Abstract
The structural knowledge about protein kinase CK2 is dominated by crystal structures of human CK2α, the catalytic subunit of human CK2, and the product of the CSNK2A1 gene. In contrast, far fewer structures of CK2α′, its paralogous isoform and the product of the CSNK2A2 gene, have been published. However, according to a PDB survey, CK2α′ is the superior alternative for crystallographic studies because of the inherent potential of the single mutant CK2α′Cys336Ser to provide crystal structures with atomic resolution. In particular, a triclinic crystal form of CK2α′Cys336Ser is a robust tool to determine high-quality enzyme-ligand complex structures via soaking. In this work, further high-resolution CK2α′Cys336Ser structures in complex with selected ligands emphasizing this trend are described. In one of these structures, the “N-terminal segment site”, a small-molecule binding region never found in any eukaryotic protein kinase and holding the potential for the development of highly selective substrate-competitive CK2 inhibitors, was discovered. In order to also address the binding site for the non-catalytic subunit CK2β, which is inaccessible in these triclinic CK2α′Cys336Ser crystals for small molecules, a reliable path to a promising monoclinic crystal form of CK2α′Cys336Ser is presented. In summary, the quality of CK2α′Cys336Ser as an exquisite crystallographic tool is solidified.
1. Introduction
Towards the middle of 2023, the protein structure database PDB [,] contains 315 entries with homologues of CK2α, the catalytic subunit of protein kinase CK2. 41 of them belong to CK2α from Zea mays (UNIPROT_ID P28523) [,], which provided the first CK2α crystal structure [] and dominated the early years of CK2 structural biology. With the crystal structure of the CK2 holoenzyme (heterotetrameric complex of CK2α with the regulatory subunit CK2β) from Homo sapiens [], human CK2α (UNIPROT_ID P68400; product of the CSNK2A1 gene) entered the PDB in 2001; it is now included in 231 entries, the majority of them being released since 2016 (Figure 1a) and belonging to complexes of human CK2α with ATP-competitive inhibitors [,,,].
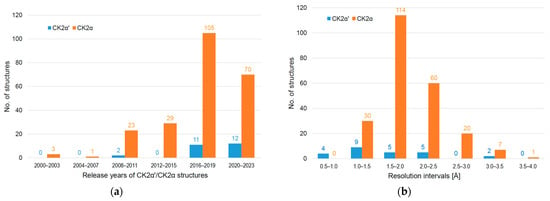
Figure 1.
Distributions of CK2α and CK2α′ crystal structures based on data extracted from the PDB [,]. (a) Structures clustered into intervals of release years; (b) structures clustered into intervals of resolution.
In comparison, the PDB contains only 25 entries of human CK2α′, the paralogous isoform of human CK2α (UNIPROT_ID P19784; product of the CSNK2A2 gene), albeit with an increasing trend (Figure 1a). A main reason for this backlog were solubility issues occurring with wild-type CK2α′ after recombinant expression in bacteria [], but also in insect cells []. The chemical background of this problem became apparent after an inspection of the sequences of human CK2α and CK2α′—they are to 82% identical but differ completely in the C-terminal regions—and the construction of the chimeric variant CK2α_α′ containing its final 24 residues from CK2α′ []. This chimera has a conspicuous tendency to form intermolecular disulphide bridges, which drew attention to Cys336 as part of the CK2α′-derived tail; replacing this cysteine with serine led to the full-length variant CK2α′Cys336Ser, which was well soluble and applicable for enzymatic, calorimetric, and crystallographic studies []. Alternatively, truncating the C-terminal segment, including Cys336, led to CK2α′ constructs that could be successfully crystallized as well [,].
However, CK2α′Cys336Ser was still somewhat problematic for the purpose of crystallization because of its propensity to crystallize as extremely tiny needles under a large variety of conditions. Although these crystal needles grow slightly thicker in the presence of 3-(4,5,6,7-tetrabromo-1H-benzotriazol-1-yl)propan-1-ol (MB002; Scheme 1) [,] and certain other ATP-competitive inhibitors [,], which led to the first crystal structures of CK2α′Cys336Ser [,,], a real breakthrough in growth behaviour only came with a significant increase of the lithium chloride concentration in the crystallization droplets []. This—combined with a sophisticated seeding and soaking strategy to replace the originally bound MB002 ligand from the ATP site—resulted in large, compact, and robust CK2α′Cys336Ser crystals. They led to a number of atomic-resolution structures of CK2α′Cys336Ser [], among them a 1.04-Å complex structure with the benchmark CK2 inhibitor CX-4945 [,,,].
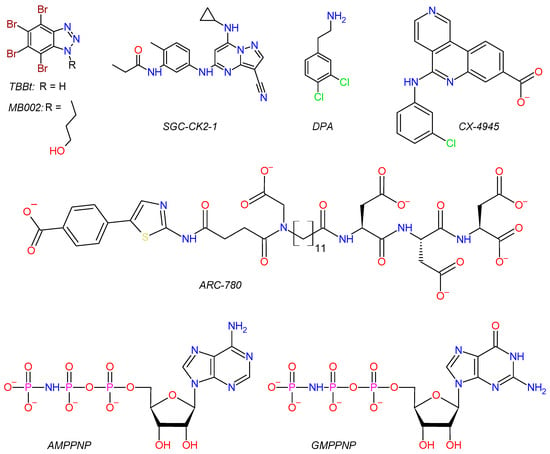
Scheme 1.
Structures of CK2 ligands used in this study.
Subsequently, these CK2α′Cys336Ser crystals together with the optimized soaking protocol [] were systematically exploited:
- For some members of a series of halogenated triazolo pyridines, the exact binding mode at the ATP site could only be clarified via high-resolution complex structures with CK2α′Cys336Ser [];
- In the case of some 2-aminothiazole compounds supposed to occupy an allosteric binding site [,], CK2α′Cys336Ser complex structures revealed that in fact the ATP cavity harboured these inhibitory ligands []; with resolutions of 0.833 Å (PDB_ID 6TGU) and 0.922 Å (PDB_ID 6TE2), two of these structures are currently the best resolved among about 4500 X-ray diffraction entries in the PDB [,] with protein kinase chains;
- The power of the approach became particularly obvious when the αD pocket, an allosteric site originally discovered with CK2α [,], could be occupied via soaking of CK2α′Cys336Ser/MB002 crystals, although this required extensive local conformational rearrangements in the crystalline state of the protein [].
The excellent properties of CK2α′Cys336Ser crystals are responsible for the fact that the resolution distribution of CK2α′ structures is significantly shifted to the high-resolution side compared to CK2α (Figure 1b). In this work, we supplement the collection of CK2α′Cys336Ser/ligand complex structures with further cases. Partially, they were obtained via the crystallization and soaking protocol originally published [], but we also present selected structural examples resulting from efforts to overcome certain limitations of those crystals and procedures.
2. Results and Discussion
2.1. Complex Structures Determined with Triclinic CK2α′Cys336Ser Crystals
2.1.1. CK2α′Cys336Ser Structure in Complex with the ATP-Site Ligands SGC-CK2-1
The size-optimized CK2α′Cys336Ser/MB002 crystals used by Lindenblatt et al. [] to determine atomic resolution complex structures belong to the triclinic crystal system (space group P1). In the corresponding crystal packing, the ATP-site is well accessible, so that in particular, high-affinity ligands like CX-4945 can easily displace the crystallization helper compound MB002 [].
We expected a similar performance for SGC-CK2-1 (Scheme 1), an ATP-competitive CK2 inhibitor with IC50 values for CK2α/CK2α′ inhibition in the low nanomolar range and with an extremely high selectivity [,]. SGC-CK2-1 has the potential to compete with CX-4945 for the role of benchmark CK2 inhibitor and superior CK2-sensitive tool in cell biology studies, as already demonstrated [,,]. In fact, soaking of CK2α′Cys336Ser/MB002 crystals in 2.5 mM SGC-CK2-1 led to a high-quality CK2α′Cys336Ser/SGC-CK2-1 complex structure refined to 1.352 Å resolution (Table 1, Figure 2a).

Table 1.
X-ray diffraction data and refinement statistics for triclinic CK2α′Cys336Ser crystals.
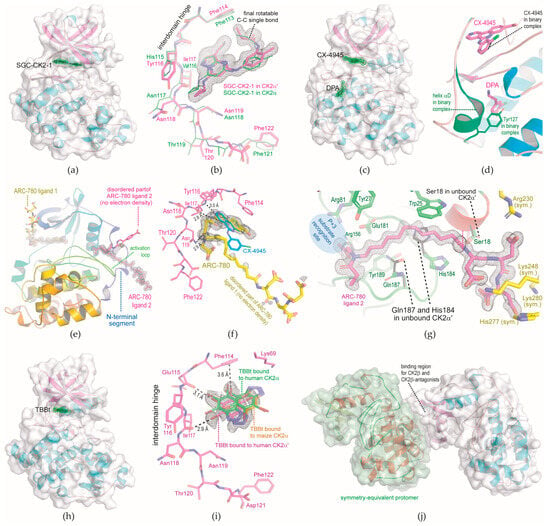
Figure 2.
CK2α′Cys336Ser structures in complex with various ligands obtained in triclinic crystal form. The cutoff level for electron density illustration in this figure is generally 1 σ. The structure pictures were produced with PyMol []. (a) Overview of the CK2α′Cys336Ser/SGC-CK2-1 complex structure. (b) SGC-CK2-1 occupying the ATP site in CK2α′Cys336Ser (magenta-coloured C-atoms) and for comparison in CK2α (green C-atoms; PDB_ID 6Z83 []). (c) Overview of the CK2α′Cys336Ser structure in complex with CX-4945 and DPA. (d) Hinge/helix αD region of the ternary CK2α′Cys336Ser/CX-4945/DPA complex (this work) and an overlaid binary CK2α′Cys336Ser/CX-4945 structure (PDB_ID 6HMB []). (e) Overview of the CK2α′Cys336Ser structure in complex with two molecules of ARC-780. (f) ARC-780 ligand 1 occupying the ATP site of CK2α′Cys336Ser; the lengths of three H-bonds between the ligand and the enzyme’s hinge region (black dotted lines) are given in Å; for comparison, CX-4945 is drawn from PDB_ID 6HMB [] after 3D-fit of the protein matrices. (g) The ordered part of ARC-780 ligand 2 occupying the novel N-terminal segment site of CK2α′Cys336Ser; side chains of a neighbouring symmetry mate were drawn with yellow C-atoms; Ser18, His184, and Gln187 from unbound (ARC-780-free) CK2α′Cys336Ser were depicted with black C-atoms. (h) Overview of the CK2α′Cys336Ser/TBBt complex structure. (i) Zoom of TBBt bound to the ATP site of CK2α′Cys336Ser (magenta-coloured C-atoms); for comparison, TBBt was drawn as bound to maize CK2α (orange-coloured C-atoms; PDB_ID 1J91 []) or to human CK2α at pH 8.5 (green C-atoms; PDB_ID 7QGE []). (j) Involvement of the CK2β binding site of CK2α′Cys336Ser in a crystal contact.
For comparison, a CK2α/SGC-CK2-1 complex structure was published with a resolution of 2.17 Å []. An overlay of this structure on the novel CK2α′Cys336Ser/SGC-CK2-1 structure (Figure 2b) reveals that SGC-CK2-1 is bound in a similar way to the two paralogs, albeit with a noteworthy conformational difference in the terminal propionamide moiety: the torsion angle around the final C-C-bond as indicated in Figure 2b is −23.14° in CK2α′Cys336Ser-bound SGC-CK2-1, but 35.24° or 59.65°, respectively, in the two crystallographically independent copies of the CK2α/SGC-CK2-1 structure []. This difference is not caused by the direct protein environment around the propionamide moiety since it is identical in the two isoforms. Rather, Figure 2b suggests that the structural background lies in the interdomain hinge where CK2α and CK2α′ have their only sequence difference in the ATP-site region: Tyr116 of CK2α′ is more voluminous than its equivalent His115 of CK2α so that SGC-CK2-1 can approach the hinge less closely in CK2α′. This causes a shift visible at the cyclopropyl group and a constraint at the other side of the molecule, namely at the aforementioned propionamide moiety, which penetrates deeper into the ATP site of CK2α′ than into that of CK2α (Figure 2b). This subtle effect correlates well with the fact that the IC50 value of SGC-CK2-1 for CK2α′ inhibition is only half as large as for CK2α inhibition [], and it could be a first step on the way to the attractive goal of generating isoform-specific CK2 inhibitors [,].
2.1.2. Ternary Complex Structure of CK2α′Cys336Ser, CX-4945, and DPA
The αD pocket of CK2α and CK2α′ is a so-called “cryptic site” []; this designation was introduced for “hidden” pockets becoming visible and accessible only after conformational changes induced in the presence of a suitable ligand. In the case of the αD pocket, this ligand was 3,4-dichloro phenethylamine (DPA; Scheme 1), which led to the discovery of the αD pocket in 2016 in a fragment-based screen with CK2α crystals [,], while in all CK2α structures published before, it was blocked by either a phenylalanine or a tyrosine side chain from the eponymous helix αD. After its discovery, the αD pocket was used to develop the bivalent inhibitor CAM4066 [,].
For CK2α′, the existence of an αD pocket was confirmed via soaking DPA into a CK2α′Cys336Ser/MB002 crystal []. In the resulting ternary complex structure, the two ligands are 6.9 Å away from each other, a distance that could be chemically bridged and led to the bivalent and highly selective inhibitor KN2 []. This success induced the hypothesis that the ATP site and the αD pocket of crystalline CK2α′Cys336Ser can be occupied independently of each other, so that CK2α′Cys336Ser structures in complex with arbitrary pairs of ligands can be obtained via soaking CK2α′Cys336Ser/MB002 crystals.
To test this, we soaked a CK2α′Cys336Ser/MB002 crystal simultaneously with CX-4945 and DPA. In the resulting crystal structure with a resolution below 1.1 Å (Table 1), both ligands were well defined by electron density (Figure 2c), meaning CX-4945 had completely replaced MB002 and DPA had opened and occupied the αD pocket. A 3D-fit of this ternary CK2α′Cys336Ser/CX-4945/DPA complex on the binary CK2α′Cys336Ser/CX-4945 complex structure (Figure 2d) illustrates again the large conformational adaptations occurring in the αD region during soaking. In summary, this experiment confirms that the ATP site and the αD pocket of crystalline CK2α′Cys336Ser cannot only be occupied independently but even simultaneously.
2.1.3. N-Terminal Segment Site: A Novel Ligand Binding Site Discovered in a CK2α′Cys336Ser Structure in Complex with the Bisubstrate Inhibitor ARC-780
ARC-780 (Scheme 1) is a follow-up compound of a series of CK2 bisubstrate inhibitors [] occupying the ATP site and addressing the recognition region for peptide or protein substrates as well, so that the corresponding Ki values are partly in the picomolar range []. ARC-780 contains a long aliphatic linker of 11 methylen groups (Scheme 1), resulting in high hydrophobicity and many rotational degrees of freedom. Therefore, we extended the soaking time of initial CK2α′Cys336Ser/MB002 crystals in ARC-780 containing solutions to eight months. In this time, the concentrations of salts were stepwise decreased from 900 mM LiCl, 500 mM NaCl, and 110 mM Tris/HCl to finally 50 mM LiCl and 10 mM Tris/HCl, while in parallel, the concentration of DMSO was increased from 0 to 30% (v/v), and the concentration of the precipitant PEG6000 was adopted to saturation.
In spite of these extreme alterations of the surrounding solution, the CK2α′Cys336Ser crystals retained their integrity, and one of them led to a 1.255 Å crystal structure (structure no. 3 in Table 1). Surprisingly, in this structure, two large patches of non-protein electron density were visible. They could be interpreted unambiguously with two separate ARC-780 molecules, and in both cases, large, albeit different, parts of ARC-780 were not covered with significant electron density (Figure 2e).
The first ARC-780 ligand occupies the ATP site (Figure 2f, Video S1): its terminal carboxy phenyl moiety binds next to the Phe114 side chain, and after 3D-fit of the according protein matrices, it overlaps entirely with the corresponding sub-structure of CX-4945. Using its triazole ring and the two subsequent amide bonds, the first ARC-780 ligand forms three close hydrogen bonds with the enzyme’s hinge region, while the aliphatic linker and the final peptidic part are completely disordered (Figure 2f).
The second ARC-780 ligand—or at least a considerable part of it—is bound to a region (Figure 2g, Video S1) that, to our knowledge, has never been described before as a small-molecule binding site in CK2α, CK2α′, or any other member of the superfamily of eukaryotic protein kinases (EPKs). We call it the “N-terminal segment site” here because it is located close to an N-terminal extension of the EPK-typical N-lobe domain that attaches the likewise EPK-typical activation loop and folds back to the C-lobe domain (Figure 2e). This intramolecular interaction between the N-terminal segment and the activation loop is a distinguishing feature of CK2α/CK2α′ among the EPK superfamily and the main structural basis of CK2′s constitutive activity since it mimics the intermolecular coordination of the activation loop of cyclin-dependent kinases by activatory cyclin proteins [].
Just like the αD site, the N-terminal segment site is a cryptic site []. Its access for a ligand requires large conformational changes in the side chains of Ser18, His184, and Gln187, which are indicated in Figure 2g and visualized in Video S1. The four carboxy groups in the peptidic part of ARC-780 form ionic interactions with some positively charged side chains of a crystalline CK2α′Cys336Ser neighbour (Figure 2g). Thus, the fact that this peptidic part is well defined by electron density in strong contrast to what has been observed in CK2α complex structures with precursor bisubstrate inhibitors [] is most likely an artifact of the crystal packing. Insofar, it must remain open in the moment how far the peptidic part of ARC-780 contributes to the compound’s affinity to the N-terminal segment site of the enzyme at all.
In contrast to that, the N-linked carboxy methyl group of ARC-780 (Scheme 1) is likely to be an important binding determinant because it interacts ionically with Arg156 (Figure 2g, time point 12 s in Video S1). Together with Arg81, Arg156 forms the P + 3 recognition site for CK2 substrates, which means that this carboxy methyl group would interfere with substrate binding. In the future, this fact could pave the way to developing highly selective, substrate-competitive CK2 inhibitors from the sub-structure of ARC-780 (Figure 2g) that binds to the novel N-terminal segment site with its unique molecular environment.
2.1.4. CK2α′Cys336Ser Structure in Complex with TBBt
MB002 [] (Scheme 1) is an efficient additive for the crystallization of CK2α′Cys336Ser [,], but it has the disadvantage of not being commercially available. We found its head group 4,5,6,7-tetrabromobenzotriazole (TBBt; Scheme 1) to be a suitable and inexpensive substitute: co-crystallization of CK2α′Cys336Ser with TBBt followed by micro- and macro-seeding led to CK2α′Cys336Ser/TBBt crystals, which are isomorphous to the triclinic CK2α′Cys336Ser/MB002 crystals used before, and resulted in a CK2α′Cys336Ser/TBBt complex structure of 1.033 Å resolution (structure no. 4 in Table 1; Figure 2h). For comparison, previously published complex structures with TBBt have resolutions of 2.22 Å (with maize CK2α []), 1.88 Å (with human CK2α at pH 7.5 []), and 2.27 Å (with human CK2α at pH 8.5 []).
For TBBt, a pKA value of 4.78 was reported [], so that it is monoanionic at pH 7 and above. Therefore, a “competition between electrostatic interactions and halogen bonding” [] exists for the interaction of TBBt with the ATP site of CK2α. In fact, in a human CK2α/TBBt complex structure obtained at pH 8.5, two alternative binding modes of TBBt were observed [] (Figure 2i). In one of them, which had been found before in a maize CK2α/TBBt complex structure as well [] (Figure 2i), electrostatic forces dominate and direct TBBt via its anionic triazole moiety into the proximity of the highly conserved side chain of Lys68 (which is equivalent to Lys69 of CK2α′ drawn in Figure 2i) []. In the other binding mode, however, halogen bonds force TBBt close to the interdomain hinge of human CK2α. It is this second binding mode that we observed exclusively in the CK2α′Cys336Ser/TBBt complex (Figure 2i). Apparently, there, the three halogen bonds from the bromo substituents of TBBt to the carbonyl oxygen atoms of Glu115 and Ile117, as well as to the phenyl ring of Phe114, dominate clearly over the electrostatic term.
Noteworthy, the salt concentration during crystallization of the CK2α′Cys336Ser/TBBt complex was much higher than that reported for crystallization of the CK2α/TBBt complex []. Therefore, follow-up experiments are required to clarify if the difference in TBBt binding between human CK2α and CK2α′ is a crystallization artifact or if it reflects subtle differences between the isoenzymes themselves. In any case, atomic-resolution complex structures of CK2α′Cys336Ser with halogenated inhibitors can help to refine the sophisticated knowledge about halogen bonding in the ATP site of CK2α subunits [,,].
2.2. Complex Structures Determined with Monoclinic CK2α′Cys336Ser Crystals
In order to test if the excellent diffraction of triclinic CK2α′Cys336Ser crystals is restricted to this particular crystal packing, an alternative crystal packing of CK2α′Cys336Ser had to be found. A further motivation to pursue this goal came from the observation that the outer surface of the enzyme’s N-lobal anti-parallel CK2β-sheet, which serves as an interface for the binding of the non-catalytic subunit CK2β [], is involved in a crystal contact in the triclinic CK2α′Cys336Ser crystals (Figure 2j): it is thus inaccessible for small CK2β-antagonistic molecules in soaking approaches and also for co-crystallization because the crystal packing is incompatible with any occupation of the CK2β binding site.
Small peptidic or non-peptidic molecules that compete with CK2β for its binding site and disturb the assembly of the CK2 holoenzyme have been repeatedly described [,,,,,,,] and, in several cases, structurally characterized in recent years, but exclusively for CK2α. Structures of CK2α′ in complex with such ligands do not exist at present; in order to create the prerequisite for them, a reliable pathway to an alternative crystal form of CK2α′Cys336Ser is desirable.
Since it was demonstrated with lithium sulfate [] or lithium chloride [] that the crystallization behavior of CK2α′ constructs is very sensitive to the content of lithium ions, we systematically varied the lithium chloride concentration in CK2α′Cys336Ser crystallization experiments between 500 mM and 900 mM with otherwise unchanged conditions. Already 810 mM LiCl—i.e., a slight reduction of the LiCl concentration compared to the 900 mM in crystallization solutions of the triclinic CK2α′Cys336Ser crystals (Table 1)—led to CK2α′Cys336Ser crystals with monoclinic symmetry (Table 2). In this new crystal form, two CK2α′Cys336Ser protomers per asymmetric unit are present. In one of them, the CK2β binding site is used to form the contact interface to a neighbouring symmetry mate (Figure 3a) in a similar way as it was observed for the triclinic crystals (Figure 2j). In the second protomer, however, the CK2β binding site, although also surrounded by two crystalline neighbours, is basically accessible to ligands of small and medium size (Figure 3b).

Table 2.
X-ray diffraction data and refinement statistics for monoclinic CK2α′Cys336Ser crystals.
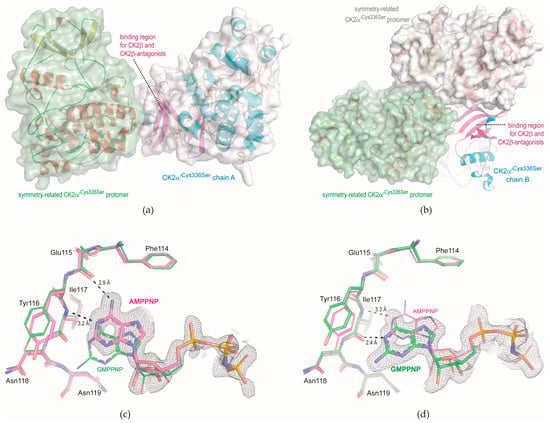
Figure 3.
CK2α′Cys336Ser structures obtained with monoclinic crystals. The asymmetric unit contains two CK2α′Cys336Ser chains. (a) Crystal contact of the N-terminal domain of CK2α′Cys336Ser chain A with a symmetry-related CK2α′Cys336Ser protomer. (b) Crystal packing environment of the N-terminal domain of CK2α′Cys336Ser chain B. (c) Cosubstrate binding site of the CK2α′Cys336Ser/AMPPNP complex structure (magenta-coloured C-atoms); for comparison, the CK2α′Cys336Ser/GMPPNP complex structure was drawn with green C-atoms. (d) Cosubstrate binding site of the CK2α′Cys336Ser/GMPPNP complex structure (green C-atoms); for comparison, the CK2α′Cys336Ser/AMPPNP complex structure was drawn with magenta-coloured C-atoms. The cutoff level for electron density illustration in this figure is generally 1 σ. The structure pictures were produced with PyMol [].
The X-ray diffraction capacity of the monoclinic CK2α′Cys336Ser crystals (Table 2) is similar to that of the triclinic crystals (Table 1), i.e., structures with atomic resolution can be achieved and expected. We demonstrate this here with two CK2α′Cys336Ser complex structures with the co-substrate analogues AMPPNP or GMPPNP (Scheme 1), both of which are well visible in electron density at the ATP site (Figure 3c,d), at least in one of the two protomers, respectively.
The ability of CK2 to use GTP as well as ATP as cosubstrate for the kinase reaction has been known since the early 1960s []. The first evidence that this dual-cosubstrate specificity is a special property of CK2 compared to other protein kinases emerged in the 1970s [], but how exceptional this feature of CK2α and CK2α′ really is among EPKs has only been demonstrated in recent years by affinity profiling []. Its structural background was clarified with maize CK2α structures in complex of AMPPNP or GMPPNP [], which revealed essentially what is visible in Figure 3c,d again: the triphospho moieties of the two ligands overlap exactly, but in the region of the purine bases, the different hydrogen-bonding potentials of adenine and guanine enforce a shift of GMPPNP compared to AMPPNP so that finally the pattern of hydrogen bonds between the purine base and the hinge region is shifted by one peptide bond toward the C-terminus.
3. Materials and Methods
3.1. Protein Expression and Purification
For any crystallization experiment shown in this work, we used the point mutant CK2α′Cys336Ser with an N-terminal His-tag []. The corresponding construct in a pETDuet-1 vector (Merck) was transformed into either chemically competent E. coli Gen-XTM (DE3) or chemically competent BL21 Star (DE3) cells. A single clone, respectively, was selected for a preculture. For this, the cells were grown overnight at 30 °C in 100 mL LB medium (10 g/L yeast extract, 20 g/L tryptone, and 20 g/L NaCl) supplemented with 100 µg/mL ampicilin under constant agitation at 180 rpm. An amount of 10 mL of the preculture was used to inocculate 600 mL LB medium.
The subsequent main cultures were grown to an OD600 of 0.6 at 37 °C under constant agitation at 180 rpm. Upon reaching an OD600 of 0.6, the temperature was lowered to 18 °C, and isopropyl-β-thiogalactosid (IPTG) was added to a final concentration of 0.5 mM. Expression was subsequently carried out for 18 h. Afterwards, the cells were harvested via centrifugation at 4 °C and 6200× g for 30 min. The pellet was washed with a 0.9% (w/v) sodium chloride solution and afterwards stored at −80 °C. For lysis, the pellet was thawed and then incubated with lysis buffer composed of 500 mM NaCl, 25 mM TRIS-HCl, pH 8.5, 10 µg/mL DNase1, and 1 mg/mL lysozyme for 1 h on ice. Sonification (40% amplitude, 2 s on/4 s off, 4 °C, 3 min total sonification) followed. The lysate was cleared by ultracentrifugation (186,000× g, 4 °C, 30 min). The supernatant was filtered and then filled into a superloop for application onto a 5 mL HisTrap FF column (Cytiva) mounted on an ÄKTA Explorer System. Application and washing (15 CV) were carried out using buffer A (40 mM imidazole, 500 mM NaCl, and 25 mM TRIS-HCl. pH 8.5). Afterwards, the protein was eluted using a linear gradient from 0 to 100% buffer B (250 mM imidazole, 500 mM NaCl, 25 mM TRIS-HCl, pH 8.5) over 15 CV. The flowrate was 0.8 mL/min. Elution was monitored via absorption at 280 nm. All fractions within a peak occurring during elution were tested using SDS-PAGE. Fractions containing pure CK2α′Cys336Ser were pooled, rebuffered into a standard assay buffer (500 mM NaCl, 25 mM TRIS-HCl, pH 8.5), and concentrated to 5 mg/mL by ultrafiltration using AMICON® tubes with a cut-off of 30 kDa.
3.2. Preparation of ARC-780
While all other CK2 ligands used in this work were either commercially available or (in the case of MB002) were provided by Prof. Maria Bretner, Warsaw, Poland, the substance ARC-780, which had been planned to be a bisubstrate inhibitor, had to be synthesized. For this, (L-Asp)3 peptide with tert-butyl-protected side chains was synthesized on 2-chlorotrityl chloride (2CTC) polystyrene resin (Iris Biotech GmbH) according to common Fmoc-peptide synthesis protocols. Succinic acid and the precursors of residues of 12-[(carboxymethyl)amino]dodecanoic acid and 4-(2-amino-1,3-thiazol-5-yl)benzoic acid were synthesized and attached to the peptide as it was described previously []. The compound was cleaved from the resin with a 3 h treatment with TFA:TIPS:H2O (95:2.5:2.5, v:v:v); subsequent RP-HPLC purification [Shimadzu Prominence LC Solution HPLC system with SPD M20A PDA detector; Phenomenex Gemini C18 RP column, 250 × 4.6 mm, particle size 5 μm, eluted with MeCN/H2O gradient (0.1% TFA) at flow rate of 1 mL/min; gradient of 20–62% MeCN/14 min, Rt of the title compound: 10.7 min, λmax = 322 nm; Figure S1] yielded ARC-780 with a purity of more than 96.5% (Peak Table insert in Figure S1). The structure was verified using high-resolution mass spectrometry (HRMS) with combined Varian 910-FT-ICR and Varian J-320 3Q spectrometers used in ESI positive ion mode (Figure S2; found: 921.31740 Da [M + H+]; m/z calcd. C40H52N6O17S [M + H+]: 921.31824 Da).
3.3. Protein Crystallization
3.3.1. General Procedure
All crystals were grown by applying the sitting drop variant of the vapor diffusion method. For this, 24-well plates (Cryschem) were used. The volume of the reservoir solution was initially 700 µL. All plates were sealed with transparent adhesive film and kept at 20 °C. All microseeding steps were carried out by destroying one crystal from a previous experiment and streaking a loop first through the resulting suspension of crystal seeds and afterwards through the drop that should be inocculated.
3.3.2. Preparation of CK2α′Cys336Ser/SGC-CK2-1 Binary Complex Crystals and of CK2α′Cys336Ser/CX-4945/DPA Ternary Complex Crystals
Co-crystals of CK2α′Cys336Ser and MB002 (Scheme 1; provided by Maria Bretner, Warsaw, Poland) were grown according to the previously published protocol []. For this, a 5 mg/mL stock solution of CK2α′Cys336Ser was mixed in a 1:10 ratio with a 10 mM stock solution of MB002 in DMSO. After incubation on ice for 30 min and centrifugation (16,100× g for 2 min, room temperature), 6 µL of the supernatant were mixed with 3 µL of the reservoir solution (900 mM LiCl, 28% (w/v) PEG 6000, 100 mM TRIS-HCl, pH 8.5). After equilibration, crystal growth was induced by microseeding. Crystals were then optimized via macroseeding. The resulting co-crystals of CK2α′Cys336Ser and MB002 were purged with reservoir solution (900 mM LiCl, 28% (w/v) PEG 6000, 100 mM TRIS-HCl, pH 8.5).
SGC-CK2-1 (Sigma-Aldrich) was solved in DMSO at a concentration of 10 mM. This solution was added to CK2α′Cys336Ser/MB002 complex crystals to a concentration of 2.5 mM for extensive soaking.
DPA (Sigma-Aldrich, St. Louis, MO, USA) was solved in DMSO at a concentration of 50 mM and CX-4945 (TargetMol, Wellesley Hills, MA, USA) to a concentration of 10 mM likewise in DMSO. 1 µL of each of these solutions, respectively, was added to a CK2α′Cys336Ser/MB002 complex crystals for extensive soaking.
3.3.3. Preparation of CK2α′Cys336Ser/ARC-780 Complex Crystals
Co-crystals of CK2α′Cys336Ser and MB002 were grown, optimized by micro- and macroseeding, and purged as described above. Then, ARC-780 was added to an initial concentration of 1 mM by adding 1 µL of a 10 mM ARC-780 solution in DMSO in a 1:10 ratio to the droplet. After extensive initial soaking, the salt concentration was successively lowered over a period of 8 months, while in parallel, the concentrations of DMSO and PEG6000 were increased. This was conducted by gradually replacing the reservoir and the droplet solution until any NaCl (which came from the storage solution of the protein) was removed, the LiCl concentration was reduced to 50 mM, the PEG 6000 concentration was increased to saturation, and a DMSO content of 30% was reached. Meanwhile, the ARC-780 ligand was included in any desalting step reaching saturation after the final dilution step.
3.3.4. Co-Crystallization of CK2α′Cys336Ser and TBBt
For this crystallization attempt, the initial co-crystallization with MB002 was avoided. Rather, a 5 mg/mL stock solution of CK2α′Cys336Ser was mixed in a 1:10 ratio with a 20 mM solution of TBBt (Sigma-Aldrich) in DMSO. After incubation on ice for 30 min and centrifugation (16,100× g for 2 min, room temperature), 10 µL of the supernatant were mixed with 5 µL of the reservoir solution (900 mM LiCl, 28% (w/v) PEG 6000, 100 mM TRIS-HCl, pH 8.5). After equilibration, microseeding was carried out to induce crystal growth. Small crystals appeared after one week. One of them was used as a macroseed by transferring it to a second drop prepared in the same manner as the first one.
3.3.5. Growth of CK2α′Cys336Ser Crystals in Complex with AMPPNP or GMPPNP
Monoclinic crystals of CK2α′Cys336Ser were grown in a slightly altered manner compared to the triclinic CK2α′Cys336Ser crystals. 5 mg/mL CK2α′Cys336Ser in storage buffer composed of 500 mM NaCl and 25 mM TRIS-HCl, pH 8.5, were centrifuged immediately after thawing and without mixing it with any inhibitor. 10 µL of the supernatant were mixed with 5 µL of a reservoir solution containing a slightly lowered LiCl concentration of 810 mM, 28% (w/v) PEG 6000, 100 mM TRIS-HCl, pH 8.5. After equilibration, crystallization was initiated by microseeding, and then crystals were optimized via macroseeding. The ATP-analogue AMPPNP or the GTP-analogue GMPPNP were combined with these crystals by socking. In both cases, a 20 mM solution in 60 mM MgCl2 was prepared. For soaking, 3 µL of the crystal mother liquor were removed and replaced with 3 µL of either 20 mM AMPPNP, 60 mM MgCl2 or 20 mM GMPPNP, 60 mM MgCl2.
3.4. X-ray Diffraction Data Collection and Processing followed by Structure Determination
To prepare X-ray diffraction data collection, any CK2α′Cys336Ser/ligand crystal except for the CK2α′Cys336Ser/ARC-780 co-crystals was cryo-protected before vitrification. For this, 1.4 µL of the reservoir solution and 0.6 µL ethylene glycol were mixed, and the crystals were incubated in this solution for a few seconds. Next, the crystals were flash frozen in liquid nitrogen. Data collections were carried out at beamlines ID 23-1 and MASSIF-3 of the European Synchrotron Radiation Facility (ESRF) in Grenoble (France) or at beamline P13 of the European Molecular Biology Laboratory (EMBL) outstation at DESY in Hamburg (Germany). The temperature during diffraction data collection was 100 K.
The raw diffraction data were processed with the autoPROC toolbox [] using default settings. The autoPROC pipeline comprises XDS [] for indexing and integration, POINTLESS [] and AIMLESS [] from the CCP4 suite [] for symmetry determination and scaling, and finally STARANISO [] for anisotropy analysis. In all cases, the ellipsoidal output data set of STARANISO [] was used for the further procedure.
All structures were solved by molecular replacement using the CK2α′Cys336Ser/MB002 structure [] (PDB_ID 6HMQ) as a starting model. The search calculations were performed with PHASER [] from Phenix []. For the subsequent structure refinement, we used the phenix.refine module [] of Phenix [] in alternation with Coot [] for manual model building. Small-molecule ligands were parametrized with eLBOW [] within the Phenix [] platform.
4. Conclusions
In this work, six crystal structures of CK2α′Cys336Ser in complex with various ligands and obtained with two different crystal forms are described. In all cases, the high-resolution limit of the underlying X-ray diffraction data is in the range from 1.03 to 1.35 Å. This demonstrates the qualification of CK2α′Cys336Ser as an exquisite tool for CK2 crystallography and emphasizes a tendency to atomic-resolution structures, as indicated already in a survey of the PDB (Figure 1b). In particular, the remarkable robustness of the longer-known triclinic CK2α′Cys336Ser crystals is demonstrated here with an eight-month soaking experiment with ARC-780, a 920 Da molecule designed as a bisubstrate inhibitor, in which the crystals were acclimated to low-salt, DMSO-rich conditions by repeated replacement. In the end, this led to the discovery of the N-terminal segment site, a ligand binding site never observed before in the EPK superfamily that offers the potential to develop highly selective substrate-competitive CK2 inhibitors.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/kinasesphosphatases1040018/s1, Figure S1: RP-HPLC purification of ARC-780; Figure S2: HRMS spectrum of ARC-780; Video S1: binding of ARC-780 to the ATP site and to the novel N-terminal segment site of CK2α′Cys336Ser.
Author Contributions
Conceptualization, C.W., D.L., K.V., A.U. and K.N.; methodology and investigation, C.W., D.L. and K.V.; validation, C.W. and K.N.; data curation, C.W. and K.N.; visualization, D.L. and K.N.; writing—original draft preparation, C.W., K.V. and K.N.; writing—review and editing, C.W., D.L., K.V., A.U. and K.N.; project administration, K.N.; funding acquisition, K.N., K.V. and A.U. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Deutsche Forschungsgemeinschaft (DFG), grants nos. NI 643/4-1 and NI 643/11-1 to K.N., and by the Estonian Research Council, grant PRG454 to K.V. and A.U.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The atomic coordinates and the structure factors of the six CK2α′Cys336Ser crystal structures documented in Table 1 and Table 2 are available from the PDB [] under the following accession codes: 8Q9S, CK2α′Cys336Ser/SGC-CG2-1 complex structure; 8QBU, CK2α′Cys336Ser/CX-4945/DPA complex structure; 8Q77, CK2α′Cys336Ser/ARC-780 complex structure; 8QCD, CK2α′Cys336Ser/TBBt complex structure; 8QCG, CK2α′Cys336Ser/AMPPNP complex structure; and 8QF1, CK2α′Cys336Ser/GMPPNP complex structure. The digital object identifier (DOI) for the raw X-ray diffraction data underlying the CK2α′Cys336Ser/TBBt structure is 10.15151/ESRF-ES-705660733.
Acknowledgments
We thank Professor Ulrich Baumann (University of Cologne) for access to the protein crystallography infrastructure at the Cologne Crystallization facility (C2f) and Professor Maria Bretner (Warsaw Technical University) for MB002 (Scheme 1). We are grateful to the staff of the European Synchrotron Radiation Facility (ESRF) in Grenoble (France) and of the European Molecular Biology Laboratory at DESY in Hamburg (Germany) for their assistance with the collection of diffraction data.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- wwPDB consortium. Protein Data Bank: The single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2019, 47, D520–D528. [Google Scholar] [CrossRef] [PubMed]
- Dobrowolska, G.; Boldyreff, B.; Issinger, O.G. Cloning and sequencing of the casein kinase 2 α subunit from Zea mays. Biochim. Biophys. Acta 1991, 1129, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Boldyreff, B.; Meggio, F.; Dobrowolska, G.; Pinna, L.A.; Issinger, O.G. Expression and characterization of a recombinant maize CK-2 α subunit. Biochim. Biophys. Acta 1993, 1173, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Niefind, K.; Guerra, B.; Pinna, L.A.; Issinger, O.G.; Schomburg, D. Crystal structure of the catalytic subunit of protein kinase CK2 from Zea mays at 2.1 Å resolution. EMBO J. 1998, 17, 2451–2462. [Google Scholar] [CrossRef] [PubMed]
- Niefind, K.; Guerra, B.; Ermakowa, I.; Issinger, O. Crystal structure of human protein kinase CK2: Insights into basic properties of the CK2 holoenzyme. EMBO J. 2001, 20, 5320–5331. [Google Scholar] [CrossRef]
- Battistutta, R. Protein kinase CK2 in health and disease: Structural bases of protein kinase CK2 inhibition. Cell. Mol. Life Sci. 2009, 66, 1868–1889. [Google Scholar] [CrossRef]
- Cozza, G.; Pinna, L.A.; Moro, S. Protein kinase CK2 inhibitors: A patent review. Expert Opin. Ther. Pat. 2012, 22, 1081–1097. [Google Scholar] [CrossRef]
- Atkinson, E.L.; Iegre, J.; Brear, P.D.; Zhabina, E.A.; Hyvönen, M.; Spring, D.R. Downfalls of Chemical Probes Acting at the Kinase ATP-Site: CK2 as a Case Study. Molecules 2021, 26, 1977. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Y.; Wang, J.; Zhou, Z.; Cao, S.; Zhang, J. Strategies of Targeting CK2 in Drug Discovery: Challenges, Opportunities, and Emerging Prospects. J. Med. Chem. 2023, 66, 2257–2281. [Google Scholar] [CrossRef]
- Olsen, B.; Boldyreff, B.; Niefind, K.; Issinger, O. Purification and characterization of the CK2α’-based holoenzyme, an isozyme of CK2α: A comparative analysis. Protein Expr. Purif. 2006, 47, 651–661. [Google Scholar] [CrossRef] [PubMed]
- Olsen, B.; Rasmussen, T.; Niefind, K.; Issinger, O. Biochemical characterization of CK2 α and α’ paralogues and their derived holoenzymes: Evidence for the existence of a heterotrimeric CK2 α’-holoenzyme forming trimeric complexes. Mol. Cell. Biochem. 2008, 316, 37–47. [Google Scholar] [CrossRef]
- Bischoff, N.; Olsen, B.; Raaf, J.; Bretner, M.; Issinger, O.G.; Niefind, K. Structure of the human protein kinase CK2 catalytic subunit CK2α’ and interaction thermodynamics with the regulatory subunit CK2β. J. Mol. Biol. 2011, 407, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Nakaniwa, T.; Kinoshita, T.; Sekiguchi, Y.; Tada, T.; Nakanishi, I.; Kitaura, K.; Suzuki, Y.; Ohno, H.; Hirasawa, A.; Tsujimoto, G. Structure of human protein kinase CK2α2 with a potent indazole-derivative inhibitor. Acta Crystallogr. F Struct. Biol. Commun. 2009, 65, 75–79. [Google Scholar] [CrossRef]
- Tsuyuguchi, M.; Nakaniwa, T.; Kinoshita, T. Crystal structures of human CK2α2 in new crystal forms arising from a subtle difference in salt concentration. Acta Crystallogr. F Struct. Biol. Commun. 2018, 74, 288–293. [Google Scholar] [CrossRef] [PubMed]
- Bretner, M.; Najda-Bernatowicz, A.; Łebska, M.; Muszyńska, G.; Kilanowicz, A.; Sapota, A. New inhibitors of protein kinase CK2, analogues of benzimidazole and benzotriazole. Mol. Cell. Biochem. 2008, 316, 87–89. [Google Scholar] [CrossRef] [PubMed]
- Hochscherf, J.; Lindenblatt, D.; Witulski, B.; Birus, R.; Aichele, D.; Marminon, C.; Bouaziz, Z.; Le Borgne, M.; Jose, J.; Niefind, K. Unexpected Binding Mode of a Potent Indeno[1,2-b]indole-Type Inhibitor of Protein Kinase CK2 Revealed by Complex Structures with the Catalytic Subunit CK2α and Its Paralog CK2α’. Pharmaceuticals 2017, 10, 98. [Google Scholar] [CrossRef] [PubMed]
- Niefind, K.; Bischoff, N.; Golub, A.G.; Bdzhola, V.G.; Balanda, A.O.; Prykhod’ko, A.O.; Yarmoluk, S.M. Structural Hypervariability of the Two Human Protein Kinase CK2 Catalytic Subunit Paralogs Revealed by Complex Structures with a Flavonol- and a Thieno[2,3-d]pyrimidine-Based Inhibitor. Pharmaceuticals 2017, 10, 9. [Google Scholar] [CrossRef]
- Lindenblatt, D.; Nickelsen, A.; Applegate, V.M.; Hochscherf, J.; Witulski, B.; Bouaziz, Z.; Marminon, C.; Bretner, M.; Le Borgne, M.; Jose, J.; et al. Diacritic binding of an indenoindole inhibitor by CK2α paralogs explored by a reliable path to atomic resolution CK2α′ structures. ACS Omega 2019, 4, 5471–5478. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui-Jain, A.; Drygin, D.; Streiner, N.; Chua, P.; Pierre, F.; O’Brien, S.E.; Bliesath, J.; Omori, M.; Huser, N.; Ho, C.; et al. CX-4945, an orally bioavailable selective inhibitor of protein kinase CK2, inhibits prosurvival and angiogenic signaling and exhibits antitumor efficacy. Cancer Res. 2010, 70, 10288–10298. [Google Scholar] [CrossRef] [PubMed]
- Battistutta, R.; Cozza, G.; Pierre, F.; Papinutto, E.; Lolli, G.; Sarno, S.; O’Brien, S.E.; Siddiqui-Jain, A.; Haddach, M.; Anderes, K.; et al. Unprecedented selectivity and structural determinants of a new class of protein kinase CK2 inhibitors in clinical trials for the treatment of cancer. Biochemistry 2011, 50, 8478–8488. [Google Scholar] [CrossRef]
- Pierre, F.; Chua, P.C.; O’Brien, S.E.; Siddiqui-Jain, A.; Bourbon, P.; Haddach, M.; Michaux, J.; Nagasawa, J.; Schwaebe, M.K.; Stefan, E.; et al. Pre-clinical characterization of CX-4945, a potent and selective small molecule inhibitor of CK2 for the treatment of cancer. Mol. Cell. Biochem. 2011, 356, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Pierre, F.; Chua, P.C.; O’Brien, S.E.; Siddiqui-Jain, A.; Bourbon, P.; Haddach, M.; Michaux, J.; Nagasawa, J.; Schwaebe, M.K.; Stefan, E.; et al. Discovery and SAR of 5-(3-chlorophenylamino)benzo[c][2,6]naphthyridine-8-carboxylic Acid (CX-4945), the First Clinical Stage Inhibitor of Protein Kinase CK2 for the Treatment of Cancer. J. Med. Chem. 2011, 54, 635–654. [Google Scholar] [CrossRef] [PubMed]
- Chojnacki, K.; Lindenblatt, D.; Wińska, P.; Wielechowska, M.; Toelzer, C.; Niefind, K.; Bretner, M. Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase. Bioorganic Chem. 2021, 106, 104502. [Google Scholar] [CrossRef]
- Bestgen, B.; Krimm, I.; Kufareva, I.; Kamal, A.A.M.; Seetoh, W.G.; Abell, C.; Hartmann, R.W.; Abagyan, R.; Cochet, C.; Le Borgne, M.; et al. 2-Aminothiazole Derivatives as Selective Allosteric Modulators of the Protein Kinase CK2. 1. Identification of an Allosteric Binding Site. J. Med. Chem. 2019, 62, 1803–1816. [Google Scholar] [CrossRef] [PubMed]
- Bestgen, B.; Kufareva, I.; Seetoh, W.; Abell, C.; Hartmann, R.W.; Abagyan, R.; Le Borgne, M.; Filhol, O.; Cochet, C.; Lomberget, T.; et al. 2-Aminothiazole Derivatives as Selective Allosteric Modulators of the Protein Kinase CK2. 2. Structure-Based Optimization and Investigation of Effects Specific to the Allosteric Mode of Action. J. Med. Chem. 2019, 62, 1817–1836. [Google Scholar] [CrossRef]
- Lindenblatt, D.; Nickelsen, A.; Applegate, V.M.; Jose, J.; Niefind, K. Structural and Mechanistic Basis of the Inhibitory Potency of Selected 2-Aminothiazole Compounds on Protein Kinase CK2. J. Med. Chem. 2020, 63, 7766–7772. [Google Scholar] [CrossRef]
- Brear, P.; De Fusco, C.; Georgiou, K.H.; Francis-Newton, N.J.; Stubbs, C.J.; Sore, H.F.; Venkitaraman, A.R.; Abell, C.; Spring, D.R.; Hyvönen, M. Specific inhibition of CK2α from an anchor outside the active site. Chem. Sci. 2016, 7, 6839–6845. [Google Scholar] [CrossRef] [PubMed]
- De Fusco, C.; Brear, P.; Iegre, J.; Georgiou, K.H.; Sore, H.F.; Hyvönen, M.; Spring, D.R. A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066. Bioorganic Med. Chem. 2017, 25, 3471–3482. [Google Scholar] [CrossRef]
- Lindenblatt, D.; Applegate, V.; Nickelsen, A.; Klußmann, M.; Neundorf, I.; Götz, C.; Jose, J.; Niefind, K. Molecular Plasticity of Crystalline CK2α’ Leads to KN2, a Bivalent Inhibitor of Protein Kinase CK2 with Extraordinary Selectivity. J. Med. Chem. 2022, 65, 1302–1312. [Google Scholar] [CrossRef]
- Wells, C.I.; Drewry, D.H.; Pickett, J.E.; Tjaden, A.; Krämer, A.; Müller, S.; Gyenis, L.; Menyhart, D.; Litchfield, D.W.; Knapp, S.; et al. Development of a potent and selective chemical probe for the pleiotropic kinase CK2. Cell Chem. Biol. 2021, 28, 546–558.e10. [Google Scholar] [CrossRef] [PubMed]
- Davis-Gilbert, Z.W.; Krämer, A.; Dunford, J.E.; Howell, S.; Senbabaoglu, F.; Wells, C.I.; Bashore, F.M.; Havener, T.M.; Smith, J.L.; Hossain, M.A.; et al. Discovery of a Potent and Selective Naphthyridine-Based Chemical Probe for Casein Kinase 2. ACS Med. Chem. Lett. 2023, 14, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Pack, M.; Götz, C.; Wrublewsky, S.; Montenarh, M. SGC-CK2-1 Is an Efficient Inducer of Insulin Production and Secretion in Pancreatic β-Cells. Pharmaceutics 2021, 14, 19. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Kinoshita, C.; Axtman, A.D.; Young, J.E. Evaluation of a Selective Chemical Probe Validates That CK2 Mediates Neuroinflammation in a Human Induced Pluripotent Stem Cell-Derived Mircroglial Model. Front. Mol. Neurosci. 2022, 15, 824956. [Google Scholar] [CrossRef] [PubMed]
- Boewe, A.S.; Wemmert, S.; Kulas, P.; Schick, B.; Götz, C.; Wrublewsky, S.; Montenarh, M.; Menger, M.D.; Laschke, M.W.; Ampofo, E. Inhibition of CK2 Reduces NG2 Expression in Juvenile Angiofibroma. Biomedicines 2022, 10, 966. [Google Scholar] [CrossRef] [PubMed]
- PyMOL. The PyMOL Molecular Graphics System, version 1.7; Schrödinger, LLC: New York, NY, USA, 2013. [Google Scholar]
- Battistutta, R.; De Moliner, E.; Sarno, S.; Zanotti, G.; Pinna, L.A. Structural features underlying selective inhibition of protein kinase CK2 by ATP site-directed tetrabromo-2-benzotriazole. Protein Sci. 2001, 10, 2200–2206. [Google Scholar] [CrossRef]
- Winiewska-Szajewska, M.; Czapinska, H.; Kaus-Drobek, M.; Fricke, A.; Mieczkowska, K.; Dadlez, M.; Bochtler, M.; Poznański, J. Competition between electrostatic interactions and halogen bonding in the protein-ligand system: Structural and thermodynamic studies of 5,6-dibromobenzotriazole-hCK2α complexes. Sci. Rep. 2022, 12, 18964. [Google Scholar] [CrossRef]
- Tickle, I.J.; Flensburg, C.; Keller, P.; Paciorek, W.; Sharff, A.; Vonrhein, C.; Bricogne, G. STARANISO; Global Phasing Ltd.: Cambridge, UK, 2018. [Google Scholar]
- Janeczko, M.; Orzeszko, A.; Kazimierczuk, Z.; Szyszka, R.; Baier, A. CK2α and CK2α’ subunits differ in their sensitivity to 4,5,6,7-tetrabromo- and 4,5,6,7-tetraiodo-1H-benzimidazole derivatives. Eur. J. Med. Chem. 2012, 47, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, A.; Tsuyuguchi, M.; Kitagawa, D.; Sawa, M.; Nakamura, S.; Nakanishi, I.; Kinoshita, T. Bivalent binding mode of an amino-pyrazole inhibitor indicates the potentials for CK2α1-selective inhibitors. Biochem. Biophys. Res. Commun. 2022, 630, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Cimermancic, P.; Weinkam, P.; Rettenmaier, T.J.; Bichmann, L.; Keedy, D.A.; Woldeyes, R.A.; Schneidman-Duhovny, D.; Demerdash, O.N.; Mitchell, J.C.; Wells, J.A.; et al. CryptoSite: Expanding the Druggable Proteome by Characterization and Prediction of Cryptic Binding Sites. J. Mol. Biol. 2016, 428, 709–719. [Google Scholar] [CrossRef]
- Pietsch, M.; Viht, K.; Schnitzler, A.; Ekambaram, R.; Steinkrüger, M.; Enkvist, E.; Nienberg, C.; Nickelsen, A.; Lauwers, M.; Jose, J.; et al. Unexpected CK2β-antagonistic functionality of bisubstrate inhibitors targeting protein kinase CK2. Bioorganic Chem. 2020, 96, 103608. [Google Scholar] [CrossRef] [PubMed]
- Czapinska, H.; Winiewska-Szajewska, M.; Szymaniec-Rutkowska, A.; Piasecka, A.; Bochtler, M.; Poznański, J. Halogen Atoms in the Protein-Ligand System. Structural and Thermodynamic Studies of the Binding of Bromobenzotriazoles by the Catalytic Subunit of Human Protein Kinase CK2. J. Phys. Chem. B 2021, 125, 2491–2503. [Google Scholar] [CrossRef] [PubMed]
- Wąsik, R.; Wińska, P.; Poznański, J.; Shugar, D. Synthesis and physico-chemical properties in aqueous medium of all possible isomeric bromo analogues of benzo-1H-triazole, potential inhibitors of protein kinases. J. Phys. Chem. B 2012, 116, 7259–7268. [Google Scholar] [CrossRef] [PubMed]
- Battistutta, R.; Mazzorana, M.; Cendron, L.; Bortolato, A.; Sarno, S.; Kazimierczuk, Z.; Zanotti, G.; Moro, S.; Pinna, L.A. The ATP-binding site of protein kinase CK2 holds a positive electrostatic area and conserved water molecules. Chembiochem 2007, 8, 1804–1809. [Google Scholar] [CrossRef]
- Winiewska-Szajewska, M.; Maciejewska, A.M.; Speina, E.; Poznański, J.; Paprocki, D. Synthesis of Novel Halogenated Heterocycles Based on o-Phenylenediamine and Their Interactions with the Catalytic Subunit of Protein Kinase CK2. Molecules 2021, 26, 3163. [Google Scholar] [CrossRef]
- Laudet, B.; Barette, C.; Dulery, V.; Renaudet, O.; Dumy, P.; Metz, A.; Prudent, R.; Deshiere, A.; Dideberg, O.; Filhol, O.; et al. Structure-based design of small peptide inhibitors of protein kinase CK2 subunit interaction. Biochem. J. 2007, 408, 363–373. [Google Scholar] [CrossRef]
- Laudet, B.; Moucadel, V.; Prudent, R.; Filhol, O.; Wong, Y.S.; Royer, D.; Cochet, C. Identification of chemical inhibitors of protein-kinase CK2 subunit interaction. Mol. Cell. Biochem. 2008, 316, 63–69. [Google Scholar] [CrossRef]
- Brear, P.; North, A.; Iegre, J.; Hadje Georgiou, K.; Lubin, A.; Carro, L.; Green, W.; Sore, H.F.; Hyvönen, M.; Spring, D.R. Novel non-ATP competitive small molecules targeting the CK2 α/β interface. Bioorganic Med. Chem. 2018, 26, 3016–3020. [Google Scholar] [CrossRef]
- Kufareva, I.; Bestgen, B.; Brear, P.; Prudent, R.; Laudet, B.; Moucadel, V.; Ettaoussi, M.; Sautel, C.F.; Krimm, I.; Engel, M.; et al. Discovery of holoenzyme-disrupting chemicals as substrate-selective CK2 inhibitors. Sci. Rep. 2019, 9, 15893. [Google Scholar] [CrossRef]
- Lindenblatt, D.; Horn, M.; Götz, C.; Niefind, K.; Neundorf, I.; Pietsch, M. Design of CK2β-Mimicking Peptides as Tools To Study the CK2α/CK2β Interaction in Cancer Cells. ChemMedChem 2019, 14, 833–841. [Google Scholar] [CrossRef]
- Tang, S.; Zhang, N.; Zhou, Y.; Cortopassi, W.A.; Jacobson, M.P.; Zhao, L.J.; Zhong, R.G. Structure-based Discovery of Novel CK2α-Binding Cyclic Peptides with Anti-cancer Activity. Mol. Infor. 2019, 38, e1800089. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, E.L.; Iegre, J.; D’Amore, C.; Brear, P.; Salvi, M.; Hyvönen, M.; Spring, D.R. Development of small cyclic peptides targeting the CK2α/β interface. Chem. Commun. 2022, 58, 4791–4794. [Google Scholar] [CrossRef] [PubMed]
- Rodnight, R.; Lavin, B.E. Phosvitin kinase from brain: Activation by ions and subcellular distribution. Biochem. J. 1964, 93, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Hathaway, G.M.; Traugh, J.A. Cyclic nucleotide-independent protein kinases from rabbit reticulocytes. Purification of casein kinases. J. Biol. Chem. 1979, 254, 762–768. [Google Scholar] [CrossRef] [PubMed]
- Becher, I.; Savitski, M.M.; Savitski, M.F.; Hopf, C.; Bantscheff, M.; Drewes, G. Affinity profiling of the cellular kinome for the nucleotide cofactors ATP, ADP, and GTP. ACS Chem. Biol. 2013, 8, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Niefind, K.; Pütter, M.; Guerra, B.; Issinger, O.G.; Schomburg, D. GTP plus water mimic ATP in the active site of protein kinase CK2. Nat. Struct. Biol. 1999, 6, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Rahnel, H.; Viht, K.; Lavogina, D.; Mazina, O.; Haljasorg, T.; Enkvist, E.; Uri, A. A Selective Biligand Inhibitor of CK2 Increases Caspase-3 Activity in Cancer Cells and Inhibits Platelet Aggregation. ChemMedChem 2017, 12, 1723–1736. [Google Scholar] [CrossRef]
- Vonrhein, C.; Flensburg, C.; Keller, P.; Sharff, A.; Smart, O.; Paciorek, W.; Womack, T.; Bricogne, G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Struct. Biol. 2011, 67, 293–302. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. D Struct. Biol. 2010, 66, 125–132. [Google Scholar] [CrossRef]
- Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D Struct. Biol. 2006, 62, 72–82. [Google Scholar] [CrossRef]
- Evans, P.R.; Murshudov, G.N. How good are my data and what is the resolution? Acta Crystallogr. D Struct. Biol. 2013, 69, 1204–1214. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Struct. Biol. 2011, 67, 235–242. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Struct. Biol. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Struct. Biol. 2012, 68, 352–367. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Struct. Biol. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Moriarty, N.W.; Grosse-Kunstleve, R.W.; Adams, P.D. Electronic Ligand Builder and Optimization Workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Crystallogr. D Struct. Biol. 2009, 65, 1074–1080. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).