Abstract
Prostate cancer (PCa) is one of the most common malignancies in men, where early and accurate detection is crucial. While PSA testing has been the diagnostic standard, its limited specificity leads to unnecessary biopsies and missed significant cancers. Urinary biomarkers such as PCA3 and TMPRSS2-ERG and multi-marker assays (MyProstateScore, SelectMDx, and ExoDx) offer a promising alternative. This narrative review examines their diagnostic performance and clinical utility with the aim of understanding whether they can be integrated with the established tests and exams already in use. A literature search of PubMed, Scopus, and Medline identified some relevant recent studies (2010–2025). The findings show that PCA3 and TMPRSS2-ERG improve specificity over PSA, while multi-marker tests enhance risk stratification and reduce unnecessary procedures. MPS integrates urinary biomarkers with PSA, achieving over 95% sensitivity and negative predictive value for clinically significant cancers. SelectMDx demonstrates ~90% negative predictive value, and ExoDx assesses urinary exosomes to predict aggressive disease. Despite their advantages, challenges persist, including variability in performance, cost, and accessibility. Urinary biomarkers represent a major step toward more precise, less invasive diagnostics, with future research needed to optimize clinical integration and cost-effectiveness.
Keywords:
prostate cancer; urinary biomarkers; PSA; PCA3; TMPRSS2-ERG; MyProstateScore; SelectMDx; ExoDx; non-invasive diagnostics 1. Introduction
Prostate cancer (PCa) is among the most frequent cancers in men, where early detection plays a crucial role in improving survival rates and guiding treatment decisions. The current gold standard for PCa screening is prostate-specific antigen (PSA) testing, widely used for decades. However, PSA testing has significant limitations, primarily its lack of specificity [1]. Elevated PSA levels can result from benign prostatic hyperplasia (BPH) or prostatitis, leading to false positives. Furthermore, many patients with elevated PSA levels do not have PCa, while some with normal PSA levels may harbor clinically significant disease. These limitations underscore the urgent need for more accurate diagnostic tools to reduce unnecessary biopsies and better identify high-risk patients [1].
In recent years, urinary biomarkers have emerged as a promising alternative to PSA for PCa detection. These biomarkers, which are typically non-invasive, provide more specific information about the presence and aggressiveness of PCa. They improve diagnostic accuracy and guide treatment decisions while reducing unnecessary procedures such as prostate biopsies. Two key biomarkers, Prostate Cancer Gene 3 (PCA3) and Transmembrane protease serine 2 (TMPRSS2-ERG), have been extensively studied [2].
PCA3 is a long non-coding RNA overexpressed in PCa cells, making it an ideal candidate for PCa detection. Urine-based analysis of PCA3 offers valuable insights, particularly for men with prior negative biopsies and elevated PSA levels. Similarly, TMPRSS2-ERG, a gene fusion frequently found in PCa cells, serves as a complementary biomarker to PCA3 in enhancing diagnostic accuracy.
Beyond individual biomarkers, multi-marker assays have been developed to further refine PCa diagnostics. This narrative review examines three widely used tests: the ExoDx Prostate (EPI) test, the SelectMDx test, and the MyProstateScore (MPS) test.
The ExoDx Prostate test evaluates PCA3, the ERG gene (TMPRSS2-ERG), and the SPEDF gene (SAM Pointed Domain Containing ETS Transcription Factor) in urinary exosomes to estimate the risk of aggressive PCa. The SelectMDx test measures DLX1 and HOXC6 mRNA levels in urine collected post-prostate massage, helping to identify high-risk PCa in patients being considered for biopsy. The MyProstateScore test integrates PCA3 and TMPRSS2-ERG with serum PSA levels to provide a comprehensive assessment of high-grade tumors, reducing unnecessary biopsies and enhancing patient management.
These urinary biomarkers and their associated tests represent a significant advance in PCa diagnostics, offering more personalized and accurate assessments while minimizing invasive procedures like biopsies [3].
As far as more recent studies have demonstrated, the incorporation of various bio-markers and diagnostic tools, such as blood-based markers and urine-based markers, offers a more comprehensive assessment of the PCa disease [4,5].
The United States Food and Drug Administration (FDA) and Clinical Laboratory Improvement Amendments (CLIA) have recognized tests such as the PCA3, MPS, SelectMDx, and EPI tests for clinical use, which are also recommended by the NCCN and American Urological Association/Society of Urologic Oncology guidelines [5].
In this narrative review, we explore the diagnostic potential, clinical utility, and limitations of urinary biomarkers such as PCA3 and TMPRSS2-ERG and their associated tests. We will also examine their potential to complement or replace traditional PSA testing, contributing to a more personalized and less invasive approach to managing PCa.
1.1. Prostate Cancer Gene 3 (PCA3)
PCA3, a long non-coding RNA identified in 1999, is highly overexpressed in PCa tissues and encoded by the DD3 gene on chromosome 9 [6]. Its high specificity for PCa, with negligible expression in normal prostate tissues, makes it an invaluable biomarker for distinguishing malignant from benign conditions [7]. Unlike PSA, PCA3 levels are unaffected by benign prostatic hyperplasia (BPH) or prostatitis, which enhances its diagnostic reliability. PCA3 is particularly useful in patients with elevated PSA levels or prior negative biopsies, where PSA often fails to reliably indicate the presence of malignancy [8,9]. PCA3 testing is performed on urine samples collected after a digital rectal exam (DRE). High PCA3 scores correlate with an increased risk of PCa, while low scores reduce the likelihood of malignancy, sparing patients unnecessary biopsies and their associated complications [10].
In a meta-analysis involving 46 clinical trials and including 12,295 subjects, Yong Cui et al. analyzed the diagnostic performance of PCA3 [11]. The essential selection criteria were a “case-control or cohort design”, “diagnostic test using PCA3 itself or in combination with other biomarkers”, and “prostate biopsy as the gold standard”. The test demonstrated moderate accuracy, with the sensitivity at 65% and specificity at 73% (AUC = 0.75). PCA3 was particularly beneficial for guiding initial biopsies, with a recommended threshold of 35 to balance sensitivity and specificity.
Jihua et al. conducted an experimental study (2022) involving a total of 63 PCa patients and 61 healthy individuals in the validation cohort [12]. They collected urine samples from January to June 2019 to investigate the diagnostic potential of PCA3 combined with other markers, such as PSMA. The inclusion criteria specified that patients had undergone prostate needle biopsy and were diagnosed with PCa based on histological detection. When combined with additional biomarkers, PCA3 achieved an AUC of 0.870, demonstrating strong potential for differentiating PCa patients from healthy individuals. Furthermore, PCA3 levels in urinary exosomes correlated with a longer time to failure of androgen-deprivation therapy (ADT), particularly in patients with castration-resistant prostate cancer (CRPC).
In a retrospective study, Yulian et al. (2023) validated PCA3 as a significant urinary marker for PCa detection [13]. The study included 240 participants: 150 patients with confirmed PCa, 30 patients with benign prostatic hyperplasia (BPH), 30 patients with active chronic prostatitis, and 30 healthy volunteers. PCA3 scores were significantly higher in PCa patients compared to those with benign prostatic hyperplasia or chronic prostatitis (p < 0.001). Additionally, PCA3 scores strongly correlated with tumor aggressiveness, with higher values observed in clinically significant prostate cancer (csPCa), which means ISUP grades of 2–5. The ROC analysis demonstrated that a PCA3 threshold of 50 provided a sensitivity and specificity of 93% for identifying csPCa (AUC = 0.966). These findings reinforce PCA3’s role in predicting high-grade malignancies and guiding biopsy decisions in high-risk cases.
Among the tests available on market, the FDA-approved Progensa PCA3 Test was designed to assist in determining the need for repeat biopsies in men aged ≥50 with prior negative biopsy results [3]. The assay, which detects PCA3 and PSA mRNA in urine, has shown moderate diagnostic performance (sensitivity 0.65 and specificity 0.73). The optimal PCA3 score threshold remains debated, with 35 commonly used for balancing sensitivity and specificity. Despite its promise, its challenges include threshold variability and mRNA instability, underscoring the importance of precise sample handling.
A recent development (2024) is the study by Dou et al., in which they introduced a novel method for PCA3 detection [14]. This innovative approach utilizes urease-powered nanomotor probes, enabling ultrasensitive, non-invasive analysis of urine samples with a detection limit of 0.6 pM and a rapid detection time of just 11 min. Additionally, the probes allow real-time imaging of PCa cells, providing a precise and efficient platform for early PCa diagnosis and improving upon conventional methods (see Table 1).

Table 1.
Comparison of the main studies about PCA3.
Ongoing research aims to validate PCA3’s role in PCa diagnostics and enhance its utility by integrating it with other molecular markers or imaging techniques, potentially improving detection and prognosis. Liquid biopsy methods for detecting biomarkers in urine or blood represent a promising frontier in this effort. However, PCA3 testing faces limitations, such as variable sensitivity and specificity, and its expression is not consistently elevated in all PCas, which restricts its effectiveness in detecting early-stage or indolent tumors [15]. Additionally, the accuracy of PCA3 testing depends on obtaining adequate urine samples post-DRE, which can introduce variability. As such, PCA3 is best used in combination with other diagnostic tools to guide tailored treatment plans within a broader clinical context [16].
1.2. TMPRSS2-ERG
Tomlins et al. [17] were the first to describe the PCa fusion gene TMPRSS2-ERG, which involves the gene for the prostate-specific, androgen-responsive transmembrane serine protease TMPRSS2 and the ERG transcription factor from the Ets family. This genetic aberration represents an early molecular event in prostate carcinogenesis, suggesting its role in tumor initiation. The overexpression of ERG, driven by this fusion, disrupts pathways involved in cell proliferation, differentiation, and apoptosis.
Since its discovery in 2005, TMPRSS2-ERG has been extensively studied as a biomarker for PCa diagnosis, prognosis, and treatment decision-making [18].
Although the TMPRSS2-ERG fusion has been reported in approximately 40–70% of PCa cases in Western populations, it shows considerable ethnic and geographical variability [19]. Studies indicate that this particular fusion is notably less prevalent in Asian cohorts [20]. For instance, a Chinese study found a prevalence of approximately 16% in PCa tissues, while a systematic review of Asian data reported an overall positive rate of 27% [20,21]. These differences may be attributed to genetic, lifestyle, and environmental factors.
TMPRSS2-ERG can be detected in tissue biopsies and urine samples post-digital rectal examination (DRE), either by reverse transcription polymerase chain reaction (RT-PCR), fluorescence in situ hybridization (FISH), or immunohistochemistry (IHC).
Non-invasive approaches, particularly urinary TMPRSS2-ERG testing combined with PCA3, have shown improved diagnostic accuracy for the identification of csPCa. Some studies suggest that urinary TMPRSS2-ERG combined with PCA3 has a higher sensitivity and specificity, surpassing serum PSA testing alone [22].
Despite this, the prognostic value of TMPRSS2-ERG remains controversial. This genetic alteration has also been related to tumor size, aggressive PCa phenotypes, higher Gleason score, Gleason score upgrade upon prostatectomy, and increased progression risk, in addition to predicting malignancy at prostatic biopsy [23].
Font-Tello et al. observed that high TMPRSS2-ERG expression correlates with Gleason scores ≥ 8 and stage T3–T4 tumors, emphasizing that its expression levels, not just presence, are key indicators of aggressiveness [24]. However, other studies report no significant associations between TMPRSS2-ERG and clinical outcomes in patients treated by radical prostatectomy [25].
TMPRSS2-ERG can be incorporated into diagnostic algorithms to stratify high-risk PCa patients in the very early stages. Its detection in urine samples provides a non-invasive tool for early diagnosis and active surveillance [26]. Despite that, TMPRSS2-ERG has high variability between different populations and it may have less effectiveness as a diagnostic/prognostic biomarker for PCa for some ethnicities (like Asians) because of its low prevalence and insignificant correlation with clinical parameters.
Quantitative assessments of TMPRSS2-ERG mRNA or protein expression levels may be useful as biomarkers for diagnosis of aggressive disease subtypes. However, the prognostic relevance of TMPRSS2-ERG remains debated. Some studies report no clear association with survival outcomes, suggesting that the pure presence of this gene aberration may be less critical than its quantitative expression levels or co-occurrence with other genetic alterations [24,27].
1.3. MyProstateScore (MPS)
The MyProstateScore (MPS) is a urinary biomarkers test designed to refine PCa diagnostics by detecting csPCa. By integrating serum PSA levels with urinary PCA3 and TMPRSS2:ERG, MPS offers an advanced risk stratification tool to guide biopsy decisions.
In a prospective study, Tosoian et al. (2022) validated the MyProstateScore (MPS) test in a cohort of 1525 biopsy-naïve men [28]. The study tested MPS in several cohorts and established that a MPS threshold of ≤10 provided 97% sensitivity and 98% negative predictive value (NPV) for detecting csPCa, reducing unnecessary biopsies by 33% while missing only 3% of significant cancers. This threshold was also effective in guideline-concordant cases (PSA 3–10 ng/mL or PSA < 3 with suspicious DRE), achieving 96% sensitivity and 97% NPV, thereby minimizing invasive procedures while maintaining diagnostic accuracy.
Further insights were provided in a prospective study by Tosoian et al. [29] in patients with PI-RADS 3 lesions, where it outperformed PSA density (PSAD) in predicting Grade Group ≥ 2 cancers (AUC: 0.73 vs. 0.62). Urinary specimens were collected prior to biopsy at the University of Michigan from 2008. The study included 540 men (2015–2019) with mpMRI and MPS available. The prevalence of GG ≥ 2 cancer was 13% for PI-RADS 3, 56% for PI-RADS 4, and 87% for PI-RADS 5. MPS was significantly higher in men with GG ≥ 2 cancer compared to those with a negative or GG1 biopsy (p < 0.001). In PI-RADS 3 patients (n = 121), the AUC for predicting GG ≥ 2 cancer was 0.55 for PSA, 0.62 for PSAD, and 0.73 for MPS. MPS provided the highest net clinical benefit, highlighting its value as an adjunct to mpMRI, particularly in equivocal cases.
A prospective cohort validation study by Tosoian et al. [30] also demonstrated the utility of dual MyProstateScore (MPS) thresholds in men with prior negative biopsies. A lower MPS ≤ 15 ruled out clinically significant GG ≥ 2 cancer with 100% sensitivity and NPV, preventing 23% of unnecessary biopsies, while MPS > 40 identified high-risk patients, avoiding 67% of repeat biopsies. The primary cohort included 422 men with previous negative biopsies, and the external validation cohort included 268 men scheduled for repeat biopsies. MPS values of 15 and 40 were clinically actionable, aiding decision-making on repeat biopsies.
The test can be also used in biopsy-naïve patients, and it can drive clinical decisions. Amir H. Lebastchi et al. [31] examined MPS in 248 men referred for biopsy. MPS significantly influenced biopsy decisions, with a 46% reduction in procedures, and demonstrated higher predictive accuracy than the Prostate Cancer Prevention Trial Risk Calculator (PCPTrc). The test maintained its predictive strength in mpMRI-integrated settings, reinforcing its role as a first-line, non-invasive tool for biopsy decision-making.
Collectively, these studies establish MPS as a pivotal advance in PCa diagnostics, effectively reducing unnecessary biopsies while preserving the detection of csPCa (Table 2). Future research will refine its clinical integration and explore its synergy with emerging diagnostic modalities.

Table 2.
Comparison of the main studies about MPS.
1.4. SelectMDx
SelectMDx is a non-invasive urine-based test that offers a promising alternative by integrating molecular and clinical data to identify csPCa while reducing unnecessary biopsies. The SelectMDx test measures urinary levels of two PCa genomic biomarkers (DLX1 and HOXC6 mRNA), combined with clinical parameters such as age, PSA levels, DRE, family history of PCa, and prostate volume.
Meta-analyses prove that the SelectMDx test has high sensitivity (82%) but moderate specificity (56%) in detecting csPCa [32,33]. The negative predictive value (NPV) of SelectMDx is about 90% [30] and ensures that a negative test result reliably excludes csPCa, potentially reducing the need for invasive prostate biopsies by up to 42%, particularly in low-risk patients [34].
SelectMDx proves valuable in several key clinical areas. In pre-biopsy risk stratification, the integration of genetic markers, such as HOXC6 and DLX1 mRNA, with clinical data enables SelectMDx to identify patients who would benefit most from biopsy. This helps to spare low-risk individuals from unnecessary procedures, reducing the risk of overdiagnosis of indolent PCa [32]. In the context of active surveillance, SelectMDx can assist in avoiding aggressive interventions in selected patients, favoring non-invasive monitoring strategies instead [16]. Furthermore, when integrating imaging data, combining SelectMDx with multiparametric MRI enhances diagnostic accuracy, particularly for patients with intermediate PSA levels (3–10 ng/mL) [35]. This combined approach can reduce the need for unnecessary procedures, thus improving patient management.
Additionally, SelectMDx represents a non-invasive solution for improving PCa diagnostics. In a meta-analysis (2022), Hanting Wu et al. identified 14 studies (involving a total of 2579 patients) and demonstrated how this test has significant diagnostic value for the detection of high-grade PCa [32]. Its high sensitivity and high NPM make it a valuable tool in identifying csPCa, reducing unnecessary biopsies, and supporting the diagnostic pathway in repeat or initial biopsy settings and active surveillance [33].
Several limitations must be considered when evaluating SelectMDx. One issue is its variable performance between different populations. Test results may vary due to ethnic and demographic factors, which means that population-specific validation is necessary for accurate results across diverse groups. Another limitation is related to its cost and availability. The high cost of testing and its limited availability in certain regions pose significant barriers to widespread adoption. A recent systematic review and diagnostic meta-analysis highlighted that one of the key aspects of an ideal biomarker is its cost-effectiveness. Currently, SelectMDx costs around USD 500 when purchased directly from pharmaceutical companies in the USA [33]. Furthermore, the test’s clinical data association can be a limitation, as its effectiveness is maximized when integrated with imaging or other diagnostic tools. However, this may not always be feasible in resource-limited settings where access to such technologies is restricted.
1.5. ExoDx Prostate Intelliscore (EPI) Test
The ExoDx Prostate Intelliscore (EPI) test (Exosome Diagnostics, Waltham, MA, USA) [36] is a noninvasive liquid biopsy that quantifies three RNA targets in first-catch urine exosomes: PCA3, ERG, and SPDEF. It provides a risk score predicting whether a patient presenting for their first biopsy with an equivocal PSA from 2 to 10 ng/mL is likely to have GG2 or greater (HGPCa). Unlike traditional approaches, the EPI algorithm is independent of clinical variables such as DRE findings, which are often part of biopsy decision-making [37]. The EPI test has been included in the National Comprehensive Cancer Network guidelines for prostate cancer early detection since 2019.
In a prospective, blinded, randomized clinical utility study, Tutrone et al. (2023) included 1049 men aged ≥50 years with PSA levels between 2 and 10 ng/mL, and compared outcomes between patients whose biopsy decisions were guided by EPI results and those managed with standard-of-care (SOC) protocols [37]. A validated cutoff score of 15.6 was used to stratify patients into low or high risk for HGPC. The EPI-guided group showed a notable increase in HGPC detection rates compared to the SOC group. Importantly, patients classified as low risk by the EPI test deferred biopsies and maintained a very low risk of HGPC progression over a 2.5-year follow-up, highlighting the test’s role in avoiding unnecessary biopsies while maintaining diagnostic accuracy.
A meta-analysis combining data from three independent prospective validation studies evaluated the EPI test’s performance in detecting HGPC. This pooled analysis included 1212 men aged ≥50 years with PSA levels between 2 and 10 ng/mL who were undergoing their first biopsy. The EPI test demonstrated an AUC of 0.70, significantly outperforming PSA alone (AUC: 0.56) and other risk calculators such as the Prostate Cancer Prevention Trial Risk Calculator (AUC: 0.62) and the European Randomized Study of Screening for Prostate Cancer Risk Calculator (AUC: 0.59). Using a cutoff score of 15.6, the EPI test achieved a NPV of 90%, avoiding 30% of unnecessary (true-negative) biopsies while maintaining sensitivity for ≥GG2 disease (92.3%) and ≥GG3 disease (92.9%) [38].
Additional research further underscores the test’s clinical relevance. In a cohort of over 2000 men, the EPI score surpassed traditional multivariate risk calculators in predicting HGPC. Among the 310 men who proceeded to radical prostatectomy, 111 had Grade Group 1 cancers at biopsy and could have been candidates for active surveillance, suggesting the test’s potential in guiding treatment decisions [39].
These outcomes underscore the EPI test’s effectiveness in reducing unnecessary biopsies and their associated morbidities while maintaining robust detection rates for clinically significant prostate cancer. Despite these advantages, limitations exist. The EPI test’s predictive accuracy, while superior to some existing methods, is not absolute, and false negatives can occur. Additionally, accessibility and cost considerations may limit widespread adoption. The test is priced at approximately USD 800, which could pose a barrier for some patients or healthcare systems depending on insurance coverage and regional healthcare policies. Future perspectives include integrating the EPI test with other diagnostic modalities, such as multiparametric MRI, to enhance predictive accuracy. Ongoing studies aim to validate its utility in diverse populations and explore its role in monitoring disease progression, potentially broadening its application in PCa management.
2. Discussion
PCa diagnosis has long relied on PSA testing, which, despite its accessibility and low cost, has well-documented limitations, particularly its low specificity and inability to differentiate between benign and malignant conditions. The emergence of molecular diagnostics has introduced urinary biomarkers as promising tools to enhance detection and risk stratification. Among these, PCA3, TMPRSS2-ERG, and multiparametric tests such as MyProstateScore (MPS), SelectMDx, and ExoDx (EPI) have gained attention for their potential to refine diagnostic pathways and reduce unnecessary biopsies (Figure 1).
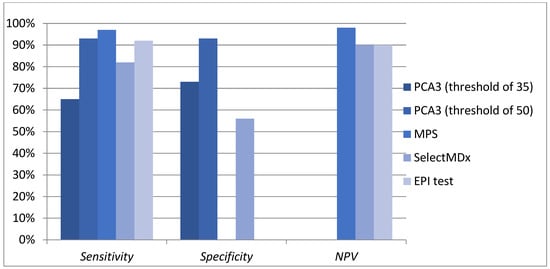
Figure 1.
Diagnostic performance of urinary markers and tests.
PCA3 offers greater specificity than PSA, making it particularly useful in men with prior negative biopsies or ambiguous PSA levels. TMPRSS2-ERG, despite its geographic and ethnic variability, provides additional diagnostic and prognostic insights when used alongside PCA3. Advanced biomarker panels like MyProstateScore and SelectMDx integrate multiple molecular and clinical parameters to improve the detection of csPCa, while ExoDx (EPI) offers a non-invasive approach to first-line risk stratification, helping to reduce unnecessary biopsies without compromising diagnostic accuracy (Table 3).

Table 3.
Summary of main features of urinary markers and tests.
However, several challenges hinder the widespread adoption of these biomarkers. Their performance varies across different populations due to genetic, demographic, and environmental factors, highlighting the need for population-specific validation. Cost and accessibility remain significant barriers, particularly in resource-limited settings, and their integration into clinical practice requires alignment with imaging modalities like multiparametric MRI to optimize diagnostic accuracy.
Despite the increasing interest in urinary biomarkers, their inclusion in clinical guidelines remains limited. Organizations such as the European Association of Urology (EAU), the National Comprehensive Cancer Network (NCCN), and the American Urological Association (AUA) recognize their potential in risk stratification and biopsy decision-making, yet they remain adjunct tools rather than standard diagnostic methods. While PCA3 is FDA-approved for guiding repeat biopsy decisions, and SelectMDx and ExoDx contribute to HGPC risk assessment, further large-scale validation is needed before they can be fully integrated into routine practice.
Future efforts should focus on refining biomarker thresholds, enhancing cost-effectiveness, and validating their performance across diverse populations. Cost and accessibility remain significant barriers to widespread adoption, particularly in resource-limited settings. However, while the initial cost may be high, some studies suggest that early diagnosis can lead to long-term savings by reducing future expenses and improving quality-adjusted life years (QALYs) [40].
Combining urinary biomarkers with advanced imaging and genetic profiling could revolutionize prostate cancer management, leading to a more personalized, non-invasive, and efficient diagnostic approach. Addressing current limitations and fostering clinical adoption will be crucial to enhancing patient care, minimizing overdiagnosis and overtreatment, and ensuring timely detection of HGPC.
3. Conclusions
Despite PSA remaining the standard screening tool for PCa, its lack of specificity often leads to unnecessary invasive procedures and the risk of missing csPCa. To overcome these limits, several urinary biomarkers have been developed and validated, demonstrating improved diagnostic accuracy and risk stratification capabilities.
The PCA3 test has shown high specificity and predictive value, particularly in patients with elevated PSA and prior negative biopsies. It reduces the need for repeated invasive procedures by accurately identifying those at higher risk of significant malignancy.
TMPRSS2-ERG could be another promising biomarker, enhancing the specificity of PCa detection when combined with PCA3. This association provides a more targeted approach to diagnosis and has been related to more aggressive cancer phenotypes.
Multi-marker assays like MyProstateScore (MPS), SelectMDx, and EPI integrate multiple urinary biomarkers with clinical parameters to refine risk assessment. MPS combines PCA3, TMPRSS2-ERG, and PSA levels, achieving over 95% sensitivity and NPV for csPCa. Use of SelectMDx as a stand-alone test demonstrates approximately 90% NPV. The ExoDx test assesses urinary exosomes to predict the likelihood of aggressive disease, providing a non-invasive alternative with promising diagnostic performance (Table 3).
Despite these advances, this review notes ongoing challenges such as variability in biomarker performance, cost, and accessibility. Further research and clinical integration are essential to optimize these tools’ effectiveness and ensure broader application in routine PCa management.
In conclusion, urinary biomarkers represent a significant step toward more precise, less invasive PCa diagnostics. Their ability to reduce unnecessary biopsies, improve risk stratification, and identify high-risk patients offers promising help for more personalized and effective patient care.
Author Contributions
Conceptualization, A.P., G.L. and P.U.; methodology, C.G.C., A.P., S.S. and R.D.; validation, G.L. and P.U.; formal analysis, A.P.; investigation, C.G.C., A.P., S.S. and R.D.; data curation, C.G.C., S.S. and R.D.; writing—original draft preparation, C.G.C., S.S. and R.D.; writing—review and editing, C.G.C., A.P., S.S. and R.D.; supervision, A.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this review.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| MDPI | Multidisciplinary Digital Publishing Institute |
| DOAJ | Directory of open access journals |
| PSA | Prostate-specific antigen |
| PCa | Prostate cancer |
| csPCa | Clinically significant prostate cancer |
| HGPC | High-grade prostate cancer |
| NPV | Negative predictive value |
| AUC | Area under the curve |
| MPS | MyProstateScore |
| EPI | ExoDx Prostate Intelliscore |
References
- Merriel, S.W.D.; Pocock, L.; Gilbert, E.; Creavin, S.; Walter, F.M.; Spencer, A.; Hamilton, W. Systematic Review and Meta-Analysis of the Diagnostic Accuracy of Prostate-Specific Antigen (PSA) for the Detection of Prostate Cancer in Symptomatic Patients. BMC Med. 2022, 20, 54. [Google Scholar] [CrossRef]
- Salagierski, M.; Schalken, J.A. PCA3 and TMPRSS2-ERG: Promising Biomarkers in Prostate Cancer Diagnosis. Cancers 2010, 2, 1432–1440. [Google Scholar] [CrossRef]
- Chen, J.-Y.; Wang, P.-Y.; Liu, M.-Z.; Lyu, F.; Ma, M.-W.; Ren, X.-Y.; Gao, X.-S. Biomarkers for Prostate Cancer: From Diagnosis to Treatment. Diagnostics 2023, 13, 3350. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.H.-M.; Ko, I.C.-H.; Ng, C.F. Liquid Biomarkers in Prostate Cancer: Recent Advancements and Future Directions. Curr. Opin. Urol. 2025, 35, 3–12. [Google Scholar] [CrossRef]
- Liu, Y.; Hatano, K.; Nonomura, N. Liquid Biomarkers in Prostate Cancer Diagnosis: Current Status and Emerging Prospects. World J. Mens Health 2025, 43, 8. [Google Scholar] [CrossRef]
- Bussemakers, M.J.G.; van Bokhoven, A.; Verhaegh, G.W.; Smit, F.P.; Karthaus, H.F.M.; Schalken, J.A.; Debruyne, F.M.J.; Ru, N.; Isaacs, W.B. Adrie van Bokhoven DD3: A New Prostate-Specific Gene, Highly Overexpressed in Prostate Cancer. Cancer Res. 1999, 59, 5975–5979. [Google Scholar] [PubMed]
- Ferreira, L.B.; Palumbo, A.; De Mello, K.D.; Sternberg, C.; Caetano, M.S.; De Oliveira, F.L.; Neves, A.F.; Nasciutti, L.E.; Goulart, L.R.; Gimba, E.R.P. PCA3 Noncoding RNA Is Involved in the Control of Prostate-Cancer Cell Survival and Modulates Androgen Receptor Signaling. BMC Cancer 2012, 12, 507. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Gou, X.; Huang, P.; Mou, C. The PCA3 Test for Guiding Repeat Biopsy of Prostate Cancer and Its Cut-off Score: A Systematic Review and Meta-Analysis. Asian J. Androl. 2014, 16, 487. [Google Scholar] [CrossRef]
- Marks, L.S.; Bostwick, D.G. Prostate Cancer Specificity of PCA3 Gene Testing: Examples from Clinical Practice. Rev. Urol. 2008, 10, 175. [Google Scholar]
- Haese, A.; De La Taille, A.; Van Poppel, H.; Marberger, M.; Stenzl, A.; Mulders, P.F.A.; Huland, H.; Abbou, C.-C.; Remzi, M.; Tinzl, M.; et al. Clinical Utility of the PCA3 Urine Assay in European Men Scheduled for Repeat Biopsy. Eur. Urol. 2008, 54, 1081–1088. [Google Scholar] [CrossRef]
- Cui, Y.; Cao, W.; Li, Q. Evaluation of Prostate Cancer Antigen 3 for Detecting Prostate Cancer: A Systematic Review and Meta-Analysis. Nature 2016, 6, 25776. [Google Scholar] [CrossRef]
- Gan, J.; Zeng, X.; Wang, X.; Wu, Y.; Lei, P.; Wang, Z.; Yang, C.; Hu, Z. Effective Diagnosis of Prostate Cancer Based on mRNAs From Urinary Exosomes. Front. Med. 2022, 9, 736110. [Google Scholar] [CrossRef] [PubMed]
- Mytsyk, Y.; Nakonechnyi, Y.; Dosenko, V.; Kowal, P.; Pietrus, M.; Gazdikova, K.; Labudova, M.; Caprnda, M.; Prosecky, R.; Dragasek, J.; et al. The Performance and Limitations of PCA3, TMPRSS2:ERG, HOXC6 and DLX1 Urinary Markers Combined in the Improvement of Prostate Cancer Diagnostics. Clin. Biochem. 2023, 116, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Dou, P.; Liu, Q.; Chen, M.; Xu, W.; Zhou, H.; Zhang, X.; Jiang, C.; Zhang, Y.; Li, S.; Mao, L.; et al. Urease-Powered Nanomotor Probe for PCA3 Ultrasensitive Detection in Human Urine and Controllable Imaging in Live Cells. Sens. Actuators B Chem. 2025, 426, 137051. [Google Scholar] [CrossRef]
- Tomlins, S.A.; Day, J.R.; Lonigro, R.J.; Hovelson, D.H.; Siddiqui, J.; Kunju, L.P.; Dunn, R.L.; Meyer, S.; Hodge, P.; Groskopf, J.; et al. Urine TMPRSS2:ERG Plus PCA3 for Individualized Prostate Cancer Risk Assessment. Eur. Urol. 2016, 70, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Fiorella, D.; Marenco, J.L.; Mascarós, J.M.; Borque-Fernando, Á.; Esteban, L.M.; Calatrava, A.; Pastor, B.; López-Guerrero, J.A.; Rubio-Briones, J. Role of PCA3 and SelectMDx in the Optimization of Active Surveillance in Prostate Cancer. Actas Urológicas Españolas 2021, 45, 439–446. [Google Scholar] [CrossRef]
- Tomlins, S.A.; Rhodes, D.R.; Perner, S.; Dhanasekaran, S.M.; Mehra, R.; Sun, X.-W.; Varambally, S.; Cao, X.; Tchinda, J.; Kuefer, R. Recurrent Fusion of TMPRSS2 and ETS Transcription Factor Genes in Prostate Cancer. Science 1979, 310, 644–648. [Google Scholar] [CrossRef]
- Sanguedolce, F.; Cormio, A.; Brunelli, M.; D’Amuri, A.; Carrieri, G.; Bufo, P.; Cormio, L. Urine TMPRSS2: ERG Fusion Transcript as a Biomarker for Prostate Cancer: Literature Review. Clin. Genitourin. Cancer 2016, 14, 117–121. [Google Scholar] [CrossRef]
- Väänänen, R.-M.; Lilja, H.; Kauko, L.; Helo, P.; Kekki, H.; Cronin, A.M.; Vickers, A.J.; Nurmi, M.; Alanen, K.; Bjartell, A.; et al. Cancer-Associated Changes in the Expression of TMPRSS2-ERG, PCA3, and SPINK1 in Histologically Benign Tissue From Cancerous vs Noncancerous Prostatectomy Specimens. Urology 2014, 83, 511.e1–511.e7. [Google Scholar] [CrossRef]
- Kong, D.-P.; Chen, R.; Zhang, C.-L.; Zhang, W.; Xiao, G.-A.; Wang, F.-B.; Ta, N.; Gao, X.; Sun, Y.-H. Prevalence and Clinical Application of TMPRSS2-ERG Fusion in Asian Prostate Cancer Patients: A Large-Sample Study in Chinese People and a Systematic Review. Asian J. Androl. 2020, 22, 200–207. [Google Scholar] [CrossRef]
- Warli, S.M.; Warli, M.H.; Prapiska, F.F. PCA3 and TMPRSS2: ERG Urine Level as Diagnostic Biomarker of Prostate Cancer. Res. Rep. Urol. 2023, 15, 149–155. [Google Scholar] [CrossRef]
- Sanda, M.G.; Feng, Z.; Howard, D.H.; Tomlins, S.A.; Sokoll, L.J.; Chan, D.W.; Regan, M.M.; Groskopf, J.; Chipman, J.; Patil, D.H.; et al. Association Between Combined TMPRSS2:ERG and PCA3 RNA Urinary Testing and Detection of Aggressive Prostate Cancer. JAMA Oncol 2017, 3, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Tomlins, S.A.; Aubin, S.M.J.; Siddiqui, J.; Lonigro, R.J.; Sefton-Miller, L.; Miick, S.; Williamsen, S.; Hodge, P.; Meinke, J.; Blase, A.; et al. Urine TMPRSS2:ERG Fusion Transcript Stratifies Prostate Cancer Risk in Men with Elevated Serum PSA. Sci. Transl. Med. 2011, 3, 94ra72. [Google Scholar] [CrossRef] [PubMed]
- Font-Tello, A.; Juanpere, N.; de Muga, S.; Lorenzo, M.; Lorente, J.A. Association of ERG and TMPRSS2-ERG with Grade, Stage, and Prognosis of Prostate Cancer Is Dependent on Their Expression Levels. Prostate 2015, 75, 1216–1226. [Google Scholar] [CrossRef] [PubMed]
- Gopalan, A.; Leversha, M.A.; Satagopan, J.M.; Zhou, Q.; Al-Ahmadie, H.A.; Fine, S.W.; Eastham, J.A.; Scardino, P.T.; Scher, H.I.; Tickoo, S.K.; et al. TMPRSS2-ERG Gene Fusion Is Not Associated with Outcome in Patients Treated by Prostatectomy. Cancer Res. 2009, 69, 1400–1406. [Google Scholar] [CrossRef]
- García-Perdomo, H.A.; Chaves, M.J.; Osorio, J.C.; Sanchez, A. Association between TMPRSS2:ERG Fusion Gene and the Prostate Cancer: Systematic Review and Meta-Analysis. Cent. Eur. J. Urol. 2018, 71, 410. [Google Scholar] [CrossRef]
- Krstanoski, Z.; Vokac, N.K.; Zagorac, A.; Pospihalj, B.; Munda, M.; Dzeroski, S.; Golouh, R. TMPRSS2:ERG Gene Aberrations May Provide Insight into pT Stage in Prostate Cancer. BMC Urol. 2016, 16, 35. [Google Scholar] [CrossRef]
- Tosoian, J.J.; Trock, B.J.; Morgan, T.M.; Salami, S.S.; Tomlins, S.A.; Spratt, D.E.; Siddiqui, J.; Kunju, L.P.; Botbyl, R.; Chopra, Z.; et al. Use of the MyProstateScore Test to Rule Out Clinically Significant Cancer: Validation of a Straightforward Clinical Testing Approach. J. Urol. 2021, 205, 732–739. [Google Scholar] [CrossRef]
- Tosoian, J.J.; Singhal, U.; Davenport, M.S.; Wei, J.T.; Montgomery, J.S.; George, A.K.; Salami, S.S.; Mukundi, S.G.; Siddiqui, J.; Kunju, L.P.; et al. Urinary MyProstateScore (MPS) to Rule out Clinically-Significant Cancer in Men with Equivocal (PI-RADS 3) Multiparametric MRI: Addressing an Unmet Clinical Need. Urology 2022, 164, 184–190. [Google Scholar] [CrossRef]
- Tosoian, J.J.; Sessine, M.S.; Trock, B.J.; Ross, A.E.; Xie, C.; Zheng, Y.; Samora, N.L.; Siddiqui, J.; Niknafs, Y.; Chopra, Z.; et al. MyProstateScore in Men Considering Repeat Biopsy: Validation of a Simple Testing Approach. Prostate Cancer Prostatic Dis. 2023, 26, 563–567. [Google Scholar] [CrossRef]
- Lebastchi, A.H.; Russell, C.M.; Niknafs, Y.S.; Eyrich, N.W.; Botbyl, R.; Kabeer, R.; Osawa, T.; Siddiqui, J.; Siddiqui, R.; Davenport, M.S.; et al. Impact of the MyProstateScore (MPS) Test on the Clinical Decision to Undergo Prostate Biopsy: Results from a Contemporary Academic Practice. Urology 2020, 145, 204–210. [Google Scholar] [CrossRef]
- Wu, H.; Wu, Y.; He, P.; Liang, J.; Xu, X.; Ji, C. A Meta-Analysis for the Diagnostic Accuracy of SelectMDx in Prostate Cancer. PLoS ONE 2024, 19, e0285745. [Google Scholar] [CrossRef] [PubMed]
- Kawada, T.; Shim, S.R.; Quhal, F.; Rajwa, P.; Pradere, B.; Yanagisawa, T.; Bekku, K.; Laukhtina, E.; Von Deimling, M.; Teoh, J.Y.-C.; et al. Diagnostic Accuracy of Liquid Biomarkers for Clinically Significant Prostate Cancer Detection: A Systematic Review and Diagnostic Meta-Analysis of Multiple Thresholds. Eur. Urol. Oncol. 2024, 7, 649–662. [Google Scholar] [CrossRef]
- Hendriks, R.J.; Van Der Leest, M.M.G.; Israël, B.; Hannink, G.; YantiSetiasti, A.; Cornel, E.B.; Hulsbergen-van De Kaa, C.A.; Klaver, O.S.; Sedelaar, J.P.M.; Van Criekinge, W.; et al. Clinical Use of the SelectMDx Urinary-Biomarker Test with or without mpMRI in Prostate Cancer Diagnosis: A Prospective, Multicenter Study in Biopsy-Naïve Men. Prostate Cancer Prostatic Dis. 2021, 24, 1110–1119. [Google Scholar] [CrossRef]
- Visser, W.C.H.; De Jong, H.; Steyaert, S.; Melchers, W.J.G.; Mulders, P.F.A.; Schalken, J.A. Clinical Use of the mRNA Urinary Biomarker SelectMDx Test for Prostate Cancer. Prostate Cancer Prostatic Dis. 2022, 25, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Tutrone, R.; Donovan, M.J.; Torkler, P.; Tadigotla, V.; McLain, T.; Noerholm, M.; Skog, J.; McKiernan, J. Clinical Utility of the Exosome Based ExoDx Prostate(IntelliScore) EPI Test in Men Presenting for Initial Biopsy with a PSA 2–10 Ng/mL. Prostate Cancer Prostatic Dis. 2020, 23, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Tutrone, R.; Lowentritt, B.; Neuman, B.; Donovan, M.J.; Hallmark, E.; Cole, T.J.; Yao, Y.; Biesecker, C.; Kumar, S.; Verma, V.; et al. ExoDx Prostate Test as a Predictor of Outcomes of High-Grade Prostate Cancer—An Interim Analysis. Prostate Cancer Prostatic Dis. 2023, 26, 596–601. [Google Scholar] [CrossRef]
- Margolis, E.; Brown, G.; Partin, A.; Carter, B.; McKiernan, J.; Tutrone, R.; Torkler, P.; Fischer, C.; Tadigotla, V.; Noerholm, M.; et al. Predicting High-Grade Prostate Cancer at Initial Biopsy: Clinical Performance of the ExoDx (EPI) Prostate Intelliscore Test in Three Independent Prospective Studies. Prostate Cancer Prostatic Dis. 2022, 25, 296–301. [Google Scholar] [CrossRef]
- Borbiev, T.; Kohaar, I.; Petrovics, G. Clinical Biofluid Assays for Prostate Cancer. Cancers 2023, 16, 165. [Google Scholar] [CrossRef]
- Govers, T.M.; Caba, L.; Resnick, M.J. Cost-Effectiveness of Urinary Biomarker Panel in Prostate Cancer Risk Assessment. J. Urol. 2018, 200, 1221–1226. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).