scholar] [CrossRef]
- Vitali, B.; Minervini, G.; Rizzello, C.G.; Spisni, E.; Maccaferri, S.; Brigidi, P.; Gobbetti, M.; Di Cagno, R. Novel probiotic candidates for humans isolated from raw fruits and vegetables. Food Microbiol. 2012, 31, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Pennacchia, C.; Ercolini, D.; Blaiotta, G.; Pepe, O.; Mauriello, G.; Villani, F. Selection of Lactobacillus strains from fermented sausages for their potential use as probiotics. Meat Sci. 2004, 67, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Kirtzalidou, E.; Pramateftaki, P.; Kotsou, M.; Kyriacou, A. Screening for lactobacilli with probiotic properties in the infant gut microbiota. Anaerobe 2011, 17, 440–443. [Google Scholar] [CrossRef] [PubMed]
- Vizoso-Pinto, M.G.; Franz, C.M.; Schillinger, U.; Holzapfel, W.H. Lactobacillus spp. with in vitro probiotic properties from human faeces and traditional fermented products. Int. J. Food Microbiol. 2006, 109, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.; Jiménez, E.; Olivares, M.; Marín, M.L.; Fernández, L.; Xaus, J.; Rodríguez, J.M. Lactobacillus salivarius CECT 5713, a potential probiotic strain isolated from infant feces and breast milk of a mother-child pair. Int. J. Food Microbiol. 2006, 112, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Peres, C.M.; Peres, C.; Hernández-Mendoza, A.; Malcata, F.X. Review on fermented plant materials as carriers and sources of potentially probiotic lactic acid bacteria-With an emphasis on table olives. Trends Food Sci. Technol. 2012, 26, 31–42. [Google Scholar] [CrossRef]
- Hurtado, A.; Reguant, C.; Bordons, A.; Rozès, N. Lactic acid bacteria from fermented table olives. Food Microbiol. 2012, 31, 1–8. [Google Scholar] [CrossRef]
- Argyri, A.A.; Nisiotou, A.A.; Mallouchos, A.; Panagou, E.Z.; Tassou, C.C. Performance of two potential probiotic Lactobacillus strains from the olive microbiota as starters in the fermentation of heat shocked green olives. Int. J. Food Microbiol. 2014, 171, 68–76. [Google Scholar] [CrossRef]
- Blana, V.A.; Grounta, A.; Tassou, C.C.; Nychas, G.-J.E.; Panagou, E.Z. Inoculated fermentation of green olives with potential probiotic Lactobacillus pentosus and Lactobacillus plantarum starter cultures isolated from industrially fermented olives. Food Microbiol. 2014, 38, 208–218. [Google Scholar] [CrossRef]
- Xavier, K.B.; Bassler, B.L. LuxS quorum sensing: More than just a numbers game. Curr. Opin. Microbiol. 2003, 6, 191–197. [Google Scholar] [CrossRef]
- Lebeer, S.; Verhoeven, T.L.A.; Vélez, M.P.; Vanderleyden, J.; De Keersmaecker, S.C.J. Impact of environmental and genetic factors on biofilm formation by the probiotic strain Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 2007, 73, 6768–6775. [Google Scholar] [CrossRef] [PubMed]
- Lebeer, S.; De Keersmaecker, S.C.J.; Verhokomeven, T.L.A.; Fadda, A.A.; Marchal, K.; Vanderleyden, J. Functional Analysis of luxS in the Probiotic Strain Lactobacillus rhamnosus GG Reveals a Central Metabolic Role Important for Growth and Biofilm Formation. J. Bacteriol. 2007, 189, 860–871. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wu, R.; Zhang, J.; Li, P. Overexpression of luxS Promotes Stress Resistance and Biofilm Formation of Lactobacillus paraplantarum L-ZS9 by Regulating the Expression of Multiple Genes. Front. Microbiol. 2018, 9, 2628. [Google Scholar] [CrossRef] [PubMed]
- Moslehi-Jenabian, S.; Gori, K.; Jespersen, L. AI-2 signalling is induced by acidic shock in probiotic strains of Lactobacillus spp. Int. J. Food Microbiol. 2009, 135, 295–302. [Google Scholar] [CrossRef] [PubMed]
- O’Byrne, C.P.; Feehily, C.; Ham, R.; Karatzas, K.A.G. A modified rapid enzymatic microtiter plate assay, for the quantification of intracellular γ-aminobutyric acid and succinate semialdehyde in bacterial cells. J. Microbiol. Methods 2011, 84, 137–139. [Google Scholar] [CrossRef]
- Foster, A.C.; Kemp, J.A. Glutamate- and GABA-based CNS therapeutics. Curr. Opin. Pharmacol. 2006, 6, 7–17. [Google Scholar] [CrossRef]
- Möhler, H. The GABA system in anxiety and depression and its therapeutic potential. Neuropharmacology 2012, 62, 42–53. [Google Scholar] [CrossRef]
- Franciosi, E.; Carafa, I.; Nardin, T.; Schiavon, S.; Poznanski, E.; Cavazza, A.; Larcher, A.; Tuohy, K.M. Biodiversity and γ-Aminobutyric Acid Production by Lactic Acid Bacteria Isolated from Traditional Alpine Raw Cow’s Milk Cheeses. BioMed Res. Int. 2015, 2015, 625740. [Google Scholar] [CrossRef]
- Siragusa, S.; De Angelis, M.; Di Cagno, R.; Rizzello, C.G.; Coda, R.; Gobbetti, M. Synthesis of γ-Aminobutyric Acid by Lactic Acid Bacteria Isolated from a Variety of Italian Cheeses. Appl. Environ. Microbiol. 2007, 73, 7283–7290. [Google Scholar] [CrossRef]
- Park, K.-B.; Oh, S.-H. Production of yogurt with enhanced levels of gamma-aminobutyric acid and valuable nutrients using lactic acid bacteria and germinated soybean extract. Bioresour. Technol. 2006, 98, 1675–1679. [Google Scholar] [CrossRef]
- Ohmori, T.; Tahara, M.; Ohshima, T. Mechanism of gamma-aminobutyric acid (GABA) production by a lactic acid bacterium in yogurt-sake. Process Biochem. 2018, 74, 21–27. [Google Scholar] [CrossRef]
- Doulgeraki, A.I.; Hondrodimou, O.; Iliopoulos, V.; Panagou, E.Z. Lactic acid bacteria and yeast heterogeneity during aerobic and modified atmosphere packaging storage of natural black Conservolea olives in polyethylene pouches. Food Control 2012, 26, 49–57. [Google Scholar] [CrossRef]
- Cocolin, L.; Stella, S.; Nappi, R.; Bozzetta, E.; Cantoni, C.; Comi, G. Analysis of PCR-based methods for characterization of Listeria monocytogenes strains isolated from different sources. Int. J. Food Microbiol. 2005, 103, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Hoiseth, S.K.; Stocker, B.A. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 1981, 291, 238–239. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Hume, M.E.; Pillai, S.D. Autoinducer-2-like activity associated with foods and its interaction with food additives. J. Food Prot. 2004, 67, 1457–1462. [Google Scholar] [CrossRef] [PubMed]
- Blana, V.A.; Doulgeraki, A.I.; Nychas, G.-J.E. Autoinducer-2-like Activity in Lactic Acid Bacteria Isolated from Minced Beef Packaged under Modified Atmospheres. J. Food Prot. 2011, 74, 631–635. [Google Scholar] [CrossRef] [PubMed]
- Surette, M.G.; Bassler, B.L. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 1998, 95, 7046–7050. [Google Scholar] [CrossRef]
- Tsukatani, T.; Higuchi, T.; Matsumoto, K. Enzyme-based microtiter plate assay for γ-aminobutyric acid: Application to the screening of γ-aminobutyric acid-producing lactic acid bacteria. Anal. Chim. Acta 2005, 540, 293–297. [Google Scholar] [CrossRef]
- Karatzas, K.-A.G.; Brennan, O.; Heavin, S.; Morrissey, J.; O’Byrne, C.P. Intracellular Accumulation of High Levels of γ-Aminobutyrate by Listeria monocytogenes 10403S in Response to Low pH: Uncoupling of γ-Aminobutyrate Synthesis from Efflux in a Chemically Defined Medium. Appl. Environ. Microbiol. 2010, 76, 3529–3537. [Google Scholar] [CrossRef]
- Taranto, M.P.; Perez-Martinez, G.; Font de Valdez, G. Effect of bile acid on the cell membrane functionality of lactic acid bacteria for oral administration. Res. Microbiol. 2006, 157, 720–725. [Google Scholar] [CrossRef]
- Jacobsen, C.N.; Rosenfeldt Nielsen, V.; Hayford, A.E.; Moller, P.L.; Michaelsen, K.F.; Pærregaard, A.; Sandström, B.; Tvede, M.; Jakobsen, M. Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl. Environ. Microbiol. 1999, 65, 4949–4956. [Google Scholar] [PubMed]
- Papadimitriou, K.; Zoumpopoulou, G.; Foligné, B.; Alexandraki, V.; Kazou, M.; Pot, B.; Tsakalidou, E. Discovering probiotic microorganisms: In vitro, in vivo, genetic and omics approaches. Front. Microbiol. 2015, 6, 58. [Google Scholar] [CrossRef] [PubMed]
- Begley, M.; Gahan, C.G.M.; Hill, C. The interaction between bacteria and bile. FEMS Microbiol. 2005, 29, 625–651. [Google Scholar] [CrossRef] [PubMed]
- Morelli, L. In vitro assessment of probiotic bacteria: From survival to functionality. Int. Dairy J. 2007, 17, 1278–1283. [Google Scholar] [CrossRef]
- Begley, M.; Hill, C.; Gahan, C.G.M. Bile Salt Hydrolase Activity in Probiotics. Appl. Environ. Microbiol. 2006, 72, 1729–1738. [Google Scholar] [CrossRef]
- Liong, M.T.; Shah, N.P. Acid and Bile tolerance and the cholesterol removal ability of lactobacilli strains. J. Dairy Sci. 2005, 88, 55–66. [Google Scholar] [CrossRef]
- Zoumpopoulou, G.; Foligne, B.; Christodoulou, K.; Grangette, C.; Pot, B.; Tsakalidou, E. Lactobacillus fermentum ACA-DC 179 displays probiotic potential in vitro and protects against trinitrobenzene sulfonic acid (TNBS)-induced colitis and Salmonella infection in murine models. Int. J. Food Microbiol. 2008, 121, 18–26. [Google Scholar] [CrossRef]
- Ruiz-Moyano, S.; Martín, A.; Benito, M.J.; Casquete, R.; Serradilla, M.J.; Córdoba, M.D.G. Safety and functional aspects of pre-selected lactobacilli for probiotic use in Iberian dry-fermented sausages. Meat Sci. 2009, 83, 460–467. [Google Scholar] [CrossRef]
- Tejero-Sariňena, S.; Barlow, J.; Costabile, A.; Gibson, G.R.; Rowland, I. In vitro evaluation of the antimicrobial activity of a range of probiotics against pathogens: Evidence for the effects of organic acids. Anaerobe 2012, 18, 530–538. [Google Scholar] [CrossRef]
- Rubio, R.; Jofré, A.; Martín, B.; Aymerich, T.; Garriga, M. Characterization of lactic acid bacteria isolated from infant faeces as potential probiotic starter cultures for fermented sausages. Food Microbiol. 2014, 38, 303–311. [Google Scholar] [CrossRef]
- Park, H.; Shin, H.; Lee, K.; Holzapfel, W. Autoinducer-2 properties of kimchi are associated with lactic acid bacteria involved in its fermentation. Int. J. Food Microbiol. 2016, 225, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Surette, M.G.; Bassler, B.L. Quorum sensing in Escherichia coli, Salmonella typhimurium and Vibrio harveyi: A new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 1999, 96, 1639–1644. [Google Scholar] [CrossRef] [PubMed]
- De Keersmaecker, S.C.J.; Vanderleyden, J. Constraints on detection of autoinducer-2 (AI-2) signalling molecules using Vibrio harveyi as a reporter. Microbiology 2003, 149, 1953–1956. [Google Scholar] [CrossRef] [PubMed]
- Ammor, M.S.; Michaelidis, C.; Nychas, G.-J.E. Insights into the role of quorum sensing in food spoilage. J. Food Prot. 2008, 71, 1510–1525. [Google Scholar] [CrossRef]
- Johansen, P.; Jespersen, L. Impact of quorum sensing on the quality of fermented foods. Curr. Opin. Food Sci. 2017, 13, 16–25. [Google Scholar] [CrossRef]
- Ruiz-Barba, J.L.; Caballero-Guerrero, B.; Maldonado-Barragán, A.; Jiménez-Díaz, R. Coculture with specific bacteria enhances survival of Lactobacillus plantarum NC8, an autoinducer-regulated bacteriocin producer, in olive fermentations. Food Microbiol. 2010, 27, 413–417. [Google Scholar] [CrossRef]
- Caballero-Guerrero, B.; Lucena-Padrós, H.; Maldonado-Barragán, A.; Ruiz-Barba, J.L. High-salt brines compromise autoinducer-mediated bacteriocinogenic Lactobacillus plantarum survival in Spanish-style green olive fermentations. Food Microbiol. 2013, 33, 90–96. [Google Scholar] [CrossRef]
- Rizzello, C.G.; Filannino, P.; Di Cagno, R.; Calasso, M.; Gobbeti, M. Quorum-sensing regulation of constitutive plantaricin by Lactobacillus plantarum strains under a model system for vegetables and fruits. Appl. Environ. Microbiol. 2014, 80, 777–787. [Google Scholar] [CrossRef]
- Doulgeraki, A.I.; Paraskevopoulos, N.; Nychas, G.J.E.; Panagou, E.Z. An in vitro study of Lactobacillus plantarum strains for the presence of plantaricin genes and their potential control of the table olive microbiota. Antonie van Leeuwenhoek 2013, 103, 821–832. [Google Scholar] [CrossRef]
- Feehily, C.; O’Byrne, C.P.; Karatzas, K.-A.G. Functional γ-Aminobutyrate Shunt in Listeria monocytogenes: Role in Acid Tolerance and Succinate Biosynthesis. Appl. Environ. Microbiol. 2013, 79, 74–80. [Google Scholar] [CrossRef]
- Komatsuzaki, N.; Shima, J.; Kawamoto, S.; Momose, H.; Kimura, T. Production of γ-aminobutyric acid (GABA) by Lactobacillus paracasei isolated from traditional fermented foods. Food Microbiol. 2005, 22, 497–504. [Google Scholar] [CrossRef]
- Karatzas, K.-A.G.; Suur, L.; O’Byrne, C.P. Characterisation of the intracellular glutamate decarboxylase system: Analysis of its function, transcription, and role in the acid resistance of various strains of Listeria monocytogenes. Appl. Environ. Microbiol. 2012, 78, 3571–3579. [Google Scholar] [CrossRef] [PubMed]
- Villegas, J.M.; Brown, L.; de Giori, G.S.; Hebert, E.M. Optimization of batch culture conditions for GABA production by Lactobacillus brevis CRL 1942, isolated from quinoa sourdough. LWT Food Sci. Technol. 2016, 67, 22–26. [Google Scholar] [CrossRef]
- Yunes, R.A.; Poluektova, E.U.; Dyachkova, M.S.; Klimina, K.M.; Kovtun, A.S.; Averina, O.V.; Orlova, V.S.; Danilenko, V.N. GABA production and structure of gadB/gadC genes in Lactobacillus and Bifidobacterium strains from human microbiota. Anaerobe 2016, 42, 197–204. [Google Scholar] [CrossRef]
- Barrett, E.; Ross, R.P.; O’Toole, P.W.; Fitzgerald, G.F.; Stanton, C. γ-Aminobutyric acid production by culturable bacteria from the human intestine. J. Appl. Microbiol. 2012, 113, 411–417. [Google Scholar] [CrossRef]
- Valenzuela, J.A.; Flórez, A.B.; Vázquez, L.; Vasek, O.M.; Mayo, B. Production of γ-aminobutyric acid (GABA) by lactic acid bacteria strains isolated from traditional, starter-free dairy products made of raw milk. Benef. Microbes 2019, 10, 579–589. [Google Scholar] [CrossRef]
- Sokovic Bajic, S.; Djokic, J.; Dinic, M.; Veljovic, K.; Golic, N.; Mihajlovic, S.; Tolinacki, M. GABA-Producing Natural Dairy Isolate from Artisanal Zlatar Cheese Attenuates Gut Inflammation and Strengthens Gut Epithelial Barrier in vitro. Front. Microbiol. 2019, 10, 527. [Google Scholar] [CrossRef]
- Shan, Y.; Man, C.X.; Han, X.; Li, L.; Guo, Y.; Deng, Y.; Li, T.; Zhang, L.W.; Jiang, Y.J. Evaluation of improved γ-aminobutyric acid production in yogurt using Lactobacillus plantarum NDC75017. J. Dairy Sci. 2015, 98, 2138–2149. [Google Scholar] [CrossRef]
- Renes, E.; Linares, D.M.; González, L.; Fresno, J.M.; Tornadijo, M.E.; Stanton, C. Production of conjugated linoleic acid and gamma-aminobutyric acid by autochthonous lactic acid bacteria and detection of the genes involved. J. Funct. Foods 2017, 34, 340–346. [Google Scholar] [CrossRef]
- Zhuang, K.; Jiang, Y.; Feng, X.; Li, L.; Dang, F.; Zhang, W.; Man, C. Transcriptomic response to GABA-producing Lactobacillus plantarum CGMCC 1.2437T induced by L-MSG. PLoS ONE 2018, 13, e0199021. [Google Scholar] [CrossRef]
- Mazzoli, R.; Pessione, E. The Neuro-endocrinological role of microbial glutamate and GABA signaling. Front. Microbiol. 2016, 7, 1934. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Lee, M.Y.; Ji, G.E.; Lee, Y.S.; Hwang, K.T. Production of γ-aminobutyric acid in black raspberry juice during fermentation by Lactobacillus brevis GABA100. Int. J. Food Microbiol. 2009, 130, 12–16. [Google Scholar] [CrossRef] [PubMed]

| Species | Strain | Final Counts (log CFU/mL) |
|---|---|---|
| Lactobacillus plantarum | B355 | <1 |
| Lactobacillus plantarum | B359 | 2.57 ± 1.14 |
| Lactobacillus plantarum | B372 | 6.05 ± 0.41 |
| Lactobacillus plantarum | B373 | 7.31 ± 0.63 |
| Lactobacillus plantarum | B374 | 4.15 ± 0.15 |
| Lactobacillus plantarum | B375 | 7.00 ± 0.83 |
| Lactobacillus plantarum | B380 | 7.68 ± 0.16 |
| Lactobacillus plantarum | B384 | 6.62 ± 0.43 |
| Lactobacillus pentosus | B356 | 6.69 ± 1.08 |
| Lactobacillus pentosus | B357 | <1 |
| Lactobacillus pentosus | B360 | 1.79 ± 1.18 |
| Lactobacillus pentosus | B361 | 3.96 ± 0.73 |
| Lactobacillus pentosus | B362 | 7.08 ± 1.29 |
| Lactobacillus pentosus | B363 | 6.81 ± 1.07 |
| Lactobacillus pentosus | B364 | 6.34 ± 1.32 |
| Lactobacillus pentosus | B366 | 6.78 ± 0.22 |
| Lactobacillus pentosus | B368 | 7.01 ± 1.01 |
| Lactobacillus pentosus | B369 | 5.48 ± 0.07 |
| Lactobacillus pentosus | B370 | 7.61 ± 0.14 |
| Lactobacillus pentosus | B371 | 6.26 ± 0.52 |
| Lactobacillus pentosus | B377 | 7.85 ± 0.11 |
| Lactobacillus pentosus | B378 | 6.92 ± 0.83 |
| Lactobacillus pentosus | B383 | 4.61 ± 0.31 |
| Lactobacillus pentosus | B385 | 7.24 ± 0.73 |
| Lactobacillus pentosus | B399 | 6.48 ± 0.01 |
| Lactobacillus pentosus | B400 | 2.07 ± 0.32 |
| Lactobacillus pentosus | B401 | <1 |
| Lactobacillus pentosus | B402 | <1 |
| Lactobacillus paraplantarum | B365 | 3.22 ± 1.56 |
| Pediococcus ethanolidurans | B389 | 6.46 ± 0.25 |
| Pediococcus ethanolidurans | B397 | <1 |
| Lactobacillus coryniformis | B395 | 2.57 ± 0.11 |
| Lactobacillus coryniformis | B403 | <1 |
| Strains | Test | ||||
|---|---|---|---|---|---|
| Low pH (SR%) A | Bile Salts (SR%) B | Bile Salts Hydrolase C | Haemolytic Activity | Antimicrobial Activity | |
| Lactobacillus pentosus B356 | 72.17 | 91.38 | 1 | γ | - |
| Lactobacillus pentosus B362 | 75.05 | 87.47 | 1 | γ | - |
| Lactobacillus pentosus B363 | 72.84 | 93.42 | 1 | γ | - |
| Lactobacillus pentosus B364 | 67.55 | 91.88 | 1 | γ | - |
| Lactobacillus pentosus B366 | 76.38 | 90.83 | 1 | γ | - |
| Lactobacillus pentosus B368 | 80.16 | 96.78 | 1 | γ | - |
| Lactobacillus pentosus B370 | 87.38 | 96.83 | 1 | γ | - |
| Lactobacillus pentosus B371 | 71.86 | 90.04 | 1 | γ | - |
| Lactobacillus plantarum B372 | 66.93 | 101.29 | 1 | γ | - |
| Lactobacillus plantarum B373 | 77.40 | 98.46 | 1 | γ | - |
| Lactobacillus plantarum B375 | 74.12 | 94.60 | 1 | γ | - |
| Lactobacillus pentosus B377 | 88.19 | 97.26 | 1 | γ | - |
| Lactobacillus pentosus B378 | 76.20 | 93.88 | 0 | γ | - |
| Lactobacillus plantarum B380 | 84.88 | 93.53 | 1 | γ | - |
| Lactobacillus plantarum B384 | 72.35 | 96.76 | 1 | γ | - |
| Lactobacillus pentosus B385 | 77.41 | 95.03 | 1 | γ | - |
| Lactobacillus pentosus B399 | 70.56 | 91.32 | 0 | γ | - |
| Strains | AI-2 Activity | GABA Determination | |||
|---|---|---|---|---|---|
| Bacterial Counts A | Relative AI-2 Activity B | Bacterial Counts C | pH D | GABA E | |
| Lactobacillus pentosus B356 | 7.57 ± 0.15 | 0.68 ± 0.22 a | 9.52 ± 0.17 | 3.92 ± 0.00 | 0.30 ± 0.05 a |
| Lactobacillus pentosus B362 | 7.55 ± 0.06 | 0.57 ± 0.15 a | 9.40 ± 0.17 | 3.91 ± 0.00 | 0.16 ± 0.03 a |
| Lactobacillus pentosus B363 | 7.13 ± 0.22 | 0.70 ± 0.27 a | 9.52 ± 0.17 | 4.09 ± 0.01 | 0.52 ± 0.49 a |
| Lactobacillus pentosus B364 | 7.26 ± 0.08 | 0.58 ± 0.28 a | 9.44 ± 0.17 | 4.00 ± 0.04 | 0.40 ± 0.11 a |
| Lactobacillus pentosus B366 | 7.47 ± 0.09 | 0.48 ± 0.11 a | 9.66 ± 0.42 | 3.92 ± 0.00 | 0.30 ± 0.07 a |
| Lactobacillus pentosus B368 | 7.51 ± 0.37 | 0.50 ± 0.16 a | 9.32 ± 0.06 | 4.02 ± 0.03 | 0.27 ± 0.06 a |
| Lactobacillus pentosus B370 | 8.30 ± 0.24 | 0.56 ± 0.17 a | 9.49 ± 0.15 | 4.00 ± 0.04 | 0.39 ± 0.06 a |
| Lactobacillus pentosus B371 | 7.73 ± 0.15 | 0.58 ± 0.24 a | 9.48 ± 0.19 | 3.93 ± 0.00 | 0.46 ± 0.08 a |
| Lactobacillus plantarum B372 | 7.28 ± 0.14 | 1.39 ± 0.45 b | 9.51 ± 0.07 | 4.05 ± 0.03 | 0.66 ± 0.05 a |
| Lactobacillus plantarum B373 | 8.11 ± 0.52 | 1.28 ± 0.33 b | 9.31 ± 0.01 | 3.95 ± 0.03 | 0.56 ± 0.51 a |
| Lactobacillus plantarum B375 | 8.15 ± 0.22 | 1.34 ± 0.68 b | 9.60 ± 0.43 | 3.93 ± 0.00 | 0.53 ± 0.13 a |
| Lactobacillus pentosus B377 | 6.98 ± 0.13 | 0.53 ± 0.27 a | 9.28 ± 0.24 | 3.99 ± 0.06 | 0.30 ± 0.07 a |
| Lactobacillus pentosus B378 | 7.18 ± 0.50 | 0.32 ± 0.12 a | 9.41 ± 0.42 | 3.92 ± 0.00 | 0.14 ± 0.10 a |
| Lactobacillus plantarum B380 | 7.34 ± 0.01 | 1.57 ± 1.16 b | 9.29 ± 0.16 | 3.92 ± 0.00 | 0.40 ± 0.05 a |
| Lactobacillus plantarum B384 | 6.42 ± 0.16 | 0.38 ± 0.14 a | 9.27 ± 0.27 | 4.06 ± 0.01 | 0.42 ± 0.17 a |
| Lactobacillus pentosus B385 | 7.15 ± 0.22 | 0.54 ± 0.15 a | 9.22 ± 0.32 | 3.95 ± 0.04 | 0.34 ± 0.21 a |
| Lactobacillus pentosus B399 | 6.94 ± 0.15 | 0.48 ± 0.19 a | 9.49 ± 0.02 | 3.95 ± 0.03 | 0.23 ± 0.02 a |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavli, F.; Gkana, E.; Adebambo, O.; Karatzas, K.-A.; Panagou, E.; Nychas, G.-J.E. Ιn Vitro Screening of γ-Aminobutyric Acid and Autoinducer-2 Signalling in Lactic Acid Bacteria Exhibiting Probiotic Potential Isolated from Natural Black Conservolea Olives. Foods 2019, 8, 640. https://doi.org/10.3390/foods8120640
Pavli F, Gkana E, Adebambo O, Karatzas K-A, Panagou E, Nychas G-JE. Ιn Vitro Screening of γ-Aminobutyric Acid and Autoinducer-2 Signalling in Lactic Acid Bacteria Exhibiting Probiotic Potential Isolated from Natural Black Conservolea Olives. Foods. 2019; 8(12):640. https://doi.org/10.3390/foods8120640
Chicago/Turabian StylePavli, Foteini, Eleni Gkana, Oluwabunmi Adebambo, Kimon-Andreas Karatzas, Efstathios Panagou, and George-John E. Nychas. 2019. "Ιn Vitro Screening of γ-Aminobutyric Acid and Autoinducer-2 Signalling in Lactic Acid Bacteria Exhibiting Probiotic Potential Isolated from Natural Black Conservolea Olives" Foods 8, no. 12: 640. https://doi.org/10.3390/foods8120640
APA StylePavli, F., Gkana, E., Adebambo, O., Karatzas, K.-A., Panagou, E., & Nychas, G.-J. E. (2019). Ιn Vitro Screening of γ-Aminobutyric Acid and Autoinducer-2 Signalling in Lactic Acid Bacteria Exhibiting Probiotic Potential Isolated from Natural Black Conservolea Olives. Foods, 8(12), 640. https://doi.org/10.3390/foods8120640