Interaction Networks Explain Holoenzyme Allostery in Protein Kinase A
Abstract
1. Introduction
2. cAMP-Mediated PKA Signaling
3. PKA and Human Disease
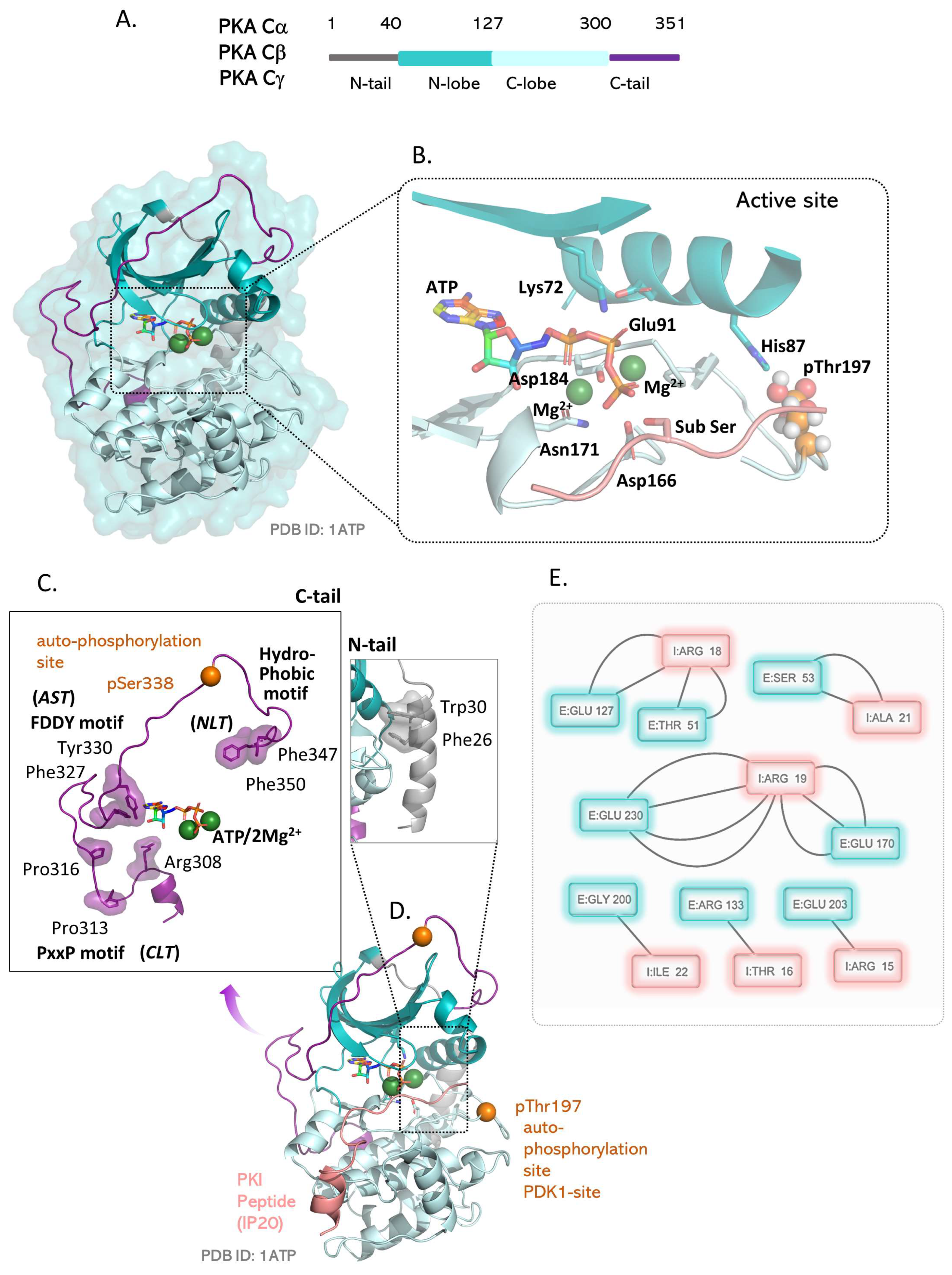
4. The PKA Catalytic Subunit
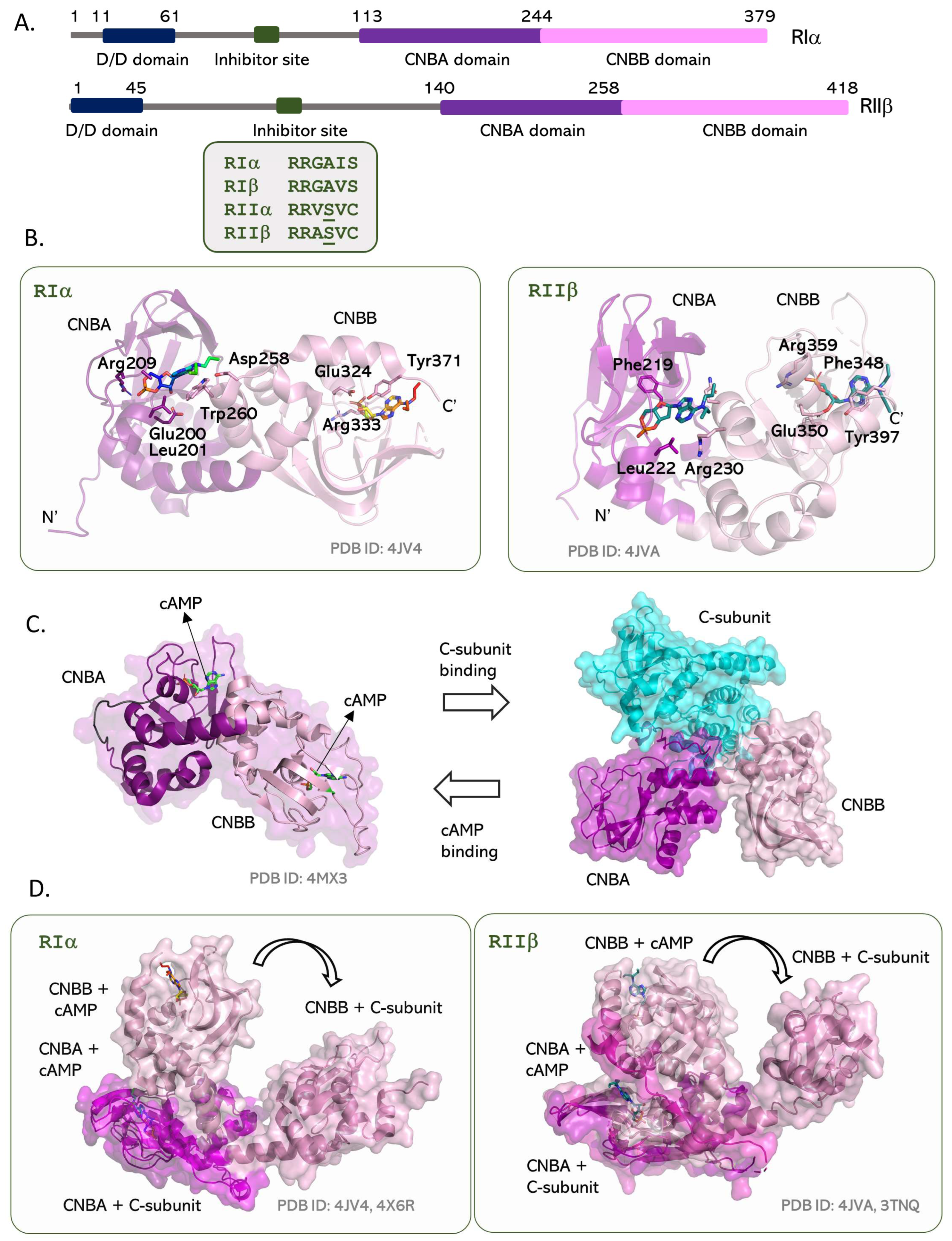
5. The PKA Regulatory Subunit
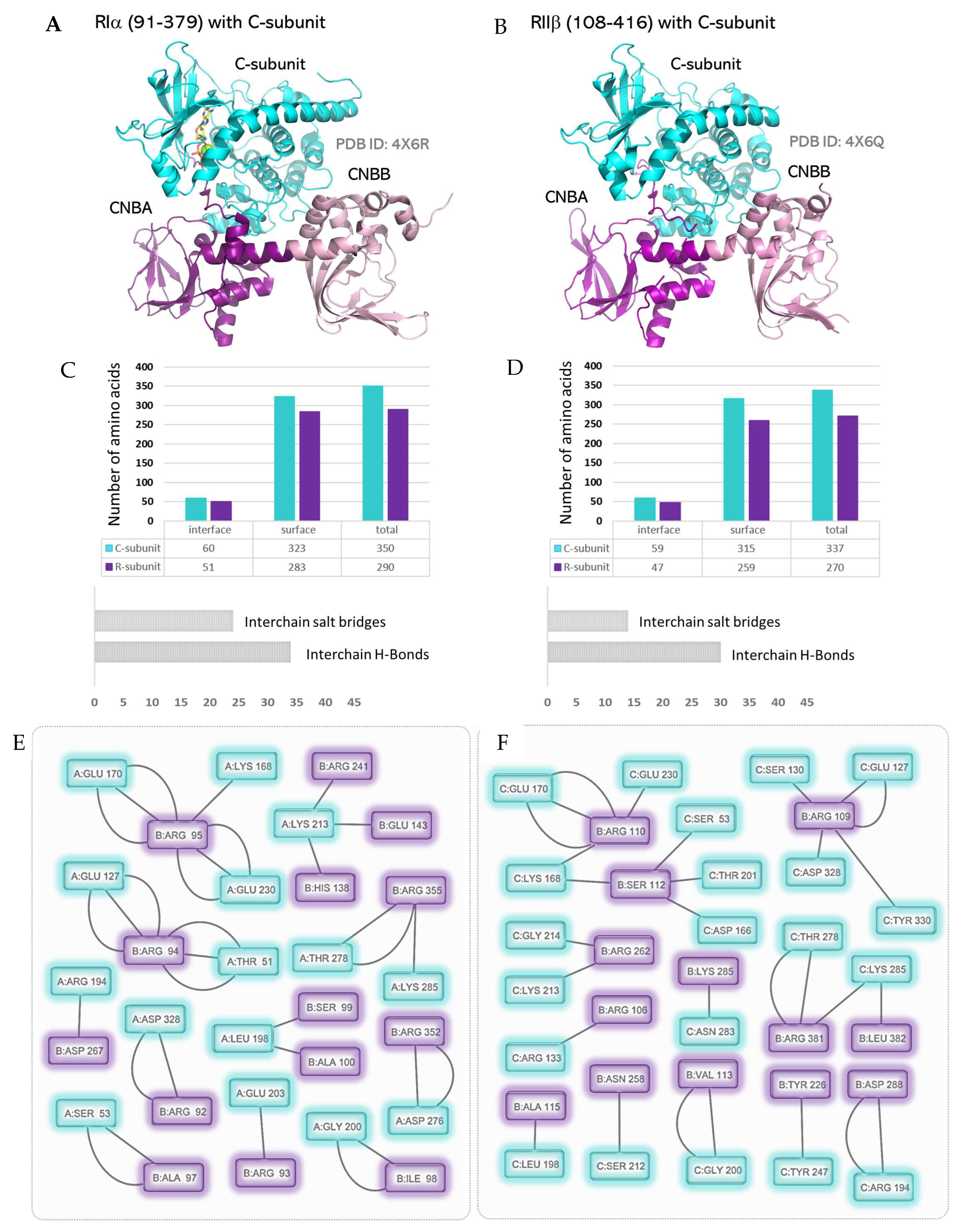
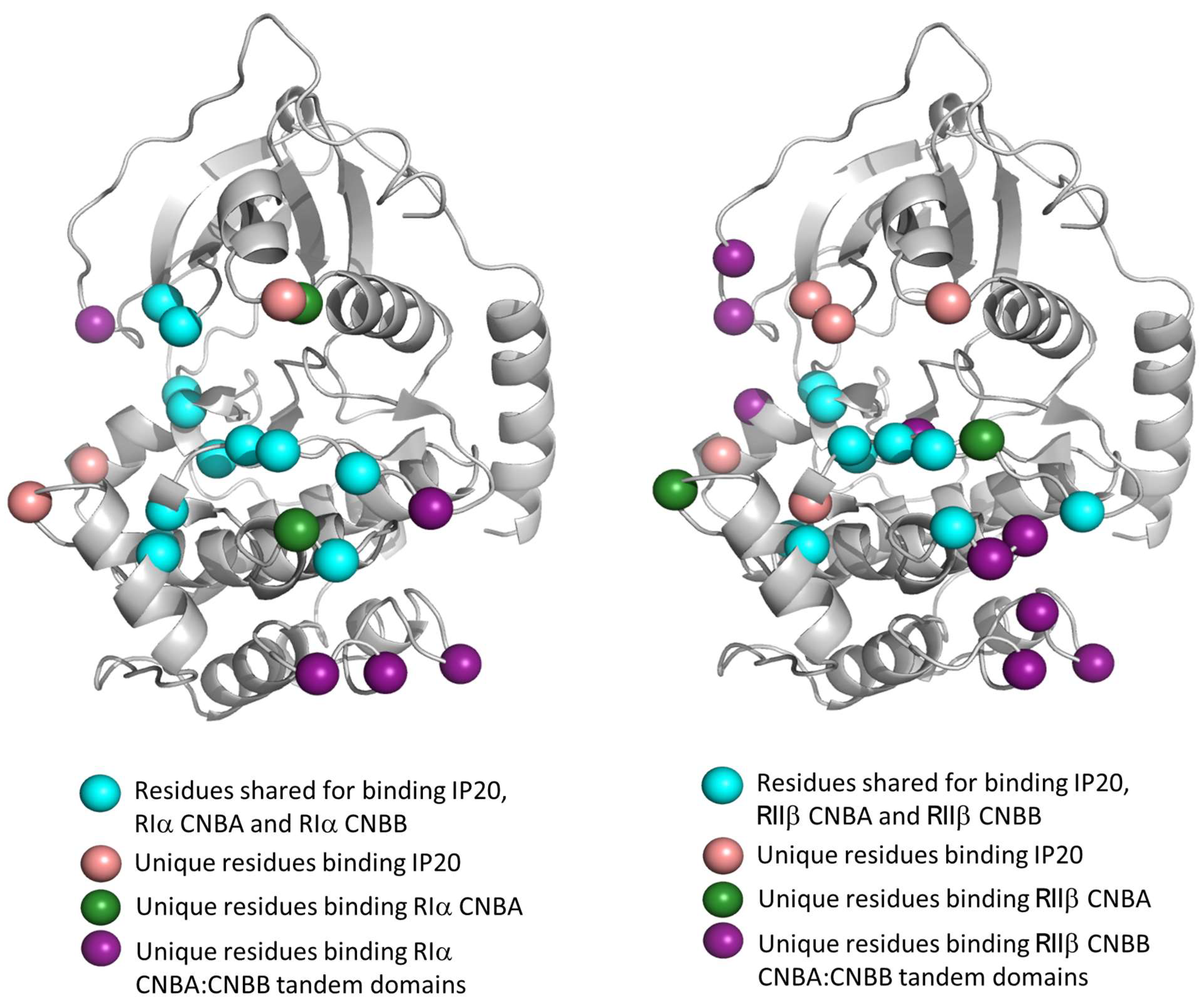
6. The PKA Holoenzymes and Contact Networks of R:C Complexes
7. Conclusions and Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fabbro, D.; Cowan-Jacob, S.W.; Moebitz, H. Ten things you should know about protein kinases: IUPHAR Review 14. Br. J. Pharmacol. 2015, 172, 2675–2700. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.; D’Souza, R.C.; Tyanova, S.; Schaab, C.; Wiśniewski, J.R.; Cox, J.; Mann, M. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 2014, 8, 1583–1594. [Google Scholar] [CrossRef] [PubMed]
- Smoly, I.; Shemesh, N.; Ziv-Ukelson, M.; Ben-Zvi, A.; Yeger-Lotem, E. An Asymmetrically Balanced Organization of Kinases versus Phosphatases across Eukaryotes Determines Their Distinct Impacts. PLoS Comput. Biol. 2017, 13, e1005221. [Google Scholar] [CrossRef] [PubMed]
- Nikolic, I.; Leiva, M.; Sabio, G. The role of stress kinases in metabolic disease. Nat. Rev. Endocrinol. 2020, 16, 697–716. [Google Scholar] [CrossRef]
- Lahiry, P.; Torkamani, A.; Schork, N.J.; Hegele, R.A. Kinase mutations in human disease: Interpreting genotype–phenotype relationships. Nat. Rev. Genet. 2010, 11, 60–74. [Google Scholar] [CrossRef]
- Kumar, A.; Cookson, M.R. Role of LRRK2 kinase dysfunction in Parkinson disease. Expert. Rev. Mol. Med. 2011, 13, e20. [Google Scholar] [CrossRef]
- Castelo-Soccio, L.; Kim, H.; Gadina, M.; Schwartzberg, P.L.; Laurence, A.; O’Shea, J.J. Protein kinases: Drug targets for immunological disorders. Nat. Rev. Immunol. 2023, 1–20. [Google Scholar] [CrossRef]
- Stenberg, K.A.; Riikonen, P.T.; Vihinen, M. KinMutBase, a database of human disease-causing protein kinase mutations. Nucleic Acids Res. 2000, 28, 369–371. [Google Scholar] [CrossRef][Green Version]
- Cohen, P.; Cross, D.; Jänne, P.A. Kinase drug discovery 20 years after imatinib: Progress and future directions. Nat. Rev. Drug Discov. 2021, 20, 551–569. [Google Scholar] [CrossRef]
- Ferguson, F.M.; Gray, N.S. Kinase inhibitors: The road ahead. Nat. Rev. Drug Discov. 2018, 17, 353–377. [Google Scholar] [CrossRef]
- Taylor, S.S.; Wu, J.; Bruystens, J.G.H.; Del Rio, J.C.; Lu, T.-W.; Kornev, A.P.; Ten Eyck, L.F. From structure to the dynamic regulation of a molecular switch: A journey over 3 decades. J. Biol. Chem. 2021, 296, 100746. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.S.; Zhang, P.; Steichen, J.M.; Keshwani, M.M.; Kornev, A.P. PKA: Lessons learned after twenty years. Biochim. Biophys. Acta 2013, 1834, 1271–1278. [Google Scholar] [CrossRef] [PubMed]
- Fischer, E.H.; Krebs, E.G. Conversion of phosphorylase b to phosphorylase a in muscle extracts. J. Biol. Chem. 1955, 216, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Krebs, E.G.; Kent, A.B.; Fischer, E.H. The muscle phosphorylase b kinase reaction. J. Biol. Chem. 1958, 231, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Rall, T.W.; Sutherland, E.W. Formation of a cyclic adenine ribonucleotide by tissue particles. J. Biol. Chem. 1958, 232, 1065–1076. [Google Scholar] [CrossRef]
- Sutherland, E.W.; Rall, T.W. Fractionation and characterization of a cyclic adenine ribonucleotide formed by tissue particles. J. Biol. Chem. 1958, 232, 1077–1091. [Google Scholar] [CrossRef]
- Cohen, P. The regulation of protein function by multisite phosphorylation—A 25 year update. Trends Biochem. Sci. 2000, 25, 596–601. [Google Scholar] [CrossRef]
- Walsh, D.A.; Perkins, J.P.; Krebs, E.G. An adenosine 3’,5’-monophosphate-dependant protein kinase from rabbit skeletal muscle. J. Biol. Chem. 1968, 243, 3763–3765. [Google Scholar] [CrossRef]
- Kuo, J.F.; Greengard, P. Cyclic nucleotide-dependent protein kinases, iv. widespread occurrence of adenosine 3′,5′-monophosphate-dependent protein kinase in various tissues and phyla of the animal kingdom. Proc. Natl. Acad. Sci. USA 1969, 64, 1349–1355. [Google Scholar] [CrossRef]
- Shoji, S.; Ericsson, L.H.; Walsh, K.A.; Fischer, E.H.; Titani, K. Amino acid sequence of the catalytic subunit of bovine type II adenosine cyclic 3’,5’-phosphate-dependent protein kinase. Biochemistry 1983, 22, 3702–3709. [Google Scholar] [CrossRef]
- Uhler, M.D.; Carmichael, D.F.; Lee, D.C.; Chrivia, J.C.; Krebs, E.G.; McKnight, G.S. Isolation of cDNA clones coding for the catalytic subunit of mouse cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA 1986, 83, 1300–1304. [Google Scholar] [CrossRef] [PubMed]
- Knighton, D.R.; Zheng, J.; Ten Eyck, L.F.; Ashford, V.A.; Xuong, N.-H.; Taylor, S.S.; Sowadski, J.M. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 1991, 253, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Dema, A.; Perets, E.; Schulz, M.S.; Deák, V.A.; Klussmann, E. Pharmacological targeting of AKAP-directed compartmentalized cAMP signalling. Cell. Signal. 2015, 27, 2474–2487. [Google Scholar] [CrossRef] [PubMed]
- Wild, A.R.; Dell’Acqua, M.L. Potential for therapeutic targeting of AKAP signaling complexes in nervous system disorders. Pharmacol. Ther. 2018, 185, 99–121. [Google Scholar] [CrossRef]
- Sapio, L.; Gallo, M.; Illiano, M.; Chiosi, E.; Naviglio, D.; Spina, A.; Naviglio, S. The natural cAMP elevating compound forskolin in cancer therapy: Is it time? J. Cell. Physiol. 2017, 232, 922–927. [Google Scholar] [CrossRef]
- Kleppe, R.; Krakstad, C.; Selheim, F.; Kopperud, R.; Ove Doskeland, S. The cAMP-dependent protein kinase pathway as therapeutic target-possibilities and pitfalls. Curr. Top. Med. Chem. 2011, 11, 1393–1405. [Google Scholar] [CrossRef]
- Oldham, W.M.; Hamm, H.E. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat. Rev. Mol. Cell Biol. 2008, 9, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Sassone-Corsi, P. The cyclic AMP pathway. Cold Spring Harb. Perspect. Biol. 2012, 4, a011148. [Google Scholar] [CrossRef]
- Grisan, F.; Iannucci, L.F.; Surdo, N.C.; Gerbino, A.; Zanin, S.; Di Benedetto, G.; Pozzan, T.; Lefkimmiatis, K. PKA compartmentalization links cAMP signaling and autophagy. Cell Death Differ. 2021, 28, 2436–2449. [Google Scholar] [CrossRef]
- Li, F.; Gangal, M.; Juliano, C.; Gorfain, E.; Taylor, S.S.; Johnson, D.A. Evidence for an internal entropy contribution to phosphoryl transfer: A study of domain closure, backbone flexibility, and the catalytic cycle of cAMP-dependent protein kinase. J. Mol. Biol. 2002, 315, 459–469. [Google Scholar] [CrossRef][Green Version]
- Zhao, L.; Liu, J.; He, C.; Yan, R.; Zhou, K.; Cui, Q.; Meng, X.; Li, X.; Zhang, Y.; Nie, Y. Protein kinase A determines platelet life span and survival by regulating apoptosis. J. Clin. Investig. 2017, 127, 4338–4351. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.; Brennesvik, E.O.; Lai, Y.-C.; Shepherd, P.R. GSK-3β regulation in skeletal muscles by adrenaline and insulin: Evidence that PKA and PKB regulate different pools of GSK-3. Cell. Signal. 2007, 19, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Tonucci, F.M.; Almada, E.; Borini-Etichetti, C.; Pariani, A.; Hidalgo, F.; Rico, M.J.; Girardini, J.; Favre, C.; Goldenring, J.R.; Menacho-Marquez, M. Identification of a CIP4 PKA phosphorylation site involved in the regulation of cancer cell invasiveness and metastasis. Cancer Lett. 2019, 461, 65–77. [Google Scholar] [CrossRef] [PubMed]
- London, E.; Bloyd, M.; Stratakis, C.A. PKA functions in metabolism and resistance to obesity: Lessons from mouse and human studies. J. Endocrinol. 2020, 246, R51–R64. [Google Scholar] [CrossRef] [PubMed]
- Moleschi, K.; Melacini, G. Signaling at crossroads: The dialogue between PDEs and PKA is spoken in multiple languages. Biophys. J. 2014, 107, 1259–1260. [Google Scholar] [CrossRef]
- Murthy, K.S.; Zhou, H.; Makhlouf, G.M. PKA-dependent activation of PDE3A and PDE4 and inhibition of adenylyl cyclase V/VI in smooth muscle. Am. J. Physiol.-Cell Physiol. 2002, 282, C508–C517. [Google Scholar] [CrossRef]
- Khannpnavar, B.; Mehta, V.; Qi, C.; Korkhov, V. Structure and function of adenylyl cyclases, key enzymes in cellular signaling. Curr. Opin. Struct. Biol. 2020, 63, 34–41. [Google Scholar] [CrossRef]
- Torres-Quesada, O.; Mayrhofer, J.E.; Stefan, E. The many faces of compartmentalized PKA signalosomes. Cell. Signal. 2017, 37, 1–11. [Google Scholar] [CrossRef]
- Zhu, Y.R.; Jiang, X.X.; Zheng, Y.; Xiong, J.; Wei, D.; Zhang, D.M. Cardiac function modulation depends on the A-kinase anchoring protein complex. J. Cell. Mol. Med. 2019, 23, 7170–7179. [Google Scholar] [CrossRef]
- Wong, W.; Scott, J.D. AKAP signalling complexes: Focal points in space and time. Nat. Rev. Mol. Cell Biol. 2004, 5, 959–970. [Google Scholar] [CrossRef]
- Amer, Y.O.; Hebert-Chatelain, E. Mitochondrial cAMP-PKA signaling: What do we really know? Biochim. et Biophys. Acta (BBA)-Bioenerg. 2018, 1859, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Westphal, R.S.; Tavalin, S.J.; Lin, J.W.; Alto, N.M.; Fraser, I.D.; Langeberg, L.K.; Sheng, M.; Scott, J.D. Regulation of NMDA receptors by an associated phosphatase-kinase signaling complex. Science 1999, 285, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Dell’Acqua, M.L.; Smith, K.E.; Gorski, J.A.; Horne, E.A.; Gibson, E.S.; Gomez, L.L. Regulation of neuronal PKA signaling through AKAP targeting dynamics. Eur. J. Cell Biol. 2006, 85, 627–633. [Google Scholar] [CrossRef] [PubMed]
- Pallien, T.; Klussmann, E. New aspects in cardiac L-type Ca2+ channel regulation. Biochem. Soc. Trans. 2020, 48, 39–49. [Google Scholar] [CrossRef]
- Scott, J. A-kinase-anchoring proteins and cytoskeletal signalling events. Biochem. Soc. Trans. 2003, 31, 87–89. [Google Scholar] [CrossRef] [PubMed]
- Rababa’h, A.; Singh, S.; Suryavanshi, S.V.; Altarabsheh, S.E.; Deo, S.V.; McConnell, B.K. Compartmentalization role of A-kinase anchoring proteins (AKAPs) in mediating protein kinase A (PKA) signaling and cardiomyocyte hypertrophy. Int. J. Mol. Sci. 2014, 16, 218–229. [Google Scholar] [CrossRef]
- Smith, F.D.; Reichow, S.L.; Esseltine, J.L.; Shi, D.; Langeberg, L.K.; Scott, J.D.; Gonen, T. Intrinsic disorder within an AKAP-protein kinase A complex guides local substrate phosphorylation. Elife 2013, 2, e01319. [Google Scholar] [CrossRef]
- Smith, F.D.; Esseltine, J.L.; Nygren, P.J.; Veesler, D.; Byrne, D.P.; Vonderach, M.; Strashnov, I.; Eyers, C.E.; Eyers, P.A.; Langeberg, L.K. Local protein kinase A action proceeds through intact holoenzymes. Science 2017, 356, 1288–1293. [Google Scholar] [CrossRef]
- Walker-Gray, R.; Stengel, F.; Gold, M.G. Mechanisms for restraining cAMP-dependent protein kinase revealed by subunit quantitation and cross-linking approaches. Proc. Natl. Acad. Sci. USA 2017, 114, 10414–10419. [Google Scholar] [CrossRef]
- Martin, B.R.; Deerinck, T.J.; Ellisman, M.H.; Taylor, S.S.; Tsien, R.Y. Isoform-specific PKA dynamics revealed by dye-triggered aggregation and DAKAP1α-mediated localization in living cells. Chem. Biol. 2007, 14, 1031–1042. [Google Scholar] [CrossRef]
- Mayr, B.; Montminy, M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2001, 2, 599–609. [Google Scholar] [CrossRef]
- De Boer, A.; Letzel, T.; Lingeman, H.; Irth, H. Systematic development of an enzymatic phosphorylation assay compatible with mass spectrometric detection. Anal. Bioanal. Chem. 2005, 381, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Dalton, G.D.; Smith, F.L.; Smith, P.A.; Dewey, W.L. Alterations in brain Protein Kinase A activity and reversal of morphine tolerance by two fragments of native Protein Kinase A inhibitor peptide (PKI). Neuropharmacology 2005, 48, 648–657. [Google Scholar] [CrossRef]
- Liu, C.; Ke, P.; Zhang, J.; Zhang, X.; Chen, X. Protein Kinase Inhibitor Peptide as a Tool to Specifically Inhibit Protein Kinase A. Front. Physiol. 2020, 11, 574030. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Yu, L.; Tu, Q.; Zhang, M.; He, H.; Chen, W.; Gao, J.; Yu, J.; Wu, Q.; Zhao, S. Cloning and mapping of human PKIB and PKIG, and comparison of tissue expression patterns of three members of the protein kinase inhibitor family, including PKIA. Biochem. J. 2000, 349, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, L.G.; Kornev, A.P.; McClendon, C.L.; Veglia, G.; Taylor, S.S. Mutation of a kinase allosteric node uncouples dynamics linked to phosphotransfer. Proc. Natl. Acad. Sci. USA 2017, 114, E931–E940. [Google Scholar] [CrossRef]
- Saldanha, S.A.; Kaler, G.; Cottam, H.B.; Abagyan, R.; Taylor, S.S. Assay principle for modulators of protein-protein interactions and its application to non-ATP-competitive ligands targeting protein kinase A. Anal. Chem. 2006, 78, 8265–8272. [Google Scholar] [CrossRef][Green Version]
- Haushalter, K.J.; Casteel, D.E.; Raffeiner, A.; Stefan, E.; Patel, H.H.; Taylor, S.S. Phosphorylation of protein kinase A (PKA) regulatory subunit RIalpha by protein kinase G (PKG) primes PKA for catalytic activity in cells. J. Biol. Chem. 2018, 293, 4411–4421. [Google Scholar] [CrossRef]
- Bruystens, J.G.H.; Wu, J.; Fortezzo, A.; Kornev, A.P.; Blumenthal, D.K.; Taylor, S.S. PKA RIα Homodimer Structure Reveals an Intermolecular Interface with Implications for Cooperative cAMP Binding and Carney Complex Disease. Structure 2014, 22, 59–69. [Google Scholar] [CrossRef]
- Walker, C.; Wang, Y.; Olivieri, C.; Karamafrooz, A.; Casby, J.; Bathon, K.; Calebiro, D.; Gao, J.; Bernlohr, D.A.; Taylor, S.S.; et al. Cushing’s syndrome driver mutation disrupts protein kinase A allosteric network, altering both regulation and substrate specificity. Sci. Adv. 2019, 5, eaaw9298. [Google Scholar] [CrossRef]
- Beuschlein, F.; Fassnacht, M.; Assié, G.; Calebiro, D.; Stratakis, C.A.; Osswald, A.; Ronchi, C.L.; Wieland, T.; Sbiera, S.; Faucz, F.R. Constitutive activation of PKA catalytic subunit in adrenal Cushing’s syndrome. N. Engl. J. Med. 2014, 370, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Cheung, J.; Ginter, C.; Cassidy, M.; Franklin, M.C.; Rudolph, M.J.; Robine, N.; Darnell, R.B.; Hendrickson, W.A. Structural insights into mis-regulation of protein kinase A in human tumors. Proc. Natl. Acad. Sci. USA 2015, 112, 1374–1379. [Google Scholar] [CrossRef] [PubMed]
- Bertherat, J.; Horvath, A.; Groussin, L.; Grabar, S.; Boikos, S.; Cazabat, L.; Libe, R.; René-Corail, F.; Stergiopoulos, S.; Bourdeau, I. Mutations in regulatory subunit type 1A of cyclic adenosine 5′-monophosphate-dependent protein kinase (PRKAR1A): Phenotype analysis in 353 patients and 80 different genotypes. J. Clin. Endocrinol. Metab. 2009, 94, 2085–2091. [Google Scholar] [CrossRef]
- Bossis, I.; Stratakis, C.A. Minireview: PRKAR1A: Normal and abnormal functions. Endocrinology 2004, 145, 5452–5458. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, L.S.; Sandrini, F.; Monbo, J.; Lin, J.P.; Carney, J.A.; Stratakis, C.A. Genetic heterogeneity and spectrum of mutations of the PRKAR1A gene in patients with the carney complex. Hum. Mol. Genet. 2000, 9, 3037–3046. [Google Scholar] [CrossRef] [PubMed]
- Stratakis, C.A.; Kirschner, L.S.; Carney, J.A. Clinical and molecular features of the Carney complex: Diagnostic criteria and recommendations for patient evaluation. J. Clin. Endocrinol. Metab. 2001, 86, 4041–4046. [Google Scholar] [CrossRef]
- Laxminarayana, D.; Kammer, G.M. mRNA mutations of type I protein kinase A regulatory subunit α in T lymphocytes of a subject with systemic lupus erythematosus. Int. Immunol. 2000, 12, 1521–1529. [Google Scholar] [CrossRef]
- Pérez-Lorenzo, M.J.; Galindo, M.; García-González, A.J.; Criado, G. Increased expression of A-kinase anchoring proteins in T cells from systemic lupus erythematosus patients. J. Transl. Med. 2010, 8, P49. [Google Scholar] [CrossRef]
- Enns, L.C.; Ladiges, W. Protein kinase A signaling as an anti-aging target. Ageing Res. Rev. 2010, 9, 269–272. [Google Scholar] [CrossRef]
- Enns, L.C.; Morton, J.F.; Mangalindan, R.S.; McKnight, G.S.; Schwartz, M.W.; Kaeberlein, M.R.; Kennedy, B.K.; Rabinovitch, P.S.; Ladiges, W.C. Attenuation of age-related metabolic dysfunction in mice with a targeted disruption of the Cβ subunit of protein kinase A. J. Gerontol. Ser. A Biomed. Sci. Med. Sci. 2009, 64, 1221–1231. [Google Scholar] [CrossRef]
- Gertler, A.; Solomon, G. Leptin-activity blockers: Development and potential use in experimental biology and medicine. Can. J. Physiol. Pharmacol. 2013, 91, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Spina, A.; Di Maiolo, F.; Esposito, A.; D’Auria, R.; Di Gesto, D.; Chiosi, E.; Sorvillo, L.; Naviglio, S. Integrating leptin and cAMP signalling pathways in triple-negative breast cancer cells. Front. Biosci. 2013, 18, 133–144. [Google Scholar] [CrossRef]
- Naviglio, S.; Di Gesto, D.; Romano, M.; Sorrentino, A.; Illiano, F.; Sorvillo, L.; Abbruzzese, A.; Marra, M.; Caraglia, M.; Chiosi, E.; et al. Leptin enhances growth inhibition by cAMP elevating agents through apoptosis of MDA-MB-231 breast cancer cells. Cancer Biol. Ther. 2009, 8, 1183–1190. [Google Scholar] [CrossRef] [PubMed]
- Naviglio, S.; Di Gesto, D.; Illiano, F.; Chiosi, E.; Giordano, A.; Illiano, G.; Spina, A. Leptin potentiates antiproliferative action of cAMP elevation via protein kinase A down-regulation in breast cancer cells. J. Cell Physiol. 2010, 225, 801–809. [Google Scholar] [CrossRef]
- Graham, R.P.; Jin, L.; Knutson, D.L.; Kloft-Nelson, S.M.; Greipp, P.T.; Waldburger, N.; Roessler, S.; Longerich, T.; Roberts, L.R.; Oliveira, A.M. DNAJB1-PRKACA is specific for fibrolamellar carcinoma. Mod. Pathol. 2015, 28, 822–829. [Google Scholar] [CrossRef]
- Turnham, R.E.; Smith, F.D.; Kenerson, H.L.; Omar, M.H.; Golkowski, M.; Garcia, I.; Bauer, R.; Lau, H.-T.; Sullivan, K.M.; Langeberg, L.K.; et al. An acquired scaffolding function of the DNAJ-PKAc fusion contributes to oncogenic signaling in fibrolamellar carcinoma. eLife 2019, 8, e44187. [Google Scholar] [CrossRef]
- Wu, K.-J.; Mattioli, M.; Morse, H.C.; Dalla-Favera, R. c-MYC activates protein kinase A (PKA) by direct transcriptional activation of the PKA catalytic subunit beta (PKA-Cβ) gene. Oncogene 2002, 21, 7872–7882. [Google Scholar] [CrossRef]
- Howe, A.K. Regulation of actin-based cell migration by cAMP/PKA. Biochim. Biophys. Acta 2004, 1692, 159–174. [Google Scholar] [CrossRef]
- Shaikh, D.; Zhou, Q.; Chen, T.; Ibe, J.C.; Raj, J.U.; Zhou, G. cAMP-dependent protein kinase is essential for hypoxia-mediated epithelial-mesenchymal transition, migration, and invasion in lung cancer cells. Cell Signal. 2012, 24, 2396–2406. [Google Scholar] [CrossRef]
- Jiang, P.; Enomoto, A.; Takahashi, M. Cell biology of the movement of breast cancer cells: Intracellular signalling and the actin cytoskeleton. Cancer Lett. 2009, 284, 122–130. [Google Scholar] [CrossRef]
- McKenzie, A.J.; Campbell, S.L.; Howe, A.K. Protein kinase A activity and anchoring are required for ovarian cancer cell migration and invasion. PLoS ONE 2011, 6, e26552. [Google Scholar] [CrossRef] [PubMed]
- Cho-Chung, Y.S. Protein kinase A-directed antisense blockade of cancer growth: Single gene-based therapeutic approach. Appl. Antisense Oligonucleotide Technol. 1998, 263–281. [Google Scholar]
- Stratakis, C.A.; Cho-Chung, Y.S. Protein kinase A and human disease. Trends Endocrinol. Metab. 2002, 13, 50–52. [Google Scholar] [CrossRef] [PubMed]
- Cho-Chung, Y.S.; Nesterova, M.V. Tumor reversion: Protein kinase A isozyme switching. Ann. N. Y. Acad. Sci. 2005, 1058, 76–86. [Google Scholar] [CrossRef]
- Tortora, G.; Ciardiello, F. Protein kinase A as target for novel integrated strategies of cancer therapy. Ann. N. Y. Acad. Sci. 2002, 968, 139–147. [Google Scholar] [CrossRef]
- Beebe, S.J.; Øyen, O.; Sandberg, M.; Frøysa, A.; Hansson, V.; Jahnsen, T. Molecular cloning of a tissue-specific protein kinase (Cγ) from human testis—Representing a third isoform for the catalytic subunit of cAMP-dependent protein kinase. Mol. Endocrinol. 1990, 4, 465–475. [Google Scholar] [CrossRef]
- Zimmermann, B.; Chiorini, J.A.; Ma, Y.; Kotin, R.M.; Herberg, F.W. PrKX is a novel catalytic subunit of the cAMP-dependent protein kinase regulated by the regulatory subunit type I. J. Biol. Chem. 1999, 274, 5370–5378. [Google Scholar] [CrossRef]
- Skalhegg, B.S.; Tasken, K. Specificity in the cAMP/PKA signaling pathway. Differential expression, regulation, and subcellular localization of subunits of PKA. Front. Biosci.-Landmark 2000, 5, D678–D693. [Google Scholar] [CrossRef]
- Schiebel, K.; Winkelmann, M.; Mertz, A.; Xu, X.; Page, D.C.; Weil, D.; Petit, C.; Rappold, G.A. Abnormal XY interchange between a novel isolated protein kinase gene, PRKY, and its homologue, PRKX, accounts for one third of all (Y+)XX males and (Y-)XY females. Hum. Mol. Genet. 1997, 6, 1985–1989. [Google Scholar] [CrossRef]
- Søberg, K.; Jahnsen, T.; Rognes, T.; Skålhegg, B.S.; Laerdahl, J.K. Evolutionary paths of the cAMP-dependent protein kinase (PKA) catalytic subunits. PLoS ONE 2013, 8, e60935. [Google Scholar] [CrossRef]
- Agustin, J.T.S.; Wilkerson, C.G.; Witman, G.B. The unique catalytic subunit of sperm cAMP-dependent protein kinase is the product of an alternative Cα mRNA expressed specifically in spermatogenic cells. Mol. Biol. Cell 2000, 11, 3031–3044. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Guthrie, C.R.; Skålhegg, B.S.; McKnight, G.S. Two novel brain-specific splice variants of the murine Cβ gene of cAMP-dependent protein kinase. J. Biol. Chem. 1997, 272, 29560–29565. [Google Scholar] [CrossRef]
- Showers, M.O.; Maurer, R.A. Cloning of cDNA for the catalytic subunit of cAMP-dependent protein kinase. Methods Enzymol. 1988, 159, 311–318. [Google Scholar] [CrossRef] [PubMed]
- San Agustin, J.T.; Leszyk, J.D.; Nuwaysir, L.M.; Witman, G.B. The catalytic subunit of the cAMP-dependent protein kinase of ovine sperm flagella has a unique amino-terminal sequence. J. Biol. Chem. 1998, 273, 24874–24883. [Google Scholar] [CrossRef]
- San Agustin, J.T.; Witman, G.B. Differential expression of the C(s) and Calpha1 isoforms of the catalytic subunit of cyclic 3’,5’-adenosine monophosphate-dependent protein kinase testicular cells. Biol. Reprod. 2001, 65, 151–164. [Google Scholar] [CrossRef]
- Reinton, N.; Orstavik, S.; Haugen, T.B.; Jahnsen, T.; Taskén, K.; Skålhegg, B.S. A novel isoform of human cyclic 3’,5’-adenosine monophosphate-dependent protein kinase, c alpha-s, localizes to sperm midpiece. Biol. Reprod. 2000, 63, 607–611. [Google Scholar] [CrossRef]
- Nolan, M.A.; Babcock, D.F.; Wennemuth, G.; Brown, W.; Burton, K.A.; McKnight, G.S. Sperm-specific protein kinase A catalytic subunit Calpha2 orchestrates cAMP signaling for male fertility. Proc. Natl. Acad. Sci. USA 2004, 101, 13483–13488. [Google Scholar] [CrossRef]
- Desseyn, J.L.; Burton, K.A.; McKnight, G.S. Expression of a nonmyristylated variant of the catalytic subunit of protein kinase A during male germ-cell development. Proc. Natl. Acad. Sci. USA 2000, 97, 6433–6438. [Google Scholar] [CrossRef]
- Skålhegg, B.S.; Huang, Y.; Su, T.; Idzerda, R.L.; McKnight, G.S.; Burton, K.A. Mutation of the Calpha subunit of PKA leads to growth retardation and sperm dysfunction. Mol. Endocrinol. 2002, 16, 630–639. [Google Scholar] [CrossRef]
- Uhler, M.D.; Chrivia, J.C.; McKnight, G.S. Evidence for a second isoform of the catalytic subunit of cAMP-dependent protein kinase. J. Biol. Chem. 1986, 261, 15360–15363. [Google Scholar] [CrossRef]
- Wiemann, S.; Kinzel, V.; Pyerin, W. Isoform C beta 2, an unusual form of the bovine catalytic subunit of cAMP-dependent protein kinase. J. Biol. Chem. 1991, 266, 5140–5146. [Google Scholar] [CrossRef] [PubMed]
- Ørstavik, S.; Reinton, N.; Frengen, E.; Langeland, B.T.; Jahnsen, T.; Skålhegg, B.S. Identification of novel splice variants of the human catalytic subunit Cbeta of cAMP-dependent protein kinase. Eur. J. Biochem. 2001, 268, 5066–5073. [Google Scholar] [CrossRef] [PubMed]
- Kvissel, A.K.; Ørstavik, S.; Øistad, P.; Rootwelt, T.; Jahnsen, T.; Skålhegg, B.S. Induction of Cbeta splice variants and formation of novel forms of protein kinase A type II holoenzymes during retinoic acid-induced differentiation of human NT2 cells. Cell. Signal. 2004, 16, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Larsen, A.C.; Kvissel, A.K.; Hafte, T.T.; Avellan, C.I.; Eikvar, S.; Rootwelt, T.; Ørstavik, S.; Skålhegg, B.S. Inactive forms of the catalytic subunit of protein kinase A are expressed in the brain of higher primates. FEBS J. 2008, 275, 250–262. [Google Scholar] [CrossRef] [PubMed]
- Beebe, S.J.; Salomonsky, P.; Jahnsen, T.; Li, Y. The C gamma subunit is a unique isozyme of the cAMP-dependent protein kinase. J. Biol. Chem. 1992, 267, 25505–25512. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Morris, G.Z.; Beebe, S.J. Characterization of the cAMP-dependent protein kinase catalytic subunit Cgamma expressed and purified from sf9 cells. Protein Expr. Purif. 2004, 35, 156–169. [Google Scholar] [CrossRef]
- Carr, S.A.; Biemann, K.; Shoji, S.; Parmelee, D.C.; Titani, K. n-Tetradecanoyl is the NH2-terminal blocking group of the catalytic subunit of cyclic AMP-dependent protein kinase from bovine cardiac muscle. Proc. Natl. Acad. Sci. USA 1982, 79, 6128–6131. [Google Scholar] [CrossRef]
- Adams, J.A.; McGlone, M.L.; Gibson, R.; Taylor, S.S. Phosphorylation modulates catalytic function and regulation in the cAMP-dependent protein kinase. Biochemistry 1995, 34, 2447–2454. [Google Scholar] [CrossRef]
- Taylor, S.S.; Radzio-Andzelm, E. Chapter 179—cAMP-Dependent Protein Kinase. In Handbook of Cell Signaling, 2nd ed.; Bradshaw, R.A., Dennis, E.A., Eds.; Academic Press: San Diego, CA, USA, 2010; pp. 1461–1469. [Google Scholar]
- Keshwani, M.M.; Klammt, C.; von Daake, S.; Ma, Y.; Kornev, A.P.; Choe, S.; Insel, P.A.; Taylor, S.S. Cotranslational cis-phosphorylation of the COOH-terminal tail is a key priming step in the maturation of cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA 2012, 109, E1221–E1229. [Google Scholar] [CrossRef]
- Moore, M.J.; Kanter, J.R.; Jones, K.C.; Taylor, S.S. Phosphorylation of the Catalytic Subunit of Protein Kinase A: AUTOPHOSPHORYLATION VERSUS PHOSPHORYLATION BY PHOSPHOINOSITIDE-DEPENDENT KINASE-1. J. Biol. Chem. 2002, 277, 47878–47884. [Google Scholar] [CrossRef]
- Carrera, A.C.; Alexandrov, K.; Roberts, T.M. The conserved lysine of the catalytic domain of protein kinases is actively involved in the phosphotransfer reaction and not required for anchoring ATP. Proc. Natl. Acad. Sci. USA 1993, 90, 442–446. [Google Scholar] [CrossRef] [PubMed]
- Meharena, H.S.; Fan, X.; Ahuja, L.G.; Keshwani, M.M.; McClendon, C.L.; Chen, A.M.; Adams, J.A.; Taylor, S.S. Decoding the Interactions Regulating the Active State Mechanics of Eukaryotic Protein Kinases. PLoS Biol. 2016, 14, e2000127. [Google Scholar] [CrossRef] [PubMed]
- Humphries, K.M.; Deal, M.S.; Taylor, S.S. Enhanced dephosphorylation of cAMP-dependent protein kinase by oxidation and thiol modification. J. Biol. Chem. 2005, 280, 2750–2758. [Google Scholar] [CrossRef] [PubMed]
- Cox, S.; Taylor, S.S. Kinetic analysis of cAMP-dependent protein kinase: Mutations at histidine 87 affect peptide binding and pH dependence. Biochemistry 1995, 34, 16203–16209. [Google Scholar] [CrossRef] [PubMed]
- Khavrutskii, I.V.; Grant, B.; Taylor, S.S.; McCammon, J.A. A transition path ensemble study reveals a linchpin role for Mg(2+) during rate-limiting ADP release from protein kinase A. Biochemistry 2009, 48, 11532–11545. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cheng, Y.; Zhang, Y.; McCammon, J.A. How Does the cAMP-Dependent Protein Kinase Catalyze the Phosphorylation Reaction: An ab Initio QM/MM Study. J. Am. Chem. Soc. 2005, 127, 1553–1562. [Google Scholar] [CrossRef]
- Adams, J.A. Kinetic and catalytic mechanisms of protein kinases. Chem. Rev. 2001, 101, 2271–2290. [Google Scholar] [CrossRef]
- Taylor, S.S.; Kim, C.; Cheng, C.Y.; Brown, S.H.; Wu, J.; Kannan, N. Signaling through cAMP and cAMP-dependent protein kinase: Diverse strategies for drug design. Biochim. et Biophys. Acta (BBA)-Proteins Proteom. 2008, 1784, 16–26. [Google Scholar] [CrossRef]
- Taylor, S.S.; Søberg, K.; Kobori, E.; Wu, J.; Pautz, S.; Herberg, F.W.; Skålhegg, B.S. The Tails of Protein Kinase A. Mol. Pharmacol. 2022, 101, 219–225. [Google Scholar] [CrossRef]
- Sastri, M.; Barraclough, D.M.; Carmichael, P.T.; Taylor, S.S. A-kinase-interacting protein localizes protein kinase A in the nucleus. Proc. Natl. Acad. Sci. USA 2005, 102, 349–354. [Google Scholar] [CrossRef]
- Kannan, N.; Haste, N.; Taylor, S.S.; Neuwald, A.F. The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proc. Natl. Acad. Sci. USA 2007, 104, 1272–1277. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.S.; Kornev, A.P.; Hu, J.; Ahuja, L.G.; Taylor, S.S. Kinases and pseudokinases: Lessons from RAF. Mol. Cell. Biol. 2014, 34, 1538–1546. [Google Scholar] [CrossRef] [PubMed]
- Kornev, A.P.; Haste, N.M.; Taylor, S.S.; Ten Eyck, L.F. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc. Natl. Acad. Sci. USA 2006, 103, 17783–17788. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, L.G.; Aoto, P.C.; Kornev, A.P.; Veglia, G.; Taylor, S.S. Dynamic allostery-based molecular workings of kinase:peptide complexes. Proc. Natl. Acad. Sci. USA 2019, 116, 15052–15061. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Manus, V.S.; Kim, J.; Li, G.; Ahuja, L.G.; Aoto, P.; Taylor, S.S.; Veglia, G. Globally correlated conformational entropy underlies positive and negative cooperativity in a kinase’s enzymatic cycle. Nat. Commun. 2019, 10, 799. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.A.; Taylor, S.S. Energetic limits of phosphotransfer in the catalytic subunit of cAMP-dependent protein kinase as measured by viscosity experiments. Biochemistry 1992, 31, 8516–8522. [Google Scholar] [CrossRef]
- Lew, J.; Taylor, S.S.; Adams, J.A. Identification of a partially rate-determining step in the catalytic mechanism of cAMP-dependent protein kinase: A transient kinetic study using stopped-flow fluorescence spectroscopy. Biochemistry 1997, 36, 6717–6724. [Google Scholar] [CrossRef]
- Adams, J.A.; Taylor, S.S. Phosphorylation of peptide substrates for the catalytic subunit of cAMP-dependent protein kinase. J. Biol. Chem. 1993, 268, 7747–7752. [Google Scholar] [CrossRef]
- Masterson, L.R.; Cembran, A.; Shi, L.; Veglia, G. Allostery and binding cooperativity of the catalytic subunit of protein kinase A by NMR spectroscopy and molecular dynamics simulations. Adv. Protein Chem. Struct. Biol. 2012, 87, 363–389. [Google Scholar] [CrossRef]
- Lodish, M.; Stratakis, C.A. A genetic and molecular update on adrenocortical causes of Cushing syndrome. Nat. Rev. Endocrinol. 2016, 12, 255–262. [Google Scholar] [CrossRef]
- Cao, Y.; He, M.; Gao, Z.; Peng, Y.; Li, Y.; Li, L.; Zhou, W.; Li, X.; Zhong, X.; Lei, Y. Activating hotspot L205R mutation in PRKACA and adrenal Cushing’s syndrome. Science 2014, 344, 913–917. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Maekawa, S.; Ishii, R.; Sanada, M.; Morikawa, T.; Shiraishi, Y.; Yoshida, K.; Nagata, Y.; Sato-Otsubo, A.; Yoshizato, T. Recurrent somatic mutations underlie corticotropin-independent Cushing’s syndrome. Science 2014, 344, 917–920. [Google Scholar] [CrossRef] [PubMed]
- Goh, G.; Scholl, U.I.; Healy, J.M.; Choi, M.; Prasad, M.L.; Nelson-Williams, C.; Kunstman, J.W.; Korah, R.; Suttorp, A.-C.; Dietrich, D. Recurrent activating mutation in PRKACA in cortisol-producing adrenal tumors. Nat. Genet. 2014, 46, 613–617. [Google Scholar] [CrossRef] [PubMed]
- Honeyman, J.N.; Simon, E.P.; Robine, N.; Chiaroni-Clarke, R.; Darcy, D.G.; Lim, I.I.P.; Gleason, C.E.; Murphy, J.M.; Rosenberg, B.R.; Teegan, L. Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science 2014, 343, 1010–1014. [Google Scholar] [CrossRef]
- Xu, L.; Hazard, F.K.; Zmoos, A.-F.; Jahchan, N.; Chaib, H.; Garfin, P.M.; Rangaswami, A.; Snyder, M.P.; Sage, J. Genomic analysis of fibrolamellar hepatocellular carcinoma. Hum. Mol. Genet. 2015, 24, 50–63. [Google Scholar] [CrossRef]
- Tasken, K.; Skålhegg, B.; Solberg, R.; Andersson, K.; Taylor, S.; Lea, T.; Blomhoff, H.; Jahnsen, T.; Hansson, V. Novel isozymes of cAMP-dependent protein kinase exist in human cells due to formation of RI alpha-RI beta heterodimeric complexes. J. Biol. Chem. 1993, 268, 21276–21283. [Google Scholar] [CrossRef]
- Amieux, P.S.; McKnight, G.S. The essential role of RIα in the maintenance of regulated PKA activity. Ann. N. Y. Acad. Sci. 2002, 968, 75–95. [Google Scholar] [CrossRef]
- Horvath, A.; Bertherat, J.; Groussin, L.; Guillaud-Bataille, M.; Tsang, K.; Cazabat, L.; Libe, R.; Remmers, E.; René-Corail, F.; Faucz, F.R. Mutations and polymorphisms in the gene encoding regulatory subunit type 1-alpha of protein kinase A (PRKAR1A): An update. Hum. Mutat. 2010, 31, 369–379. [Google Scholar] [CrossRef]
- Bruystens, J.G.; Wu, J.; Fortezzo, A.; Del Rio, J.; Nielsen, C.; Blumenthal, D.K.; Rock, R.; Stefan, E.; Taylor, S.S. Structure of a PKA RIalpha Recurrent Acrodysostosis Mutant Explains Defective cAMP-Dependent Activation. J. Mol. Biol. 2016, 428, 4890–4904. [Google Scholar] [CrossRef]
- Wong, T.H.; Chiu, W.Z.; Breedveld, G.J.; Li, K.W.; Verkerk, A.J.; Hondius, D.; Hukema, R.K.; Seelaar, H.; Frick, P.; Severijnen, L.-A. PRKAR1B mutation associated with a new neurodegenerative disorder with unique pathology. Brain 2014, 137, 1361–1373. [Google Scholar] [CrossRef]
- Schreyer, S.A.; Cummings, D.E.; McKnight, G.S.; LeBoeuf, R.e.C. Mutation of the RIIβ subunit of protein kinase A prevents diet-induced insulin resistance and dyslipidemia in mice. Diabetes 2001, 50, 2555–2562. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ringheim, G.E.; Taylor, S.S. Dissecting the domain structure of the regulatory subunit of cAMP-dependent protein kinase I and elucidating the role of MgATP. J. Biol. Chem. 1990, 265, 4800–4808. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Cheng, C.Y.; Saldanha, S.A.; Taylor, S.S. PKA-I holoenzyme structure reveals a mechanism for cAMP-dependent activation. Cell 2007, 130, 1032–1043. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.S.; Ilouz, R.; Zhang, P.; Kornev, A.P. Assembly of allosteric macromolecular switches: Lessons from PKA. Nat. Rev. Mol. Cell Biol. 2012, 13, 646–658. [Google Scholar] [CrossRef]
- Zhang, P.; Knape, M.J.; Ahuja, L.G.; Keshwani, M.M.; King, C.C.; Sastri, M.; Herberg, F.W.; Taylor, S.S. Single Turnover Autophosphorylation Cycle of the PKA RIIβ Holoenzyme. PLoS Biol. 2015, 13, e1002192. [Google Scholar] [CrossRef]
- Berman, H.M.; Ten Eyck, L.F.; Goodsell, D.S.; Haste, N.M.; Kornev, A.; Taylor, S.S. The cAMP binding domain: An ancient signaling module. Proc. Natl. Acad. Sci. USA 2005, 102, 45–50. [Google Scholar] [CrossRef]
- Herberg, F.W.; Taylor, S.S.; Dostmann, W.R. Active site mutations define the pathway for the cooperative activation of cAMP-dependent protein kinase. Biochemistry 1996, 35, 2934–2942. [Google Scholar] [CrossRef]
- Zhang, P.; Smith-Nguyen, E.V.; Keshwani, M.M.; Deal, M.S.; Kornev, A.P.; Taylor, S.S. Structure and allostery of the PKA RIIbeta tetrameric holoenzyme. Science 2012, 335, 712–716. [Google Scholar] [CrossRef]
- Kornev, A.P.; Taylor, S.S.; Ten Eyck, L.F. A generalized allosteric mechanism for cis-regulated cyclic nucleotide binding domains. PLoS Comput. Biol. 2008, 4, e1000056. [Google Scholar] [CrossRef]
- Akimoto, M.; McNicholl, E.T.; Ramkissoon, A.; Moleschi, K.; Taylor, S.S.; Melacini, G. Mapping the free energy landscape of PKA inhibition and activation: A double-conformational selection model for the tandem cAMP-binding domains of PKA RIα. PLoS Biol. 2015, 13, e1002305. [Google Scholar] [CrossRef]
- Guo, C.; Zhou, H.-X. Unidirectional allostery in the regulatory subunit RIα facilitates efficient deactivation of protein kinase A. Proc. Natl. Acad. Sci. USA 2016, 113, E6776–E6785. [Google Scholar] [CrossRef] [PubMed]
- McNicholl, E.T.; Das, R.; SilDas, S.; Taylor, S.S.; Melacini, G. Communication between tandem cAMP binding domains in the regulatory subunit of protein kinase A-Iα as revealed by domain-silencing mutations. J. Biol. Chem. 2010, 285, 15523–15537. [Google Scholar] [CrossRef] [PubMed]
- Boettcher, A.J.; Wu, J.; Kim, C.; Yang, J.; Bruystens, J.; Cheung, N.; Pennypacker, J.K.; Blumenthal, D.A.; Kornev, A.P.; Taylor, S.S. Realizing the allosteric potential of the tetrameric protein kinase A RIalpha holoenzyme. Structure 2011, 19, 265–276. [Google Scholar] [CrossRef]
- Brown, S.H.; Wu, J.; Kim, C.; Alberto, K.; Taylor, S.S. Novel isoform-specific interfaces revealed by PKA RIIbeta holoenzyme structures. J. Mol. Biol. 2009, 393, 1070–1082. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Ye, F.; Bastidas, A.C.; Kornev, A.P.; Wu, J.; Ginsberg, M.H.; Taylor, S.S. An Isoform-Specific Myristylation Switch Targets Type II PKA Holoenzymes to Membranes. Structure 2015, 23, 1563–1572. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Krissinel, E. and Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Lu, T.W.; Wu, J.; Aoto, P.C.; Weng, J.H.; Ahuja, L.G.; Sun, N.; Cheng, C.Y.; Zhang, P.; Taylor, S.S. Two PKA RIalpha holoenzyme states define ATP as an isoform-specific orthosteric inhibitor that competes with the allosteric activator, cAMP. Proc. Natl. Acad. Sci. USA 2019, 116, 16347–16356. [Google Scholar] [CrossRef]
- Day, M.E.; Gaietta, G.M.; Sastri, M.; Koller, A.; Mackey, M.R.; Scott, J.D.; Perkins, G.A.; Ellisman, M.H.; Taylor, S.S. Isoform-specific targeting of PKA to multivesicular bodies. J. Cell Biol. 2011, 193, 347–363. [Google Scholar] [CrossRef]
- Ferretti, A.C.; Tonucci, F.M.; Hidalgo, F.; Almada, E.; Larocca, M.C.; Favre, C. AMPK and PKA interaction in the regulation of survival of liver cancer cells subjected to glucose starvation. Oncotarget 2016, 7, 17815–17828. [Google Scholar] [CrossRef]




| PDB | Description | R-Subunit | C-Subunit | Reference PubMed |
|---|---|---|---|---|
| 3PVB | RIA Heterodimer | RIA (73–244) CNBA | C-Subunit with ANP + 2 Mn | 21300294 |
| 5JR7 |
RIA ACRDYS
Heterodimer | RIA (92–365) CNBA + CNBB | C-Subunit with ADP | 27825928 |
| 2QCS | RIA Heterodimer | RIA (91–379:R333K) CNBA + CNBB | C-Subunit with ANP/2 Mn | 17889648 |
| 4X6R | RIA Heterodimer | RIA (91–379:R333K) CNBA + CNBB | C-Subunit Myr | 26278174 |
| 6NO7 | PKA RIA Holoenzyme | RIA Full length CNBA + CNBB | C-Subunit in two states: Apo and ATP/2 Mg bound | 31363049 |
| 4DIN | RIB Holoenzyme | RIB full length CNBA + CNBB | C-Subunit with ATP/2 Mg | 22797896 |
| 2QVS | RIIA Heterodimer | RIIA (108–416) CNBA + CNBB | C-Subunit Apo | 17932298 |
| 4WBB | RIIB Heterodimer | RIIB (108–416) Phospho IS CNBA + CNBB | C-Subunit with ADP+ 2 Ca | 26158466 |
| 3TNQ | RIIB Holoenzyme | RIIB (108–416) Phospho IS CNBA + CNBB | C-Subunit with ATP+ 2 Mg | 22323819 |
| 3TNP | RIIB Holoenzyme | RIIB full length R230K CNBA + CNBB | C-Subunit Apo | 22323819 |
| 4X6Q | RIIB Heterodimer | RIIB (108–416) CNBA + CNBB | C-Subunit Myr | 26278174 |
| 3IDB | RIIB Heterodimer | RIIB (108–268) CNBA Only | C-Subunit with ANP + 2 Mn | 19748511 |
| 3IDC | RIIB Heterodimer | RIIB (102–265) CNBA Only | C-Subunit with ANP + 2 Mn | 19748511 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Welsh, C.L.; Conklin, A.E.; Madan, L.K. Interaction Networks Explain Holoenzyme Allostery in Protein Kinase A. Kinases Phosphatases 2023, 1, 265-287. https://doi.org/10.3390/kinasesphosphatases1040016
Welsh CL, Conklin AE, Madan LK. Interaction Networks Explain Holoenzyme Allostery in Protein Kinase A. Kinases and Phosphatases. 2023; 1(4):265-287. https://doi.org/10.3390/kinasesphosphatases1040016
Chicago/Turabian StyleWelsh, Colin L., Abigail E. Conklin, and Lalima K. Madan. 2023. "Interaction Networks Explain Holoenzyme Allostery in Protein Kinase A" Kinases and Phosphatases 1, no. 4: 265-287. https://doi.org/10.3390/kinasesphosphatases1040016
APA StyleWelsh, C. L., Conklin, A. E., & Madan, L. K. (2023). Interaction Networks Explain Holoenzyme Allostery in Protein Kinase A. Kinases and Phosphatases, 1(4), 265-287. https://doi.org/10.3390/kinasesphosphatases1040016






