CK2 Chemical Probes: Past, Present, and Future
Abstract
1. Introduction
1.1. CK2 General Structure and Small Molecule Binding Sites
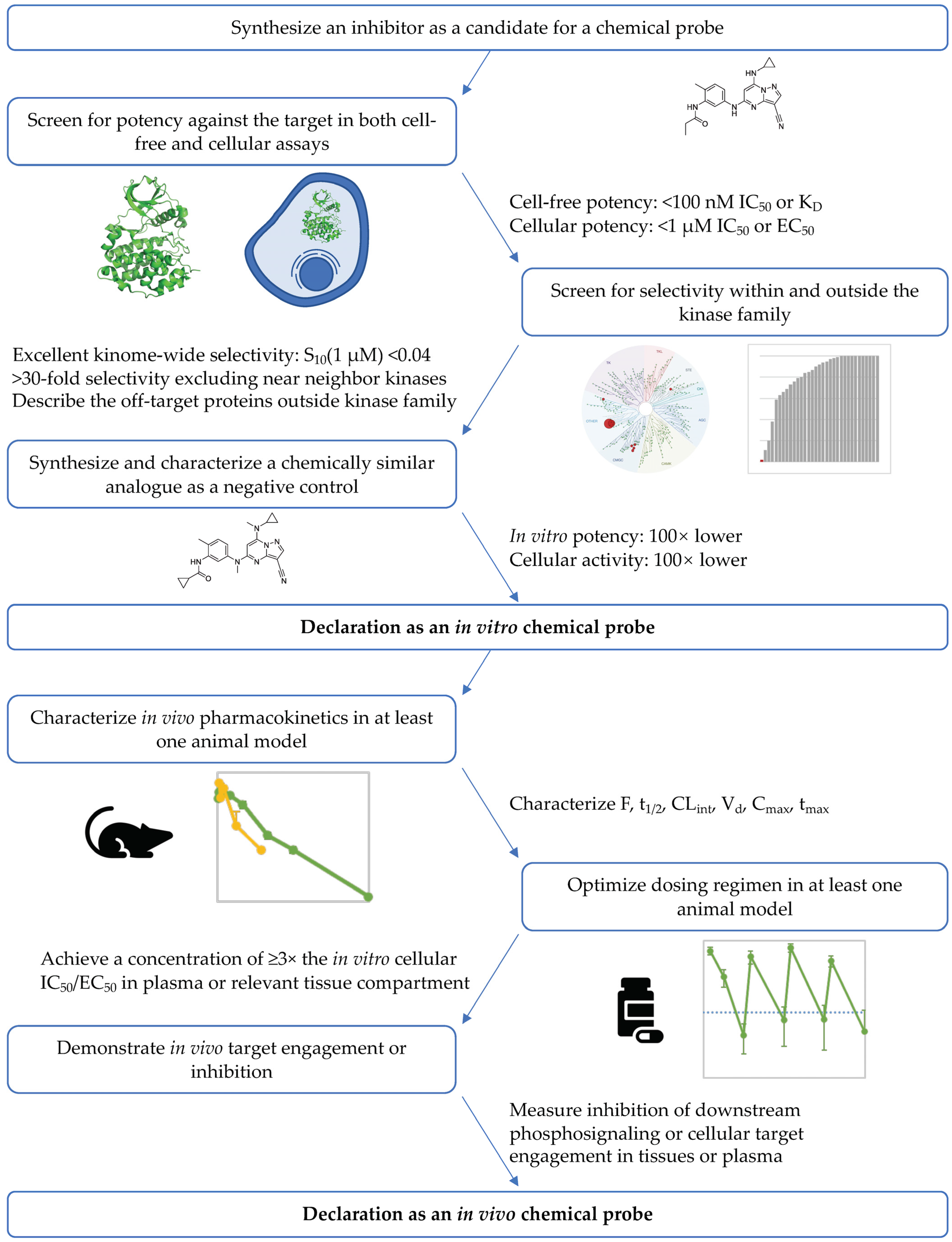
1.2. Chemical Probe Definition
| SGC Kinase Chemical Probe Criteria |
|---|
| 1. Inhibitor/agonist cell-free potency: <100 nM IC50 or KD; |
| 2. Excellent kinome-wide selectivity: S10 (1 µM) < 0.04; |
| 3. Selectivity within kinase family: >30-fold excluding near neighbor kinases; |
| 4. Selectivity outside kinase family: describe the off-target proteins; |
| 5. On target activity in cells: <1 µM IC50 or EC50; |
| 6. Negative control: in vitro potency 100× less; cellular activity 100× less. |
2. Small Molecule Inhibitors of CK2 and Chemical Probe Development
2.1. Selected Orthosteric CK2 Inhibitors and Chemical Probes
2.2. Selected Allosteric and Bivalent CK2 Inhibitors in Development
3. Use of SGC-CK2-1 by the Community in Cell-Based Studies
4. Development of an In Vivo CK2 Probe
4.1. Definition of an In Vivo Chemical Probe
4.2. Progress toward an In Vivo Chemical Probe
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Meggio, F.; Pinna, L.A. One-thousand-and-one substrates of protein kinase CK2? FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2003, 17, 349–368. [Google Scholar] [CrossRef] [PubMed]
- Menyhart, D.; Gyenis, L.; Jurcic, K.; Roffey, S.E.; Puri, A.; Jovanovic, P.; Szkop, K.J.; Pittock, P.; Lajoie, G.; Axtman, A.D.; et al. Comparison of CX-4945 and SGC-CK2-1 as inhibitors of CSNK2 using quantitative phosphoproteomics: Triple SILAC in combination with inhibitor-resistant CSNK2. Curr. Res. Chem. Biol. 2023, 3, 100041. [Google Scholar] [CrossRef]
- Salvi, M.; Borgo, C.; Pinna, L.A.; Ruzzene, M. Targeting CK2 in cancer: A valuable strategy or a waste of time? Cell Death Discov. 2021, 7, 325. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Dickmander, R.J.; Bayati, A.; Taft-Benz, S.A.; Smith, J.L.; Wells, C.I.; Madden, E.A.; Brown, J.W.; Lenarcic, E.M.; Yount, B.L.; et al. Host Kinase CSNK2 is a Target for Inhibition of Pathogenic SARS-like β-Coronaviruses. ACS Chem. Biol. 2022, 17, 1937–1950. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xia, Y.; Wang, Y.; Shentu, Y.; Zeng, K.; Mahaman, Y.A.R.; Huang, F.; Wu, M.; Ke, D.; Wang, Q.; et al. CK2 Phosphorylating I2PP2A/SET Mediates Tau Pathology and Cognitive Impairment. Front. Mol. Neurosci. 2018, 11, 146. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; McFarland, B.C.; Drygin, D.; Yu, H.; Bellis, S.L.; Kim, H.; Bredel, M.; Benveniste, E.N. Targeting Protein Kinase CK2 Suppresses Prosurvival Signaling Pathways and Growth of Glioblastoma. Clin. Cancer Res. 2013, 19, 6484–6494. [Google Scholar] [CrossRef]
- Rosenberger, A.F.N.; Morrema, T.H.J.; Gerritsen, W.H.; van Haastert, E.S.; Snkhchyan, H.; Hilhorst, R.; Rozemuller, A.J.M.; Scheltens, P.; van der Vies, S.M.; Hoozemans, J.J.M. Increased occurrence of protein kinase CK2 in astrocytes in Alzheimer’s disease pathology. J. Neuroinflamm. 2016, 13, 4. [Google Scholar] [CrossRef]
- Perez, D.I.; Gil, C.; Martinez, A. Protein kinases CK1 and CK2 as new targets for neurodegenerative diseases. Med. Res. Rev. 2011, 31, 924–954. [Google Scholar] [CrossRef] [PubMed]
- Borgo, C.; D’Amore, C.; Sarno, S.; Salvi, M.; Ruzzene, M. Protein kinase CK2: A potential therapeutic target for diverse human diseases. Signal Transduct. Target. Ther. 2021, 6, 183. [Google Scholar] [CrossRef]
- Litchfield, D.W.; Gyenis, L.; Menyhart, D.; Roffey, S.E. Towards the CSNK2 phosphoproteome—With lessons from the COVID-19 pandemic to revealing the secrets of CSNK2 and its promise as a therapeutic target. Biochim. Biophys. Acta (BBA)—Gen. Subj. 2023, 1867, 130441. [Google Scholar] [CrossRef]
- Litchfield, D.W.; Bosc, D.G.; Canton, D.A.; Saulnier, R.B.; Vilk, G.; Zhang, C. Functional specialization of CK2 isoforms and characterization of isoform-specific binding partners. Mol. Cell. Biochem. 2001, 227, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Pinna, L.A. Protein kinase CK2: A challenge to canons. J. Cell Sci. 2002, 115, 3873–3878. [Google Scholar] [CrossRef] [PubMed]
- Bibby, A.C.; Litchfield, D.W. The multiple personalities of the regulatory subunit of protein kinase CK2: CK2 dependent and CK2 independent roles reveal a secret identity for CK2beta. Int. J. Biol. Sci. 2005, 1, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Iegre, J.; Atkinson, E.L.; Brear, P.D.; Cooper, B.M.; Hyvönen, M.; Spring, D.R. Chemical probes targeting the kinase CK2: A journey outside the catalytic box. Org. Biomol. Chem. 2021, 19, 4380–4396. [Google Scholar] [CrossRef]
- Atkinson, E.L.; Iegre, J.; Brear, P.D.; Zhabina, E.A.; Hyvönen, M.; Spring, D.R. Downfalls of Chemical Probes Acting at the Kinase ATP-Site: CK2 as a Case Study. Molecules 2021, 26, 1977. [Google Scholar] [CrossRef]
- Davis-Gilbert, Z.W.; Krämer, A.; Dunford, J.E.; Howell, S.; Senbabaoglu, F.; Wells, C.I.; Bashore, F.M.; Havener, T.M.; Smith, J.L.; Hossain, M.A.; et al. Discovery of a Potent and Selective Naphthyridine-Based Chemical Probe for Casein Kinase 2. ACS Med. Chem. Lett. 2023, 14, 432–441. [Google Scholar] [CrossRef]
- Wells, C.I.; Drewry, D.H.; Pickett, J.E.; Tjaden, A.; Krämer, A.; Müller, S.; Gyenis, L.; Menyhart, D.; Litchfield, D.W.; Knapp, S.; et al. Development of a Potent and Selective Chemical Probe for the Pleiotropic Kinase CK2. Cell Chem. Biol. 2021, 28, 546–558.e10. [Google Scholar] [CrossRef]
- Krämer, A.; Kurz, C.G.; Berger, B.-T.; Celik, I.E.; Tjaden, A.; Greco, F.A.; Knapp, S.; Hanke, T. Optimization of pyrazolo[1,5-a]pyrimidines lead to the identification of a highly selective casein kinase 2 inhibitor. Eur. J. Med. Chem. 2020, 208, 112770. [Google Scholar] [CrossRef]
- Pierre, F.; Chua, P.C.; O’Brien, S.E.; Siddiqui-Jain, A.; Bourbon, P.; Haddach, M.; Michaux, J.; Nagasawa, J.; Schwaebe, M.K.; Stefan, E.; et al. Discovery and SAR of 5-(3-chlorophenylamino)benzo[c][2,6]naphthyridine-8-carboxylic acid (CX-4945), the first clinical stage inhibitor of protein kinase CK2 for the treatment of cancer. J. Med. Chem. 2011, 54, 635–654. [Google Scholar] [CrossRef]
- Battistutta, R.; Cozza, G.; Pierre, F.; Papinutto, E.; Lolli, G.; Sarno, S.; O’Brien, S.E.; Siddiqui-Jain, A.; Haddach, M.; Anderes, K.; et al. Unprecedented selectivity and structural determinants of a new class of protein kinase CK2 inhibitors in clinical trials for the treatment of cancer. Biochemistry 2011, 50, 8478–8488. [Google Scholar] [CrossRef]
- Kufareva, I.; Bestgen, B.; Brear, P.; Prudent, R.; Laudet, B.; Moucadel, V.; Ettaoussi, M.; Sautel, C.F.; Krimm, I.; Engel, M.; et al. Discovery of holoenzyme-disrupting chemicals as substrate-selective CK2 inhibitors. Sci. Rep. 2019, 9, 15893. [Google Scholar] [CrossRef] [PubMed]
- Meggio, F.; Shugar, D.; Pinna, L.A. Ribofuranosyl-benzimidazole derivatives as inhibitors of casein kinase-2 and casein kinase-1. Eur. J. Biochem. 1990, 187, 89–94. [Google Scholar] [CrossRef]
- Laudet, B.; Moucadel, V.; Prudent, R.; Filhol, O.; Wong, Y.S.; Royer, D.; Cochet, C. Identification of chemical inhibitors of protein-kinase CK2 subunit interaction. Mol. Cell Biochem. 2008, 316, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Moucadel, V.; Prudent, R.; Sautel, C.F.; Teillet, F.; Barette, C.; Lafanechere, L.; Receveur-Brechot, V.; Cochet, C. Antitumoral activity of allosteric inhibitors of protein kinase CK2. Oncotarget 2011, 2, 997–1010. [Google Scholar] [CrossRef] [PubMed]
- De Fusco, C.; Brear, P.; Iegre, J.; Georgiou, K.H.; Sore, H.F.; Hyvönen, M.; Spring, D.R. A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066. Bioorg. Med. Chem. 2017, 25, 3471–3482. [Google Scholar] [CrossRef]
- Lindenblatt, D.; Applegate, V.; Nickelsen, A.; Klußmann, M.; Neundorf, I.; Götz, C.; Jose, J.; Niefind, K. Molecular Plasticity of Crystalline CK2α′ Leads to KN2, a Bivalent Inhibitor of Protein Kinase CK2 with Extraordinary Selectivity. J. Med. Chem. 2022, 65, 1302–1312. [Google Scholar] [CrossRef]
- Brear, P.; De Fusco, C.; Hadje Georgiou, K.; Francis-Newton, N.J.; Stubbs, C.J.; Sore, H.F.; Venkitaraman, A.R.; Abell, C.; Spring, D.R.; Hyvönen, M. Specific inhibition of CK2α from an anchor outside the active site. Chem. Sci. 2016, 7, 6839–6845. [Google Scholar] [CrossRef]
- Bancet, A.; Frem, R.; Jeanneret, F.; Mularoni, A.; Bazelle, P.; Roelants, C.; Delcros, J.-G.; Guichou, J.-F.; Pillet, C.; Coste, I.; et al. AB668, a novel highly selective protein kinase CK2 inhibitor with a distinct anti-tumor mechanism as compared to CX-4945 and SGC-CK2-1. bioRxiv 2022. [Google Scholar] [CrossRef]
- Perea, S.E.; Reyes, O.; Puchades, Y.; Mendoza, O.; Vispo, N.S.; Torrens, I.; Santos, A.; Silva, R.; Acevedo, B.; López, E.; et al. Antitumor effect of a novel proapoptotic peptide that impairs the phosphorylation by the protein kinase 2 (casein kinase 2). Cancer Res. 2004, 64, 7127–7129. [Google Scholar] [CrossRef]
- Gottardo, M.F.; Capobianco, C.S.; Sidabra, J.E.; Garona, J.; Perera, Y.; Perea, S.E.; Alonso, D.F.; Farina, H.G. Preclinical efficacy of CIGB-300, an anti-CK2 peptide, on breast cancer metastasic colonization. Sci. Rep. 2020, 10, 14689. [Google Scholar] [CrossRef]
- Enkvist, E.; Viht, K.; Bischoff, N.; Vahter, J.; Saaver, S.; Raidaru, G.; Issinger, O.-G.; Niefind, K.; Uri, A. A subnanomolar fluorescent probe for protein kinase CK2 interaction studies. Org. Biomol. Chem. 2012, 10, 8645–8653. [Google Scholar] [CrossRef] [PubMed]
- Viht, K.; Saaver, S.; Vahter, J.; Enkvist, E.; Lavogina, D.; Sinijärv, H.; Raidaru, G.; Guerra, B.; Issinger, O.G.; Uri, A. Acetoxymethyl Ester of Tetrabromobenzimidazole-Peptoid Conjugate for Inhibition of Protein Kinase CK2 in Living Cells. Bioconjugate Chem. 2015, 26, 2324–2335. [Google Scholar] [CrossRef] [PubMed]
- Tsien, R.Y. A non-disruptive technique for loading calcium buffers and indicators into cells. Nature 1981, 290, 527–528. [Google Scholar] [CrossRef]
- Vahter, J.; Viht, K.; Uri, A.; Enkvist, E. Oligo-aspartic acid conjugates with benzo[c][2,6]naphthyridine-8-carboxylic acid scaffold as picomolar inhibitors of CK2. Bioorg. Med. Chem. 2017, 25, 2277–2284. [Google Scholar] [CrossRef]
- Wells, C.I.; Al-Ali, H.; Andrews, D.M.; Asquith, C.R.M.; Axtman, A.D.; Dikic, I.; Ebner, D.; Ettmayer, P.; Fischer, C.; Frederiksen, M.; et al. The Kinase Chemogenomic Set (KCGS): An Open Science Resource for Kinase Vulnerability Identification. Int. J. Mol. Sci. 2021, 22, 566. [Google Scholar] [CrossRef] [PubMed]
- Wells, C.; Couñago, R.M.; Limas, J.C.; Almeida, T.L.; Cook, J.G.; Drewry, D.H.; Elkins, J.M.; Gileadi, O.; Kapadia, N.R.; Lorente-Macias, A.; et al. SGC-AAK1-1: A Chemical Probe Targeting AAK1 and BMP2K. ACS Med. Chem. Lett. 2019, 11, 340–345. [Google Scholar] [CrossRef]
- Besnard, J.; Ruda, G.F.; Setola, V.; Abecassis, K.; Rodriguiz, R.M.; Huang, X.P.; Norval, S.; Sassano, M.F.; Shin, A.I.; Webster, L.A.; et al. Automated design of ligands to polypharmacological profiles. Nature 2012, 492, 215–220. [Google Scholar] [CrossRef]
- Kroeze, W.K.; Sassano, M.F.; Huang, X.P.; Lansu, K.; McCorvy, J.D.; Giguère, P.M.; Sciaky, N.; Roth, B.L. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat. Struct. Mol. Biol. 2015, 22, 362–369. [Google Scholar] [CrossRef]
- Huang, X.P.; Mangano, T.; Hufeisen, S.; Setola, V.; Roth, B.L. Identification of human Ether-à-go-go related gene modulators by three screening platforms in an academic drug-discovery setting. Assay Drug Dev. Technol. 2010, 8, 727–742. [Google Scholar] [CrossRef]
- Vasta, J.D.; Corona, C.R.; Wilkinson, J.; Zimprich, C.A.; Hartnett, J.R.; Ingold, M.R.; Zimmerman, K.; Machleidt, T.; Kirkland, T.A.; Huwiler, K.G.; et al. Quantitative, Wide-Spectrum Kinase Profiling in Live Cells for Assessing the Effect of Cellular ATP on Target Engagement. Cell Chem. Biol. 2018, 25, 206–214. [Google Scholar] [CrossRef]
- Arrowsmith, C.H.; Audia, J.E.; Austin, C.; Baell, J.; Bennett, J.; Blagg, J.; Bountra, C.; Brennan, P.E.; Brown, P.J.; Bunnage, M.E.; et al. The promise and peril of chemical probes. Nat. Chem. Biol. 2015, 11, 536–541. [Google Scholar] [CrossRef] [PubMed]
- Chuaqui, C.E.; Dowling, J.E.; Lyne, P.; Pontz, T.; Ye, Q. Preparation of Cyanoarylaminocycloalkylaminopyrrolopyri Midine Derivatives for Use as Antitumor Agents; WO 2013144532A1; AstraZeneca AB: Södertälje, Sweden; AstraZeneca UK Limited: Cambridge, UK, 2013. [Google Scholar]
- Dowling, J.E.; Alimzhanov, M.; Bao, L.; Block, M.H.; Chuaqui, C.; Cooke, E.L.; Denz, C.R.; Hird, A.; Huang, S.; Larsen, N.A.; et al. Structure and Property Based Design of Pyrazolo[1,5-a]pyrimidine Inhibitors of CK2 Kinase with Activity in Vivo. ACS Med. Chem. Lett. 2013, 4, 800–805. [Google Scholar] [CrossRef] [PubMed]
- Dowling, J.E.; Alimzhanov, M.; Bao, L.; Chuaqui, C.; Denz, C.R.; Jenkins, E.; Larsen, N.A.; Lyne, P.D.; Pontz, T.; Ye, Q.; et al. Potent and Selective CK2 Kinase Inhibitors with Effects on Wnt Pathway Signaling in Vivo. ACS Med. Chem. Lett. 2016, 7, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Dowling, J.E.; Chuaqui, C.; Pontz, T.W.; Lyne, P.D.; Larsen, N.A.; Block, M.H.; Chen, H.; Su, N.; Wu, A.; Russell, D.; et al. Potent and Selective Inhibitors of CK2 Kinase Identified through Structure-Guided Hybridization. ACS Med. Chem. Lett. 2012, 3, 278–283. [Google Scholar] [CrossRef] [PubMed]
- D’Amore, C.; Borgo, C.; Sarno, S.; Salvi, M. Role of CK2 inhibitor CX-4945 in anti-cancer combination therapy—Potential clinical relevance. Cell. Oncol. 2020, 43, 1003–1016. [Google Scholar] [CrossRef] [PubMed]
- Marschke, R.F.; Borad, M.J.; McFarland, R.W.; Alvarez, R.H.; Lim, J.K.; Padgett, C.S.; Hoff, D.D.V.; O’Brien, S.E.; Northfelt, D.W. Findings from the phase I clinical trials of CX-4945, an orally available inhibitor of CK2. J. Clin. Oncol. 2011, 29, 3087. [Google Scholar] [CrossRef]
- Quezada Meza, C.P.; Ruzzene, M. Protein Kinase CK2 and SARS-CoV-2: An Expected Interplay Story. Kinases Phosphatases 2023, 1, 141–150. [Google Scholar] [CrossRef]
- Licciardello, M.P.; Workman, P. A New Chemical Probe Challenges the Broad Cancer Essentiality of CK2. Trends Pharmacol. Sci. 2021, 42, 313–315. [Google Scholar] [CrossRef]
- Mishra, S.; Kinoshita, C.; Axtman, A.D.; Young, J.E. Evaluation of a selective chemical probe validates that CK2 mediates neuroinflammation in an hiPSC-derived microglial model. Front. Mol. Neurosci. 2022, 15, 824956. [Google Scholar] [CrossRef]
- Forlenza, O.V.; Diniz, B.S.; Talib, L.L.; Mendonça, V.A.; Ojopi, E.B.; Gattaz, W.F.; Teixeira, A.L. Increased serum IL-1beta level in Alzheimer’s disease and mild cognitive impairment. Dement. Geriatr. Cogn. Disord. 2009, 28, 507–512. [Google Scholar] [CrossRef]
- Huang, Z.-B.; Sheng, G.-Q. Interleukin-1β with learning and memory. Neurosci. Bull. 2010, 26, 455–468. [Google Scholar] [CrossRef]
- Swardfager, W.; Lanctôt, K.; Rothenburg, L.; Wong, A.; Cappell, J.; Herrmann, N. A meta-analysis of cytokines in Alzheimer’s disease. Biol. Psychiatry 2010, 68, 930–941. [Google Scholar] [CrossRef] [PubMed]
- Brosseron, F.; Krauthausen, M.; Kummer, M.; Heneka, M.T. Body fluid cytokine levels in mild cognitive impairment and Alzheimer’s disease: A comparative overview. Mol. Neurobiol. 2014, 50, 534–544. [Google Scholar] [CrossRef] [PubMed]
- Scarabino, D.; Peconi, M.; Broggio, E.; Gambina, G.; Maggi, E.; Armeli, F.; Mantuano, E.; Morello, M.; Corbo, R.M.; Businaro, R. Relationship between proinflammatory cytokines (Il-1beta, Il-18) and leukocyte telomere length in mild cognitive impairment and Alzheimer’s disease. Exp. Gerontol. 2020, 136, 110945. [Google Scholar] [CrossRef]
- Lyra E Silva, N.M.; Gonçalves, R.A.; Pascoal, T.A.; Lima-Filho, R.A.S.; Resende, E.d.P.F.; Vieira, E.L.M.; Teixeira, A.L.; de Souza, L.C.; Peny, J.A.; Fortuna, J.T.S.; et al. Pro-inflammatory interleukin-6 signaling links cognitive impairments and peripheral metabolic alterations in Alzheimer’s disease. Transl. Psychiatry 2021, 11, 251. [Google Scholar] [CrossRef]
- Gyenis, L.; Menyhart, D.; Cruise, E.S.; Jurcic, K.; Roffey, S.E.; Chai, D.B.; Trifoi, F.; Fess, S.R.; Desormeaux, P.J.; Núñez de Villavicencio Díaz, T.; et al. Chemical Genetic Validation of CSNK2 Substrates Using an Inhibitor-Resistant Mutant in Combination with Triple SILAC Quantitative Phosphoproteomics. Front. Mol. Biosci. 2022, 9, 909711. [Google Scholar] [CrossRef]
- Pack, M.; Götz, C.; Wrublewsky, S.; Montenarh, M. SGC-CK2-1 Is an Efficient Inducer of Insulin Production and Secretion in Pancreatic β-Cells. Pharmaceutics 2022, 14, 19. [Google Scholar] [CrossRef] [PubMed]
- Boewe, A.S.; Wemmert, S.; Kulas, P.; Schick, B.; Götz, C.; Wrublewsky, S.; Montenarh, M.; Menger, M.D.; Laschke, M.W.; Ampofo, E. Inhibition of CK2 Reduces NG2 Expression in Juvenile Angiofibroma. Biomedicines 2022, 10, 966. [Google Scholar] [CrossRef]
- Al Shoyaib, A.; Archie, S.R.; Karamyan, V.T. Intraperitoneal Route of Drug Administration: Should it Be Used in Experimental Animal Studies? Pharm. Res. 2019, 37, 12. [Google Scholar] [CrossRef]
- Barrow, J.C.; Lindsley, C.W. The Importance of PK–PD. J. Med. Chem. 2023, 66, 4273–4274. [Google Scholar] [CrossRef]
- Molina, D.M.; Jafari, R.; Ignatushchenko, M.; Seki, T.; Larsson, E.A.; Dan, C.; Sreekumar, L.; Cao, Y.; Nordlund, P. Monitoring Drug Target Engagement in Cells and Tissues Using the Cellular Thermal Shift Assay. Science 2013, 341, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Tolvanen, T.A. Current Advances in CETSA. Front. Mol. Biosci. 2022, 9, 866764. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ong, H.W.; Dickmander, R.J.; Smith, J.L.; Brown, J.W.; Tao, W.; Chang, E.; Moorman, N.J.; Axtman, A.D.; Willson, T.M. Optimization of 3-Cyano-7-cyclopropylamino-pyrazolo[1,5-a]pyrimidines Toward the Development of an In Vivo Chemical Probe for CSNK2A. bioRxiv 2023. [Google Scholar] [CrossRef]
- Eroglu, Z.; Cowey, C.L.; Soong, J.; McCormick, D.; Fan, P.; Chen, J.; Elgendy, M.; Jang, S.; Chang, A.L.S. A phase I study of CX-4945 administered orally twice daily to patients with advanced basal cell carcinoma. J. Clin. Oncol. 2020, 38, TPS10080. [Google Scholar] [CrossRef]
- Siddiqui-Jain, A.; Chua, P.; Darjania, L.; Lim, J.; O’Brien, S.; Pierre, F.; Streiner, N.; Trent, K.; Whitten, J.; Rice, W. The discovery and characterization of CX-5011, a highly selective, potent inhibitor of Protein Kinase CK2. Cancer Res. 2008, 68, 4875. [Google Scholar]
- Siddiqui-Jain, A.; Drygin, D.; Streiner, N.; Chua, P.; Pierre, F.; O’Brien, S.E.; Bliesath, J.; Omori, M.; Huser, N.; Ho, C.; et al. CX-4945, an orally bioavailable selective inhibitor of protein kinase CK2, inhibits prosurvival and angiogenic signaling and exhibits antitumor efficacy. Cancer Res. 2010, 70, 10288–10298. [Google Scholar] [CrossRef] [PubMed]
- Pierre, F.; Chua, P.C.; O’Brien, S.E.; Siddiqui-Jain, A.; Bourbon, P.; Haddach, M.; Michaux, J.; Nagasawa, J.; Schwaebe, M.K.; Stefan, E.; et al. Pre-clinical characterization of CX-4945, a potent and selective small molecule inhibitor of CK2 for the treatment of cancer. Mol. Cell. Biochem. 2011, 356, 37–43. [Google Scholar] [CrossRef]
- Castello, J.; Ragnauth, A.; Friedman, E.; Rebholz, H. CK2—An Emerging Target for Neurological and Psychiatric Disorders. Pharmaceuticals 2017, 10, 7. [Google Scholar] [CrossRef]
- Iimoto, D.S.; Masliah, E.; DeTeresa, R.; Terry, R.D.; Saitoh, T. Aberrant casein kinase II in Alzheimer’s disease. Brain Res. 1990, 507, 273–280. [Google Scholar] [CrossRef]
- Masliah, E.; Iimoto, D.S.; Mallory, M.; Albright, T.; Hansen, L.; Saitoh, T. Casein kinase II alteration precedes tau accumulation in tangle formation. Am. J. Pathol. 1992, 140, 263–268. [Google Scholar]
- Marshall, C.A.; McBride, J.D.; Changolkar, L.; Riddle, D.M.; Trojanowski, J.Q.; Lee, V.M.Y. Inhibition of CK2 mitigates Alzheimer’s tau pathology by preventing NR2B synaptic mislocalization. Acta Neuropathol. Commun. 2022, 10, 30. [Google Scholar] [CrossRef] [PubMed]
- Baum, L.; Masliah, E.; Iimoto, D.S.; Hansen, L.A.; Halliday, W.C.; Saitoh, T. Casein kinase II is associated with neurofibrillary tangles but is not an intrinsic component of paired helical filaments. Brain Res. 1992, 573, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, K.S.; Kim, A.K.; Choi, M.; Choi, K.; Kang, M.; Chi, S.W.; Lee, M.S.; Lee, J.S.; Lee, S.Y.; et al. A chemical with proven clinical safety rescues Down-syndrome-related phenotypes in through DYRK1A inhibition. Dis. Model. Mech. 2016, 9, 839–848. [Google Scholar] [PubMed]
- Perera, Y.; Farina, H.G.; Gil, J.; Rodriguez, A.; Benavent, F.; Castellanos, L.; Gómez, R.E.; Acevedo, B.E.; Alonso, D.F.; Perea, S.E. Anticancer peptide CIGB-300 binds to nucleophosmin/B23, impairs its CK2-mediated phosphorylation, and leads to apoptosis through its nucleolar disassembly activity. Mol. Cancer Ther. 2009, 8, 1189–1196. [Google Scholar] [CrossRef]
- Perera, Y.; Ramos, Y.; Padrón, G.; Caballero, E.; Guirola, O.; Caligiuri, L.G.; Lorenzo, N.; Gottardo, F.; Farina, H.G.; Filhol, O.; et al. CIGB-300 anticancer peptide regulates the protein kinase CK2-dependent phosphoproteome. Mol. Cell Biochem. 2020, 470, 63–75. [Google Scholar] [CrossRef]
- Rosales, M.; Pérez, G.V.; Ramón, A.C.; Cruz, Y.; Rodríguez-Ulloa, A.; Besada, V.; Ramos, Y.; Vázquez-Blomquist, D.; Caballero, E.; Aguilar, D.; et al. Targeting of Protein Kinase CK2 in Acute Myeloid Leukemia Cells Using the Clinical-Grade Synthetic-Peptide CIGB-300. Biomedicines 2021, 9, 766. [Google Scholar] [CrossRef]
- Perera, Y.; Farina, H.G.; Hernández, I.; Mendoza, O.; Serrano, J.M.; Reyes, O.; Gómez, D.E.; Gómez, R.E.; Acevedo, B.E.; Alonso, D.F.; et al. Systemic administration of a peptide that impairs the protein kinase (CK2) phosphorylation reduces solid tumor growth in mice. Int. J. Cancer 2008, 122, 57–62. [Google Scholar] [CrossRef]
- Solares, A.M.; Santana, A.; Baladrón, I.; Valenzuela, C.; González, C.A.; Díaz, A.; Castillo, D.; Ramos, T.; Gómez, R.; Alonso, D.F.; et al. Safety and preliminary efficacy data of a novel Casein Kinase 2 (CK2) peptide inhibitor administered intralesionally at four dose levels in patients with cervical malignancies. BMC Cancer 2009, 9, 146. [Google Scholar] [CrossRef]
- Soriano-García, J.L.; López-Díaz, A.; Solares-Asteasuainzarra, M.; Baladrón-Castrillo, I.; Batista-Albuerne, N.; García-García, I.; González-Méndez, L.; Perera-Negrín, Y.; Valenzuela-Silva, C.M.; Pedro, A.P.; et al. Pharmacological and safety evaluation of CIGB-300, a casein kinase 2 inhibitor peptide, administered intralesionally to patients with cervical cancer stage IB2/II. J. Cancer Res. Ther. 2013, 1, 163–173. [Google Scholar] [CrossRef][Green Version]
- Sarduy, M.R.; García, I.; Coca, M.A.; Perera, A.; Torres, L.A.; Valenzuela, C.M.; Baladrón, I.; Solares, M.; Reyes, V.; Hernández, I.; et al. Optimizing CIGB-300 intralesional delivery in locally advanced cervical cancer. Br. J. Cancer 2015, 112, 1636–1643. [Google Scholar] [CrossRef]
- Cruz, L.R.; Baladrón, I.; Rittoles, A.; Díaz, P.A.; Valenzuela, C.; Santana, R.; Vázquez, M.M.; García, A.; Chacón, D.; Thompson, D.; et al. Treatment with an Anti-CK2 Synthetic Peptide Improves Clinical Response in COVID-19 Patients with Pneumonia. A Randomized and Controlled Clinical Trial. ACS Pharmacol. Transl. Sci. 2021, 4, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Sterling, J.; Baker, J.R.; McCluskey, A.; Munoz, L. Systematic literature review reveals suboptimal use of chemical probes in cell-based biomedical research. Nat. Commun. 2023, 14, 3228. [Google Scholar] [CrossRef] [PubMed]

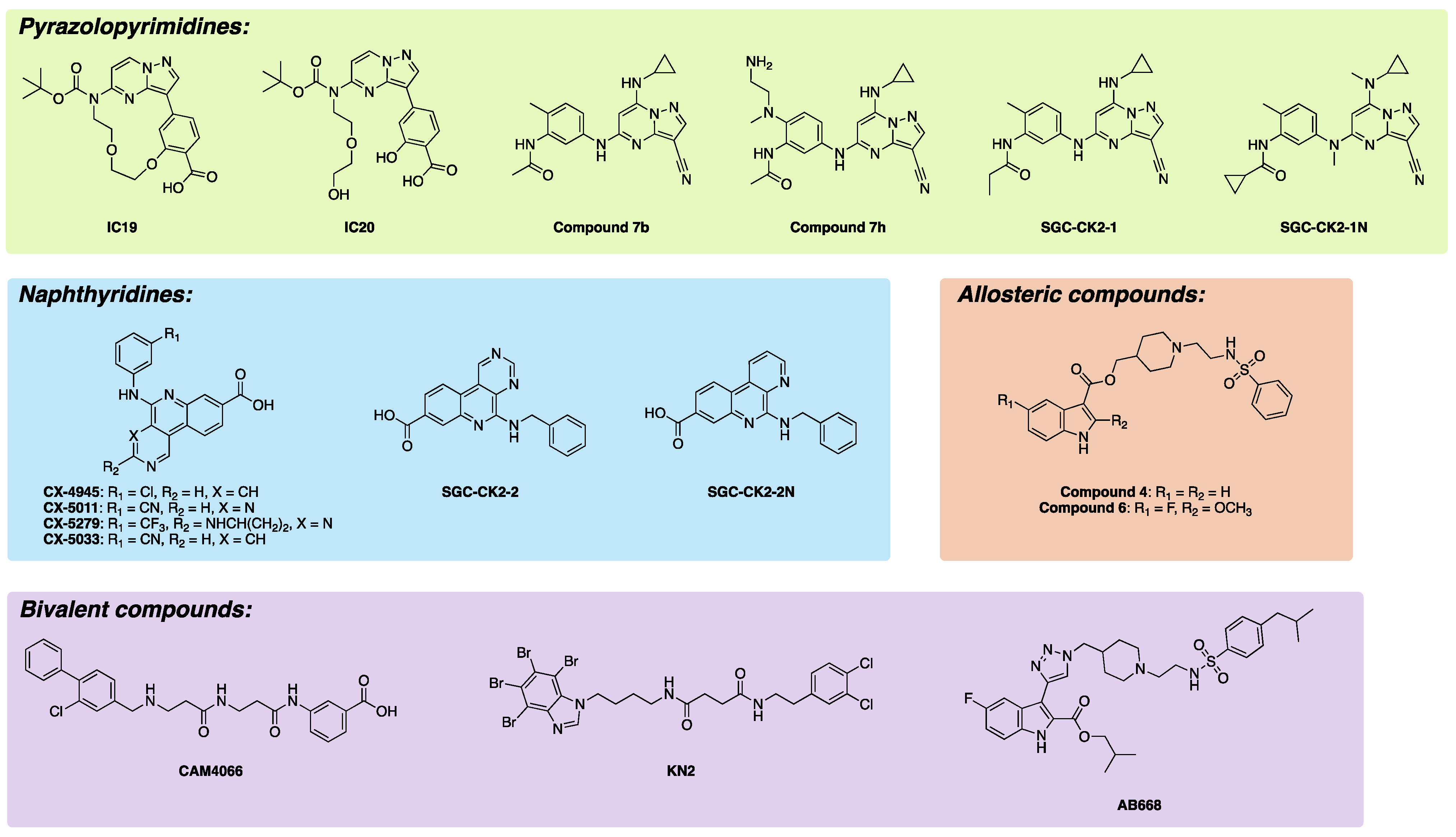
| CK2 Inhibitor | Binding Site | Core Scaffold | Biochemical CK2 Affinity | Cellular CK2 Affinity | Selectivity | Negative Control | PDB Entries | PMIDs/DOI |
|---|---|---|---|---|---|---|---|---|
| IC19 | Orthosteric | Pyrazolopyrimidine | CK2α ITC KD = 82 nM | CK2α NB 1 IC50 = 1.5 µM; CK2α’ NB IC50 = 7.4 µM | 469 kinase DiscoverX KINOMEscan S35 (1 µM) = 0.07 | - | - | 32883634 |
| IC20 | Orthosteric | Pyrazolopyrimidine | CK2α ITC KD = 12 nM | CK2α NB IC50 = 1.5 µM; CK2α’ NB IC50 = 7.6 µM | 469 kinase DiscoverX KINOMEscan S35 (1 µM) = 0.02 | - | 6YUM | 32883634 |
| Compound 7b | Orthosteric | Pyrazolopyrimidine | CK2α enzymatic IC50 < 3.0 nM | CK2α NB IC50 = 3.2 nM; CK2α’ NB IC50 = 1.8 nM | 402 kinase DiscoverX KINOMEscan S40 (0.1 µM) = 0.025; 469 kinase DiscoverX KINOMEscan S10 (1 µM) = 0.032 | - | - | 24900749, 33484635 |
| Compound 7h | Orthosteric | Pyrazolopyrimidine | CK2α enzymatic IC50 < 3.0 nM; CK2α SPR KD = 6.3 pM | CK2α NB IC50 = 5.3 nM; CK2α’ NB IC50 = 4.4 nM | 402 kinase DiscoverX KINOMEscan S50 (0.1 µM) = 0.03; 469 kinase DiscoverX KINOMEscan S10 (1 µM) = 0.042 | - | 5H8E | 26985319, 33484635 |
| SGC-CK2-1 | Orthosteric | Pyrazolopyrimidine | CK2α enzymatic IC50 = 4.2 nM; CK2α’ enzymatic IC50 = 2.3 nM | CK2α NB IC50 = 36 nM; CK2α’ NB IC50 = 16 nM | 468 kinase DiscoverX KINOMEscan S10 (1 µM) = 0.007 | SGC-CK2-1N | 6Z83 | 33484635 |
| CX-4945 | Orthosteric | Naphthyridine | CK2α enzymatic IC50 = 1 nM; CK2α’ enzymatic IC50 = 1 nM | CK2α NB IC50 = 240 nM; CK2α’ NB IC50 = 180 nM | 468 kinase DiscoverX KINOMEscan S10 (1 µM) = 0.069; 235 kinases at 0.5 µM; Gini coefficient = 0.667 | - | 3NGA, 3PE1 | 33484635, 21174434, 21870818, 37077385 |
| CX-5011 | Orthosteric | Naphthyridine | CK2α enzymatic IC50 = 2.3 nM | CK2α NB IC50 = 350 nM; CK2α’ NB IC50 = 66 nM | 235 kinases at 0.5 µM; Gini coefficient = 0.794 | - | 3PE2 | 21870818, 37077385 |
| CX-5033 | Orthosteric | Naphthyridine | CK2 holoenzyme enzymatic IC50 = 4.0 nM | CK2α NB IC50 = 4300 nM; CK2α’ NB IC50 = 1900 nM | 468 kinase DiscoverX KINOMEscan S10 (1 µM) = 0.079; 103 kinases at 0.5 µM; Gini coefficient = 0.570 | - | - | 21174434, 21870818, 37077385 |
| CX-5279 | Orthosteric | Naphthyridine | CK2α enzymatic IC50 = 0.91 nM | - | 102 kinases at 0.5 µM; Gini coefficient = 0.755 | - | 3R0T | 21870818 |
| SGC-CK2-2 | Orthosteric | Naphthyridine | CK2α enzymatic IC50 = 3.0 nM; CK2α’ enzymatic IC50 < 1.0 nM | CK2α NB IC50 = 920 nM; CK2α’ NB IC50 = 200 nM | 468 kinase DiscoverX KINOMEscan S10 (1 µM) = 0.007 | SGC-CK2-2N | 8BGC | 37077385 |
| Compound 4 | Allosteric | Indole-3-carboxylate | CK2β-dependent enzymatic IC50 = 45 µM; CK2α KD = 41 µM | - | - | - | 6FVG | 31685885 |
| Compound 6 | Allosteric | Indole-3-carboxylate | CK2β-dependent enzymatic IC50 = 22 µM; CK2α KD = 30 µM | - | - | - | 6FVF | 31685885 |
| CAM4066 | Bivalent | Biphenylmethylamine | CK2α ITC KD = 320 nM; CK2α enzymatic IC50 = 370 nM | - | 52 kinases at 2 µM; Gini coefficient = 0.82 | - | 5CU3, 5CU4, 5MO8 | 28451126 |
| KN2 | Bivalent | Benzoimidazole | CK2α2β2 holoenzyme enzymatic IC50 = 19 nM and Ki = 6.1 nM; CK2α’2β2 holoenzyme enzymatic IC50 = 16 nM and Ki = 4.0 nM | - | 83 kinases at 3 µM; Gini coefficient = 0.76 | - | 7AT5 | 34323071 |
| AB668 | Bivalent | Indole-2-carboxylate | CK2α binding assay KD = 86 nM; CK2β-dependent enzymatic Ki = 41 nM | - | 468 kinase DiscoverX KINOMEscan S10 (2 µM) = 0.007 | - | - | 10.1101/2022.12.16.520736 |
| SGC-CK2-1 | SGC-CK2-2 | |
|---|---|---|
| CK2α enzymatic IC50 | 4.2 nM | 3.0 nM |
| CK2α’ enzymatic IC50 | 2.3 nM | <1.0 nM |
| Selectivity: S10 (1 µM) | 0.007 | 0.007 |
| Selectivity window | 100-fold over DYRK2 | 200-fold over HIPK2 |
| CK2α NanoBRET IC50 | 36 nM | 920 nM |
| CK2α’ NanoBRET IC50 | 19 nM | 200 nM |
| Kinetic solubility | 3.7 µM | 211 µM |
| Proposed In Vivo Chemical Probe Criteria |
|---|
|
|
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ong, H.W.; Drewry, D.H.; Axtman, A.D. CK2 Chemical Probes: Past, Present, and Future. Kinases Phosphatases 2023, 1, 288-305. https://doi.org/10.3390/kinasesphosphatases1040017
Ong HW, Drewry DH, Axtman AD. CK2 Chemical Probes: Past, Present, and Future. Kinases and Phosphatases. 2023; 1(4):288-305. https://doi.org/10.3390/kinasesphosphatases1040017
Chicago/Turabian StyleOng, Han Wee, David H. Drewry, and Alison D. Axtman. 2023. "CK2 Chemical Probes: Past, Present, and Future" Kinases and Phosphatases 1, no. 4: 288-305. https://doi.org/10.3390/kinasesphosphatases1040017
APA StyleOng, H. W., Drewry, D. H., & Axtman, A. D. (2023). CK2 Chemical Probes: Past, Present, and Future. Kinases and Phosphatases, 1(4), 288-305. https://doi.org/10.3390/kinasesphosphatases1040017









