Simple Summary
Hepatitis E virus (HEV) infection with some Paslahepevirus balayani strains in humans is known to cause pregnancy mortality. New strains of Rocahepevirus ratti have been identified with zoonotic potential. It is currently unknown whether Rocahepevirus ratti strains can cause pregnancy mortality in humans in a similar manner as their Paslahepevirus relatives. Our presented research suggests that the emerging Rocahepevirus ratti strain LCK-3110 does not infect the human placental cell line (JEG-3), unlike Paslahepevirus balayani gt3 strains, suggesting it may not be as significant of a pregnancy risk as human HEV strains.
Abstract
Paslahepevirus balayani and Rocahepevirus ratti are genetically diverse species of hepatitis E virus [HEV]. Previously, only members of the Paslahepevirus genus were known to infect humans but recently some Rocahepevirus members have been found to be infectious to both immunocompromised and immunocompetent humans. Paslahepevirus balayani genotypes (gt) 1, 2, and 4 are known for their detrimental effects during pregnancy, causing pregnancy-related disorders. Recent findings have demonstrated the ability of Paslahepevirus balayani gt3 to replicate within placental cell lines, suggesting a direct effect on the placenta and fetus. To study whether zoonotic rat HEV strains possess a similar human-host placental tropism, we utilized JEG-3 cells to understand the replicative ability of an infectious clone of a recently reported strain of Rocahepevirus ratti, the LCK-3110 strain. Infectious cDNA clones of Pasla-, Avi-, and Rocahepevirus were transcribed and then, transduced into JEG-3 cells. Cells were harvested, and cell lysates were used for testing infectivity. Five days post-transfection or after inoculation onto naive HepG2/C3A cells, the cells were analyzed for infection. Replication in transduced JEG-3 cells and the infection potential in HepG2/C3A cells were assessed via an indirect immunofluorescence assay and a flow-cytometry assay. We found that the Rocahepevirus ratti LCK-3110 strain did not have efficient replication in JEG-3 cell cultures.
1. Introduction
Hepatitis E virus [HEV] is a positive-sense, single-stranded RNA virus and is a leading cause of acute hepatitis in humans [1,2]. HEV primarily transmits through the fecal–oral route, blood transfusion, or by vertical transmission and is known for its zoonotic potential [3,4,5,6,7,8]. Recent increases in the host range of HEV with the identification of multiple viral reservoirs highlight this emerging zoonoses. The most common animal species harboring zoonotic HEV are swine, wild boar, deer, rabbit, camel, and rat [9]. Multiple HEV spillover events have been demonstrated in more than 30 animal species, but studies confirming the zoonotic nature of each strain are still underway [9].
The Hepeviridae family consists of two orders: Orthohepevirinae and Piscihepevirinae [10]. While Piscihepevirinae has only been found to infect a few species of fish, Orthohepevirinae causes infection in a multitude of differing species. In the Orthohepevirinae, four major genera have been identified: Paslahepevirus (infects humans, pigs, deer, wild boars, rabbits, and camels), Avihepevirus (infects birds; mainly chickens), Rocahepevirus (infects rats, minks, ferrets, field mice, and humans), and Chirohepevirus (infects bats) [10]. Rocahepevirus consists of two genotypes: Rocahepevirus C1 and Rocahepevirus C2 [10].
Rocahepevirus ratti (rat HEV) was first discovered in brown rats in Germany, and since has been detected all throughout Asia, Europe, and North America [11,12,13,14]. Originally not thought pathogenic to humans due to sequence divergence when compared to Paslahepevirus strains and failure to experimentally infect non-human primates [15] and pigs [16], rat HEV strains were recently isolated from a patient in Hong Kong, then in a patient in Canada [17,18]. Rat HEV has also been discovered to initiate disease in immunocompetent individuals [18]. Due to its ability to replicate and cause pathological changes in immunocompetent individuals, further research is needed to study rat HEV. The rat HEV strain (LCK-3110; Rocahepevirus ratti genotype C1) was isolated from a 56-year-old male human liver transplant recipient in Hong Kong [17]. The patient had persistent hepatitis causing derangement of liver enzymes and lymphopenia [17]. Mild-to-moderate inflammatory infiltrate, consisting of lymphocytes in the portal tracts, was demonstrated in the liver biopsy [17].
In addition to producing chronic and debilitating infections in immunocompromised patients, there is an enigmatic relationship between HEV and pregnancy. This includes increased severity both in epidemic and sporadic disease, fulminant hepatic disease in the third trimester, vertical transmission and severe neonatal disease, and several significant obstetric events [19]. These are reported primarily in HEV-gt1 infections, while HEV-gt2 infections are less well studied but also report higher pregnancy mortality rates. High mortality is not typically documented with gt3 and gt4. However, recent data from China has shown that HEV-gt4 can cause placental injury and obstetric events, but maternal disease is usually asymptomatic and not severe [20]. Despite many HEV studies attempting to elucidate pregnancy mortality, a definitive mechanism contributing to pregnancy pathogenesis has been complex and debated [21,22]. Open reading frame 4 (ORF4) protein expressed in HEV-gt1 during endoplasmic reticulum stress has been suggested as important for enhancing viral replication and could potentially contribute to severe pregnancy morbidity [23,24,25,26]. There are data suggesting a decreased ability of HEV-gt3 to cause placental injury. However, ectopic expression of HEV-gt1 ORF4 enhances the replicative ability of HEV-gt3 [24]. Recent findings utilizing the Paslahepevirus balayani gt3 strain (Kernow-C1 P6) have shown placental cells can support replication of HEV and suggested a tissue tropism effect with higher viral replication in placental cells compared with liver cells could be a reason behind pregnancy-related disorders caused by Paslahepevirus balayani gt1 [27]. Several studies have supported this claim by the demonstration of Paslahepevirus balayani gt1 HEV in vivo and ex vivo in the placenta and maternal–fetal interface, respectively [25,28]. There is currently no information on the susceptibility and replication ability of emerging zoonotic rat HEV to human placental cells. Thus, we assessed the ability of placental cells to support infection of the rat HEV LCK-3110 strain.
Our findings demonstrate an inability of the LCK-3110 strain of Rocahepevirus ratti to replicate efficiently in a human placental cell culture system. This study is essential to understand the differential tissue tropism of Paslahepevirus balayani gt1-4 and emerging Rocahepevirus ratti strains and suggests that the current zoonotic rat HEV may lack the ability to produce pregnancy-related disorders in humans.
2. Materials and Methods
2.1. Cell Culture
JEG-3 [isolated from human placental carcinoma, ATCC number HTB-36, ATCC, Manassas, VA, USA], made by serial cloning of BeWo, and HepG2/C3A [isolated from human liver carcinoma, ATCC number HB-8065] cells were used for the experiments. JEG-3 cells were cultured in EMEM [Eagle’s Minimum Essential Medium, Thermo Fisher Scientific, Waltham, MA, USA], and HepG2/C3A cells were cultured in DMEM [Dulbecco’s Modified Essential Medium, Thermo Fisher Scientific, Waltham, MA, USA]. Supplements included 5% fetal calf serum [FCS] for both cell lines, along with 100 ug/mL of streptomycin and 100 IU/mL of penicillin [Invitrogen, Waltham, MA, USA]. HepG2/C3A cells were grown on rat-collagen-coated culture plates. Cells were kept at 37 °C in a 5% CO2 incubator.
2.2. Linearization of Plasmid DNA
For linearization, isolated plasmid DNA-encoding Paslahepevirus balayani gt3 (Kernow-C1 P6), Avihepevirus magniiecur, and Rocahepevirus ratti (LCK-3110) [29] were used. Kernow C-1 P6 was used as a positive control and Avihepevirus was used as a negative control in the study, as Avihepevirus does not replicate in mammalian cells. The restriction digest was performed at 37 °C for 2 h using MluI for Paslahepevirus balayani gt3 (Kernow-C1 P6), XhoI for Avihepevirus magniiecur, and EcoRI for Rocahepevirus ratti.
2.3. Proteinase K Digestion and Purification of Linear DNA
Proteinase K [22 mg/mL] was added to the digested product, vortexed, and incubated at 37 °C for 1 h. Afterward, 200 μL of water were added followed by 300 μL phenol/chloroform/isoamyl alcohol. The solution was then vortexed and incubated at room temperature for 5 min. The top upper aqueous phase was collected after centrifugation [13,000 rpm, room temperature] and placed into a new microcentrifuge tube. Ammonium acetate and 100% cold ethanol were added, followed by 30 min of incubation at −20 °C. The DNA was pelleted by centrifugation at 14,000× g for 30 min at 4 °C. After the ethanol [75%] wash and centrifugation at 13,000 rpm for 15 min at 4 °C, the supernatant was thrown away, and the formed DNA pellet was then air dried. The pellet was thoroughly resuspended in 15 μL of nuclease-free water. The concentration of plasmid DNA was measured using nanodrop [24].
2.4. In Vitro Transcription
Linearized DNA [Paslahepevirus balayani gt3 (Kernow-C1 P6), Avihepevirus magniiecur, and Rocahepevirus ratti] was used to construct the viral capped mRNA using a Promega Ribomax Large Scale RNA Production System T7/SP6 [PRP 1300, Promega, Madison, WI, USA] and ARCA CAP Trilink biotechnologies [N-7003, Trilink Biotechnologies, San Diego, CA, USA]. Ten µL transcription buffer 5×, 15 µL rNTP master mix, 5 µL enzyme mix, and ARCA cap 0.4 µL for each sample were mixed to prepare the master mix. A total reaction volume of 50 µL, consisting of a linearized DNA template [4 µg] with 30 µL of master mix, was prepared. Reactions were incubated for 4 h at 37 °C in a thermal cycler [Bio-Rad, Hercules, CA, USA]. Assessment of the fidelity of transcripts and normalization was achieved by gel electrophoresis on 0.8% agarose gels and visualized via ultraviolet illumination.
2.5. Transfection of JEG-3 Cells
JEG-3 cells were cultured in four 12-well plates with a seeding density of 1 × 105 cells. Sixteen µL of 1 × 105 viral RNA copies/mL of viral RNA were transfected using a Mirus Trans-IT mRNA transfection kit (Invitrogen, Waltham, MA, USA). Opti-MEM, trans-IT boost reagent, trans IT-mRNA reagent, and viral RNA were used to make the transfection complex and incubated for 4 min at 37 °C before addition to the cellular monolayer. After 48 h of transfection, cells were passaged and divided into thirds before being left undisturbed for the next 3 days at 37 °C in a humidified 5% CO2 atmosphere.
2.6. Infectivity Assay
For the HEV infection assay, HepG2/C3A cells were used [24]. Freeze-and-thaw cycles (3×) were completed with transfected JEG-3 cells at 5 days post-transfection [dpt]. Both extracellular and intracellular viruses harvested from the cell lysates were subjected to high-speed centrifugation [10,000 rpm for 5 min] to remove cellular debris, and then, the supernatant was collected. Eight hundred µL of supernatant were used to infect the HepG2/C3A cells cultured in 12 well plates. At 48 h post-inoculation, cell passage was conducted (1:3). On the 5th day after infection, harvested cells were fixed in cold methanol, permeabilized, and further stained with rabbit anti-truncated ORF2 antibody [primary] against HEV ORF2 and goat anti-rabbit IgG-PE [secondary]. The cells were analyzed via FACS for HEV ORF2 positive cells [24].
2.7. Flow Cytometry of In Vitro-Transcribed Capped Paslahepevirus balayani (Kernow-C1 P6), Avihepevirus magniiecur, Rocahepevirus ratti HEV RNA Transfected JEG-3 Cells, and Infected HEPG2/C3A Cells
Five dpt-transfected JEG-3 cells were trypsinized and pelleted at 2500 rpm for 5 min; 100% cold methanol was used to fix the cells at 4 °C for a minimum of 15 min. Cells were resuspended in phosphate-buffered saline (PBS) after the removal of methanol. Five percent non-fat dried milk was used to block the cells. Cells were treated with a primary antibody—rabbit anti-truncated ORF2 HEV diluted 1:50 in a blocking solution for 30 min at 37 °C. Cells were then washed before being subjected to a secondary antibody—goat anti-rabbit-phycoerythrin [PE] (Life Technologies, Waltham, MA, USA) at 1:200 in PBS for 30 min at 37 °C. Cells were resuspended in 200 µL of PBS after two washes. Analysis of 100,000 events using a flow cytometer (BD Accuri C6 Plus, Biosciences, San Diego, CA, USA) was conducted. Dead cells and doublet discrimination were excluded using a gating approach in mock infected cells, based on forward and side scatter profiles [24]. The same procedure was completed for the infected HepG2/C3A cells.
2.8. Immunofluorescence Assay
Transfected JEG-3 cells were fixed in 100% cold methanol and permeabilized with PBS with 0.1% tween 20 (PBST) on 5 dpt. Rabbit anti-truncated ORF2 HEV antibody (1:200 in blocking buffer) for 30 min at 37 °C was used to immunostain ORF2-encoded capsid protein. After 3 washes with PBST, a fluorescently labeled goat anti-rabbit IgG H&L antibody (Alexa Fluor 594; Abcam, Cambridge, FL, USA) was used in a dilution of 1:400 in PBS to detect bound primary antibodies. 4,6-diamidino-2-phenylindole (DAPI) was used to stain the nucleus. Wells were manually observed for specific fluorescence using a fluorescent microscope (Keyence, Itasca, IL, USA), and the presence of fluorescent foci was recorded. A fluorescent focus was defined as a minimum of one to two cells showing clear intracytoplasmic fluorescence [24].
2.9. Inoculation of Cell Culture
JEG-3 cells were cultured in 12-well plates with a seeding density of 1 × 105 cells per well in 2 mL of DMEM with 10% FBS, penicillin (100 units/mL), and streptomycin (100 g/mL) and incubated at 37 °C for 24 h. The infections were performed utilizing a 10% fecal suspension derived from intrahepatically inoculated rat HEV gnotobiotic pigs (36), diluted 1:5 in DMEM and 0.45 μm filtered. The culture media was removed, and the cells were inoculated with 1 mL of the resulting solution (1 × 107 viral RNA copies). At room temperature, plates were rocked for 1 h and then incubated at 37 °C for 6 h. The inoculum was removed, and fresh culture media was added. In addition, we also transfected the JEG-3 cells with the rat HEV LCK-3110 transcript. Sixteen µL of 1 × 105 viral RNA copies/mL of viral RNA was transfected using a Mirus Trans-IT mRNA transfection kit [Invitrogen]. Opti-MEM, trans-IT boost reagent, trans IT-mRNA reagent, and viral RNA were used to make the transfection complex and incubated for 4 min at 37 °C before addition to the cellular monolayer. After 8 h of transfection, the media was replaced with fresh media. Supernatants were collected on days 0 (6 h after inoculation or transfection), 3, 6, 9, 12, 15, 18, and 21 and were tested for rat HEV via RT-qPCR.
2.10. RNA Extraction and RT-qPCR
Trizol reagent was used to extract RNA from the harvested cell supernatant and cell lysates on day 5 post-transfection and post-infection from JEG-3 and HepG2/C3A cells, respectively. The following sets of forward and reverse primers were used for Pasla-, Avi-, and Roca-hepeviruses:
- Pasla: HEV F, 5′-GGTGGTTTCTGGGGTGAC-3′, HEV R, 5′-AGGGGTTGGTTGGATGAA-3′, and a probe 5′-FAM-TGATTCTCAGCCCTTCGC-Dabcyl-3′;
- Avi: Avian HEV F, 5′-AATGTGCTGCGGGGTGTCAA-3′, Avian HEV R, 5′-CATCTGG-TACCGTGCGAGTA-3′, and a probe 5′-FAM-CTCCCAAACGCTCCCAGCCGG A-Dabcyl-3′;
- Roca: Rat HEV F, 5′-CTTGTTGAGCTYTTCTCCCCT-3′, Rat HEV R, 5′-CTGTACCGGATGCGACCAA-3′, and a probe 5′-FAM-TGCAGCTTGTCTTTGARCCC-Dabcyl-3′.
A 10-fold serial dilution of the capped Pasla-, Avi- and Roca-HEV RNA [107 to 101 copies] was used as the standard for quantification of the viral genome copy numbers.
2.11. Statistical Analysis and Reproducibility
Quantitative data are demonstrated with the mean and standard deviation. Student’s unpaired two-tailed t test was used for the analysis of independent data. GraphPad Prism 9.4.1 was used for the statistical comparison of data between groups.
3. Results
3.1. JEG-3 Is Not Permissive to Rat HEV Replication
To confirm whether Paslahepevirus balayani gt3 [Kernow-C1 P6 strain] and Rocahepevirus ratti gtC1 [rat HEV] could replicate within JEG-3 cell cultures, the full-length capped viral RNA from the respective strains were transfected into JEG-3 cells. Transduced cells were studied for ORF2 expression via an immunofluorescence assay. The open reading frame (ORF2) is translated from sub-genomic mRNA and is only produced at later stages of HEV replication, serving as an indicator of complete viral replication [30]. To see if replication of gt3 Kernow-C1 P6, avian HEV, and rat HEV had occurred, we assessed events at the single-cell level using immunofluorescence and flow cytometry. We observed that both avian HEV and rat HEV had not successfully replicated within the JEG-3 cell cultures compared to that of the Paslahepevirus balayani gt3 Kernow-C1 P6 strain. Using antibodies directed against the ORF2 capsid protein, we detected only a small number of cells containing ORF2 protein, suggesting inefficient replication (Figure 1A,D). Our ORF2 polyclonal antibody was capable of detecting Avihepevirus and Rocahepevirus ratti ORF2 (Supplementary Figure S1) in Huh7 transduced cells. The total numbers of ORF2-expressing cells were quantified by flow cytometry on cells immunostained for ORF2-encoded capsid antigen. As depicted in Figure 1A, avian HEV and rat HEV replication are lower than Kernow-C1 P6 replication within the JEG-3 cells. Although RNA was expressed in the transfected cell supernatant and lysates (Figure 1B,C), we could not detect a significant number of ORF2-positive cells using flow-cytometry analyses.
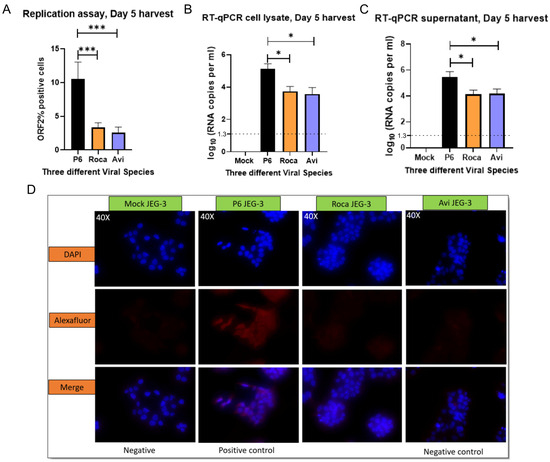
Figure 1.
Avian HEV and rat HEV do not replicate efficiently in JEG-3 cells. (A) Flow-cytometry analysis of HEV ORF2 percentage of positive cells. (B) RT-qPCR of cell lysates and supernatant (C) of transfected JEG-3 cells at 5 days post-transfection [dpt]. Even though we detected viral RNA in the supernatant and cell lysates, it could be remnants of the RNA used for initial transduction. (D) Immunofluorescence detection of HEV ORF2 antigen in methanol-fixed JEG-3 cells after 5 days post-transfection. Cells are stained with anti-HEV polyclonal serum, goat anti-rabbit IgG H&L combined with anti-rabbit Alexa fluor 594 [red] and 4′, 6-diamidino-2-phenylindole [DAPI] [blue]. Each bar (mean ± SEM) represents separate transfections stained in parallel and demonstrates the mean of three independent biological experiments with six replicates per sample (* p < 0.05, *** p < 0.001, t-test).
3.2. No Detectable Infection of Avian or Rat HEV in HEPG2/C3A Cells Using Transfected JEG-3 Cell Lysates
To determine if JEG-3 cells could produce infectious gt3 Kernow-C1 P6-, avian HEV, or rat HEV virus, transfected JEG-3 cell lysates (cells undergoing a freeze–thaw process 3×) were inoculated onto HepG2/C3A cells. To test for positive infection, the cells were probed for ORF2 capsid protein expression via an immunofluorescence assay. The cells were analyzed at the single-cell level using immunofluorescence and flow cytometry. Recapitulating the replication assay, it was observed that lysates from avian HEV and rat HEV were unable to infect the HepG2/C3A cells compared to that of the gt3 Kernow-C1 P6 strain. Antibodies directed against the ORF2 capsid protein were used to detect cells containing the ORF2 protein, suggesting that positive replication had occurred (Figure 2A,D). The quantification of the total numbers of ORF2-expressing cells was performed by staining capsid antigen that encodes ORF2 using flow-cytometer readings. As represented via viral RNA copies (Figure 2B,C), similar to the immunofluorescence results (Figure 2D), the avian HEV and rat HEV ability to infect HepG2/C3A cells was significantly lower than Kernow-C1 P6.
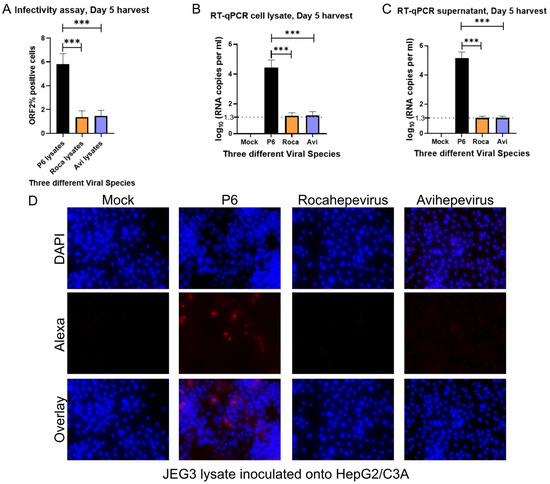
Figure 2.
Avian HEV and rat HEV infection was not observed in HepG2/C3A cells. (A) Flow-cytometry analysis of HEV ORF2 percentage of positive cells. (B) RT-qPCR of cell lysates and supernatant (C) of infected HepG2/C3A cells at 5 days post-transfection [dpt]. Even though we detect the viral RNA in the supernatant and cell lysates, it could be the remnants of the transduced RNA from the cell lysates used in the infection study. (D) Immunofluorescence detection of HEV ORF2 antigen in methanol-fixed HepG2/C3A cells after 5 days post-transfection. Cells are stained with goat anti-rabbit IgG H&L combined with anti-rabbit Alexa fluor 594 [red] and 4′, 6-diamidino-2-phenylindole [DAPI] [blue]. Each bar (mean ± SEM) represents separate transfections stained in parallel and demonstrates the mean of three independent biological experiments with six replicates per sample (*** p < 0.001, t-test).
3.3. Rat HEV LCK-3110 Does Not Replicate in JEG-3 Cells
To determine if rat HEV can replicate in JEG-3 cells if allowed to incubate for a longer period of time, we utilized a 0.45 μm filtered 10% fecal suspension derived from the rat HEV-positive gnotobiotic pigs on day 25 and diluted 1:5 in DMEM as inoculum. In addition, we utilized the rat HEV LCK-3110 capped RNA transcripts to transfect the JEG-3 cells. Cell-culture supernatants collected on days 0 and 3 from the transfected cells represent the remnants of the inoculum, as tested via RT-qPCR. Similarly, cell-culture supernatants collected from the virus-inoculated cells on days 0, 3, 6, and 9 demonstrate remnants of the inoculum. As depicted by decreasing viral RNA copies (Figure 3), the detected RNA was considered as background with the attachment of the virus/RNA to the cell surfaces. The decreasing RNA loads with time demonstrate the inability of rat HEV to replicate in the human placental JEG-3 cells.
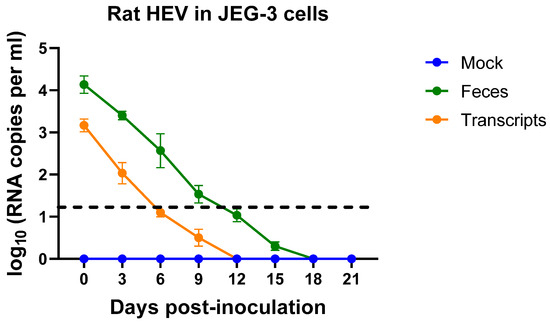
Figure 3.
Rat HEV does not replicate in JEG-3 cells. Rat HEV RNA loads in the culture supernatant of JEG-3 cell lines after inoculation with filtered 10% fecal suspension from rat HEV-inoculated gnotobiotic pigs or capped rat HEV transcripts. RNA of rat HEV in JEG-3 cells was measured on days 0, 3, 6, 9, 12, 15, 18, and 21. Independent biological experiments, mean ± SD of 3 replicates, are presented. Black dotted line represents the cut-off value.
4. Discussion
Paslahepevirus balayani gt1 HEV strains are a known cause of pregnancy mortality in humans [31,32]. Severe outcomes in the pregnant population have been attributed to HEV infection. HEV infection during pregnancy has led to fulminant hepatitis as well as increased maternal and fetal mortality [33]. Several years of studies have identified many host-related factors enhancing HEV replication during pregnancy. Hormones, immune factors, and nutritional factors have been associated with pregnancy mortality associated with HEV in the second and third trimesters of pregnancy [34]. Meanwhile, new species of HEV have emerged, resulting in serious illness in immunocompetent and immunocompromised populations [17,18]. Therefore, in our study, we determined whether an emerging zoonotic rat HEV strain possesses the ability to replicate in placental JEG-3 cells, previously demonstrated to sustain Paslahepevirus balayani gt3 Kernow-C1 P6 and gt1/Sar55 replication [27]. We demonstrated that the JEG-3 placental cells were not susceptible to rat HEV replication after viral RNA transfection, which is a typical method to study the infectiousness of specific HEV strains using infectious clones of these viruses [24,35,36]. We also demonstrated that the infectious virus could not be detected from the transfected JEG-3 cells. Like the transfection results, infected HepG2/C3A did not show any positive ORF2 staining, suggesting a lack of infectious virus in the transfected JEG-3 cells. HepG2/C3A cells have been demonstrated to support replication of the zoonotic Rocahepevirus ratti LCK-3110 strain (Supplementary Figure S2) [29] and also the non-zoonotic strain LA-B350 [37] isolated from rats. In addition, we performed long-term experiments for 3 weeks to determine whether rat HEV could adapt to JEG-3 cells. However, similar to the transfection and infection results, viral replication kinetics demonstrated an inability of rat HEV to replicate in JEG-3 cells.
JEG-3 cells have been demonstrated to support the full HEV replication cycle for Paslahepevirus balayani gt3 strains [27]. In addition, the HEV detected in tissue samples from the maternal–fetal interface and placentas from pregnant women supports the replication of Paslahepevirus balayani gt3 HEV in these extra-hepatic tissues [38]. Of the various genotypes within Paslahepevirus, only genotypes belonging to gt1 and gt2 have been found to produce pregnancy mortality in humans during natural outbreaks [20]. However, sporadic cases related to pregnancy disorders have been reported with gt3 and gt4 [7,39,40]. Placental cells are known as a site for extrahepatic HEV replication in the case of Paslahepevirus balayani gt1 and gt3, and the primary treatment for HEV, ribavirin (RBV), is contraindicated during pregnancy. Thus, it is with the utmost necessity that we understand whether emerging rat HEV could use placental cells for replication. With very limited studies within the clinical setting, there have been no data reporting on rat HEV in pregnant populations. As rat HEV continues to emerge and evolve, we may see increased numbers of human cases, requiring an in-depth knowledge of its disease potential.
Since the ability of Rocahepevirus ratti to infect human placental cell lines is not defined, we experimentally tested its ability to replicate in JEG-3 cells. The explanation behind the lower replication rate of rat HEV in JEG-3 cells is unclear at this time; however, one hypothesis is that different host factors may be necessary for effective rat HEV replication in placental cells. HEV-specific cell receptors have not been identified. However, six different receptors have been demonstrated to play roles in the spread of HEV [34,41]. In our study, we circumvented the need for receptors by initially conducting transfection studies, introducing rat HEV viral RNA directly into cells; Yet, we could not see efficient replication, as demonstrated by the single-cell flow assays. Previous studies have demonstrated that Paslahepevirus replication is unique and can only be seen in certain cell lines. Okamoto and colleagues used infected fecal materials to screen a total of 21 cell lines for their ability to support the Paslahepevirus balayani gt3 life cycle [42], out of which A549 [human lung] and PLC/PRF/5 [human liver] were the only ones shown to efficiently support infection. Similarly, Shukla et al. demonstrated that the HEV gt3 Kernow-C1 strain isolated from feces infected five human cell lines and one rhesus monkey cell line [43], with the highest titer seen in HepG2/C3A. But analogous efforts to adapt Kernow-C1 were unsuccessful [43]. We speculate the rat HEV LCK-3110 may need unique factors to fully replicate in the in vitro system. In addition, we have used the LCK-3110 rat HEV transcripts in other cell lines, such as Huh7 S10-3 liver cells, swine testicular (ST) cells, baby hamster kidney (BHK)-21, and mouse subcutaneous (LMTK) cells. ST and Huh7 S10-3 cells were the two most promising cell lines for replication studies [29]. Intrahepatic inoculation of gnotobiotic pigs with rat HEV RNA transcripts resulted in fecal viral shedding, demonstrating the functionality of the RNA transcripts [29] Thus, the rat HEV transcripts do possess replicative capability (Supplementary Figures S1 and S2) but did not show any replication in JEG-3 cells for unknown reasons.
There are several limitations in this study. To recognize the replicative ability of the virus, utilizing green fluorescent protein-producing rat HEV replicons would add additional support to our data. We were limited to just one type of placental cell-culture system. Utilizing other placental cells, such as BeWo and JAR to more thoroughly characterize the replicative ability of rat HEV would have given a clearer picture of rat HEV in placental cell-culture systems. Our primary IFA relies on polyclonal rabbit serum generated against recombinant Paslahepevirus balayani ORF2 protein, which may have differing avidities to ORF2 from Rocahepevirus and Avihepevirus strains. However, ORF2 antibodies have been shown to be cross-reactive against all HEV strains [44,45], and we have successfully utilized this rabbit anti-Paslahepevirus balayani ORF2 serum for recognizing the LCK-3110 rat HEV strain [29]. Waiting for a longer period of time post-transduction might have allowed the Rocahepevirus ratti strain additional time to overcome the barriers to restriction.
Several questions regarding rat HEV tissue tropism, life cycle, pathogenesis, and spillover ability remain to be characterized with a need to better understand the host factors regulating rat HEV replication in humans. How rat HEV infections persist and the factors that define the tissue and species tropism need to be explored further. Our presented results suggest that placental cells lack essential replication factors necessary to efficiently replicate Rocahepevirus ratti. Further in vitro studies are necessary to determine which host factors are critical for rat HEV to replicate within JEG-3 cells. Currently, there have been no reports of maternal mortality caused by Rocahepevirus ratti in either experimentally infected animal models or in human cases. Our results suggest that, unlike Paslahepevirus balayani gt3 (Kernow-C1 P6), Rocahepevirus ratti may not be capable of placental replication, presenting a reduced risk for host mortality during pregnancy. Further studies in experimentally infected rats and epidemiological monitoring in humans could confirm this hypothesis.
5. Conclusions
In conclusion, after performing both a replication assay and infectivity assay using an immuno-fluorescence-based assay and flow cytometry, we found that JEG-3 cells are not suitable for cell culture for rat HEV. While JEG-3 was able to support replication and infectivity of the Paslahepevirus balayani HEV gt3 Kernow-C1 P6 strain, it was inefficient in producing high levels of replication and infectivity for rat HEV LCK-3110.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/zoonoticdis4020012/s1, Figure S1. LCK-3110 rat HEV is replication competent in human liver cells. Figure S2. Isolation of rat HEV from infected pig feces in cell culture.
Author Contributions
K.K.Y. performed experiments, analyzed data, added to experimental conceptualization, wrote the first manuscript draft, and performed editing. J.D.H. performed experiments and visualization. S.P.K. Conceptualized the work, provided funding, supervised experiments, helped in data analysis, and revised all paper drafts. All authors have read and agreed to the published version of the manuscript.
Funding
Additional salaries and research support were provided by state and federal funds appropriated to the Ohio Agricultural Research and Development Center, The Ohio State University, and from the research funds of the National Institute of Allergy and Infectious Diseases [#R21AI151736].
Institutional Review Board Statement
Research within this manuscript was approved by the Institutional Biosafety Committee of The Ohio State University; IBC protocol 2016R00000082.
Informed Consent Statement
Not applicable.
Data Availability Statement
All the data regarding the study has been included in the manuscript.
Acknowledgments
We thank Renukaradhya Gourapura and his lab members at the Center for Food Animal Health [CFAH] for helping us with the flow-cytometry analysis. We also thank Anastasia Vlasova and her lab members at CFAH for helping us with the immunofluorescence microscopy images.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Lhomme, S.; Marion, O.; Abravanel, F.; Izopet, J.; Kamar, N. Clinical Manifestations, Pathogenesis and Treatment of Hepatitis E Virus Infections. J. Clin. Med. 2020, 9, 331. [Google Scholar] [CrossRef] [PubMed]
- Kenney, S.P.; Meng, X.J. Hepatitis E Virus Genome Structure and Replication Strategy. Cold Spring Harb. Perspect. Med. 2019, 9, a031724. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Wang, Y. Transmission of Hepatitis E Virus. Adv. Exp. Med. Biol. 2016, 948, 89–112. [Google Scholar] [CrossRef] [PubMed]
- Ticehurst, J.R.; Pisanic, N.; Forman, M.S.; Ordak, C.; Heaney, C.D.; Ong, E.; Linnen, J.M.; Ness, P.M.; Guo, N.; Shan, H.; et al. Probable transmission of hepatitis E virus (HEV) via transfusion in the United States. Transfusion 2019, 59, 1024–1034. [Google Scholar] [CrossRef] [PubMed]
- Mateos, M.L.; Camarero, C.; Lasa, E.; Teruel, J.L.; Mir, N.; Baquero, F. Hepatitis E virus: Relevance in blood donors and other risk groups. Vox Sang. 1998, 75, 267–269. [Google Scholar] [CrossRef]
- Harvala, H.; Hewitt, P.E.; Reynolds, C.; Pearson, C.; Haywood, B.; Tettmar, K.I.; Ushiro-Lumb, I.; Brailsford, S.R.; Tedder, R.; Ijaz, S. Hepatitis E virus in blood donors in England, 2016 to 2017: From selective to universal screening. Eurosurveillance 2019, 24, 1800386. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Bu, Q.; Gong, W.; Li, H.; Wang, L.; Li, S.; Sridhar, S.; Cy Woo, P.; Wang, L. Hepatitis E virus infection and its associated adverse feto-maternal outcomes among pregnant women in Qinhuangdao, China. J. Matern. Fetal Neonatal Med. 2020, 33, 3647–3651. [Google Scholar] [CrossRef] [PubMed]
- Jilani, N.; Das, B.C.; Husain, S.A.; Baweja, U.K.; Chattopadhya, D.; Gupta, R.K.; Sardana, S.; Kar, P. Hepatitis E virus infection and fulminant hepatic failure during pregnancy. J. Gastroenterol. Hepatol. 2007, 22, 676–682. [Google Scholar] [CrossRef]
- Si, F.; Widen, F.; Dong, S.; Li, Z. Hepatitis E Virus. In Hepatitis E Virus. Advances in Experimental Medicine and Biology; Wang, Y., Ed.; Springer: Singapore, 2023; Volume 1417, pp. 1–13. [Google Scholar]
- Purdy, M.A.; Drexler, J.F.; Meng, X.-J.; Norder, H.; Okamoto, H.; Van der Poel, W.H.; Reuter, G.; de Souza, W.M.; Ulrich, R.G.; Smith, D.B. ICTV virus taxonomy profile: Hepeviridae 2022. J. Gen. Virol. 2022, 103, 001778. [Google Scholar] [CrossRef] [PubMed]
- Johne, R.; Plenge-Bönig, A.; Hess, M.; Ulrich, R.G.; Reetz, J.; Schielke, A. Detection of a novel hepatitis E-like virus in faeces of wild rats using a nested broad-spectrum RT-PCR. J. Gen. Virol. 2010, 91, 750–758. [Google Scholar] [CrossRef]
- Johne, R.; Dremsek, P.; Kindler, E.; Schielke, A.; Plenge-Bönig, A.; Gregersen, H.; Wessels, U.; Schmidt, K.; Rietschel, W.; Groschup, M.H.; et al. Rat hepatitis E virus: Geographical clustering within Germany and serological detection in wild Norway rats (Rattus norvegicus). Infect. Genet. Evol. 2012, 12, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Li, T.C.; Yoshimatsu, K.; Yasuda, S.P.; Arikawa, J.; Koma, T.; Kataoka, M.; Ami, Y.; Suzaki, Y.; Mai, L.T.Q.; Hoa, N.T.; et al. Characterization of self-assembled virus-like particles of rat hepatitis E virus generated by recombinant baculoviruses. J. Gen. Virol. 2011, 92, 2830–2837. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Kunita, S.; Kawakami, M.; Kadosaka, T.; Fujita, H.; Takada, N.; Miyake, M.; Kobayashi, T.; Ohnishi, H.; Nagashima, S.; et al. First detection and characterization of rat hepatitis E Virus (HEV-C1) in Japan. Virus Res. 2022, 314, 198766. [Google Scholar] [CrossRef] [PubMed]
- Purcell, R.H.; Engle, R.E.; Rood, M.P.; Kabrane-Lazizi, Y.; Nguyen, H.T.; Govindarajan, S.; St Claire, M.; Emerson, S.U. Hepatitis E virus in rats, Los Angeles, California, USA. Emerg. Infect. Dis. 2011, 17, 2216–2222. [Google Scholar] [CrossRef] [PubMed]
- Cossaboom, C.M.; Córdoba, L.; Sanford, B.J.; Piñeyro, P.; Kenney, S.P.; Dryman, B.A.; Wang, Y.; Meng, X.J. Cross-species infection of pigs with a novel rabbit, but not rat, strain of hepatitis E virus isolated in the United States. J. Gen. Virol. 2012, 93, 1687–1695. [Google Scholar] [CrossRef]
- Sridhar, S.; Yip, C.C.Y.; Wu, S.; Cai, J.; Zhang, A.J.; Leung, K.H.; Chung, T.W.H.; Chan, J.F.W.; Chan, W.M.; Teng, J.L.L.; et al. Rat Hepatitis E Virus as Cause of Persistent Hepatitis after Liver Transplant. Emerg. Infect. Dis. 2018, 24, 2241–2250. [Google Scholar] [CrossRef] [PubMed]
- Andonov, A.; Robbins, M.; Borlang, J.; Cao, J.; Hatchette, T.; Stueck, A.; Deschambault, Y.; Murnaghan, K.; Varga, J.; Johnston, L. Rat Hepatitis E Virus Linked to Severe Acute Hepatitis in an Immunocompetent Patient. J. Infect. Dis. 2019, 220, 951–955. [Google Scholar] [CrossRef] [PubMed]
- Khuroo, M.S. Hepatitis E and Pregnancy: An Unholy Alliance Unmasked from Kashmir, India. Viruses 2021, 13, 1329. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Li, T.; Xia, Y.; Cong, C.; Chen, S.; Zhang, Y.; Gong, S.; Wang, W.; Liu, H.; Chen, D.; et al. Genotype 4 Hepatitis E virus replicates in the placenta, causes severe histopathological damage, and vertically transmits to fetuses. J. Infect. 2023, 87, 34–45. [Google Scholar] [CrossRef]
- Yadav, K.K.; Kenney, S.P. Hepatitis E Virus Zoonotic Axis. In Zoonoses: Infections Affecting Humans and Animals; Sing, A., Ed.; Springer International Publishing: Cham, Switzerland, 2022; pp. 1–28. [Google Scholar] [CrossRef]
- Yadav, K.K.; Kenney, S.P. Animal Models for Studying Congenital Transmission of Hepatitis E Virus. Microorganisms 2023, 11, 618. [Google Scholar] [CrossRef]
- Nair, V.P.; Anang, S.; Subramani, C.; Madhvi, A.; Bakshi, K.; Srivastava, A.; Shalimar Nayak, B.; Ranjith Kumar, C.T.; Surjit, M. Endoplasmic Reticulum Stress Induced Synthesis of a Novel Viral Factor Mediates Efficient Replication of Genotype-1 Hepa-titis E Virus. PLoS Pathog. 2016, 12, e1005521. [Google Scholar] [CrossRef] [PubMed]
- Yadav, K.K.; Boley, P.A.; Fritts, Z.; Kenney, S.P. Ectopic Expression of Genotype 1 Hepatitis E Virus ORF4 Increases Genotype 3 HEV Viral Replication in Cell Culture. Viruses 2021, 13, 75. [Google Scholar] [CrossRef] [PubMed]
- Gouilly, J.; Chen, Q.; Siewiera, J.; Cartron, G.; Levy, C.; Dubois, M.; Al-Daccak, R.; Izopet, J.; Jabrane-Ferrat, N.; El Costa, H. Genotype specific pathogenicity of hepatitis E virus at the human maternal-fetal interface. Nat. Commun. 2018, 9, 4748. [Google Scholar] [CrossRef]
- El-Mokhtan El-Mokhtar, M.A.; Othman, E.R.; Khashbah, M.Y.; Ismael, A.; Ghaliony, M.A.; Seddik, M.I.; Sayed, I.M. Evidence of the Extrahepatic Replication of Hepatitis E Virus in Human Endometrial Stromal Cells. Pathogens 2020, 9, 295. [Google Scholar] [CrossRef] [PubMed]
- Knegendorf, L.; Drave, S.A.; Thi, V.L.D.; Debing, Y.; Brown, R.J.; Vondran, F.W.; Resner, K.; Friesland, M.; Khera, T.; Engelmann, M. Hepatitis E virus replication and interferon responses in human placental cells. Hepatol. Commun. 2018, 2, 173–187. [Google Scholar] [CrossRef]
- Ratho, R.K.; Thakur, V.; Arya, S.; Singh, M.P.; Suri, V.; Das, A. Placenta as a site of HEV replication and inflammatory cytokines modulating the immunopathogenesis of HEV in pregnant women. J. Med. Virol. 2022, 94, 3457–3463. [Google Scholar] [CrossRef] [PubMed]
- Yadav, K.K.; Boley, P.A.; Lee, C.M.; Khatiwada, S.; Jung, K.; Laocharoensuk, T.; Hofstetter, J.; Wood, R.; Hanson, J.; Kenney, S.P. Rat hepatitis E virus (HEV) cross-species infection and transmission in pigs. bioRxiv 2023. [Google Scholar] [CrossRef]
- Graff, J.; Torian, U.; Nguyen, H.; Emerson, S.U. A bicistronic subgenomic mRNA encodes both the ORF2 and ORF3 proteins of hepatitis E virus. J. Virol. 2006, 80, 5919–5926. [Google Scholar] [CrossRef]
- Khuroo, M.S.; Kamili, S.; Khuroo, M.S. Clinical course and duration of viremia in vertically transmitted hepatitis E virus (HEV) infection in babies born to HEV-infected mothers. J. Viral Hepat. 2009, 16, 519–523. [Google Scholar] [CrossRef]
- Khuroo, M.S.; Teli, M.R.; Skidmore, S.; Sofi, M.A.; Khuroo, M.I. Incidence and severity of viral hepatitis in pregnancy. Am. J. Med. 1981, 70, 252–255. [Google Scholar] [CrossRef]
- Kumar Acharya, S.; Kumar Sharma, P.; Singh, R.; Kumar Mohanty, S.; Madan, K.; Kumar Jha, J.; Kumar Panda, S. Hepatitis E virus (HEV) infection in patients with cirrhosis is associated with rapid decompensation and death. J. Hepatol. 2007, 46, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Yadav, K.K.; Kenney, S.P. Hepatitis E Virus Immunopathogenesis. Pathogens 2021, 10, 1180. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Pierson, F.; Toth, T.; Meng, X. Construction and characterization of infectious cDNA clones of a chicken strain of hepatitis E virus (HEV), avian HEV. J. Gen. Virol. 2005, 86, 2585–2593. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.M.; LeRoith, T.; Pudupakam, R.S.; Pierson, F.W.; Huang, Y.W.; Dryman, B.A.; Meng, X.J. Construction of an infectious cDNA clone of avian hepatitis E virus (avian HEV) recovered from a clinically healthy chicken in the United States and characterization of its pathogenicity in specific-pathogen-free chickens. Vet. Microbiol. 2011, 147, 310–319. [Google Scholar] [CrossRef] [PubMed]
- Debing, Y.; Mishra, N.; Verbeken, E.; Ramaekers, K.; Dallmeier, K.; Neyts, J. A rat model for hepatitis E virus. Dis. Models Mech. 2016, 9, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Yadav, K.K.; Kenney, S.P. Extrahepatic Replication Sites of Hepatitis E Virus (HEV). Zoonotic Dis. 2023, 3, 68–84. [Google Scholar] [CrossRef]
- Anty, R.; Ollier, L.; Péron, J.M.; Nicand, E.; Cannavo, I.; Bongain, A.; Giordanengo, V.; Tran, A. First case report of an acute genotype 3 hepatitis E infected pregnant woman living in South-Eastern France. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2012, 54, 76–78. [Google Scholar] [CrossRef] [PubMed]
- Tabatabai, J.; Wenzel, J.J.; Soboletzki, M.; Flux, C.; Navid, M.H.; Schnitzler, P. First case report of an acute hepatitis E subgenotype 3c infection during pregnancy in Germany. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2014, 61, 170–172. [Google Scholar] [CrossRef] [PubMed]
- Kapur, N.; Thakral, D.; Durgapal, H.; Panda, S.K. Hepatitis E virus enters liver cells through receptor-dependent clathrin-mediated endocytosis. J. Viral Hepat. 2012, 19, 436–448. [Google Scholar] [CrossRef]
- Okamoto, H. Hepatitis E virus cell culture models. Virus Res. 2011, 161, 65–77. [Google Scholar] [CrossRef]
- Shukla, P.; Nguyen, H.T.; Torian, U.; Engle, R.E.; Faulk, K.; Dalton, H.R.; Bendall, R.P.; Keane, F.E.; Purcell, R.H.; Emerson, S.U. Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proc. Natl. Acad. Sci. USA 2011, 108, 2438–2443. [Google Scholar] [CrossRef] [PubMed]
- Sanford, B.J.; Opriessnig, T.; Kenney, S.P.; Dryman, B.A.; Córdoba, L.; Meng, X.J. Assessment of the cross-protective capability of recombinant capsid proteins derived from pig, rat, and avian hepatitis E viruses (HEV) against challenge with a genotype 3 HEV in pigs. Vaccine 2012, 30, 6249–6255. [Google Scholar] [CrossRef] [PubMed]
- Situ, J.; Hon-Yin Lo, K.; Cai, J.P.; Li, Z.; Wu, S.; Hon-Kiu Shun, E.; Foo-Siong Chew, N.; Yiu-Hung Tsoi, J.; Sze-Man Chan, G.; Hei-Man Chan, W.; et al. An immunoassay system to investigate epidemiology of Rocahepevirus ratti (rat hepatitis E virus) infection in humans. JHEP Rep. 2023, 5, 100793. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).