Abstract
Lactiplantibacillus plantarum strains are used in the food industry for their probiotic properties. Some of these bacteria have immunomodulatory effects on the host and are able to improve resistance against different pathogens, including viruses. However, to date, the bacterial genes involved in the immunomodulatory effect are not known. In this work, the complete genomes of L. plantarum MPL16, CRL1506, CRL681 and TL2766 were used to perform comparative genomics with the aim of identifying the genes involved in their differential immunomodulatory effects. L. plantarum WCFS1, a strain with proven probiotic activity, was also used for comparisons. The analysis of the genes involved in the metabolic pathways of the five strains did not reveal differences in the metabolism of amino acids, lipids, nucleotides, cofactors and vitamins, nor in the genes associated with energy metabolism or the biosynthesis of lipoproteins and teichoic acids. However, differences were found between the five strains when considering carbohydrate metabolism pathways, particularly in the presence/absence of glycosylhydrolases and glycosyltransferases. In addition, a great variability was detected in the predicted surface proteins of each L. plantarum strain. These results suggest that the surface molecules expressed in the different strains of L. plantarum could be involved in their differential ability to modulate the innate antiviral immune response.
1. Introduction
Lactiplantibacillus plantarum has one of the largest genomes known among lactic acid bacteria [1] and is therefore able to survive in a wide range of environmental niches including the gastrointestinal tract, easily colonizing the intestine of humans and other mammals [2,3]. In addition, many strains of this bacterial species have shown to possess beneficial properties for the host, including their ability to beneficially modulate the immune system [4]. Due to these properties, L. plantarum is considered one of the most widely used bacterial strains in the food industry both as a starter culture and as a probiotic [5].
Previously, we performed in vitro studies in porcine intestinal epithelial (PIE) cells and in vivo studies in mice with different L. plantarum strains (MPL16, CRL1506, CRL681 and TL2766) and we demonstrated that those strains possess a differential ability to modulate the respiratory and intestinal innate antiviral immune responses [6]. Quantitative and qualitative differences were found in the expression of genes with immunological functions in PIE cells when they were treated with the distinct L. plantarum strains before the challenge with the Toll-like receptor 3 (TLR3) agonist polyinosinic:polycytidylic acid [poly(I:C)]. The transcriptional profile performed in vitro on PIE cells challenged with poly(I:C) allowed the selection of a new immunobiotic strain, L. plantarum MPL16, with the ability to stimulate in vivo antiviral immunity not only in the intestinal mucosa, but also in the respiratory tract, when administered orally [6]. On the other hand, our studies showed that the immunomodulatory strain L. plantarum CRL1506 only stimulates antiviral immunity at the intestinal level, whereas the CRL681 and TL2766 strains did not present immunomodulatory effects in the context of the response mediated by the activation of TLR3 [6]. Therefore, having strains of L. plantarum with different abilities to modulate intestinal and respiratory antiviral immunity provides an interesting opportunity to investigate the bacterial molecule(s) that could be involved in their differential immunomodulatory activities.
With the rapid development of next-generation sequencing techniques, the complete genomes of multiple strains of L. plantarum have been sequenced. This scientific-technological advance has provided a better understanding of the relationships between genotypes and functions [7,8]. Comparative genomics allows efficient analysis of intra-strain differences in the genomes of multiple L. plantarum strains, leading to better functional analysis at the molecular level [7,8,9]. These studies revealed that L. plantarum exhibits a high conservation of genes involved in the synthesis or degradation of proteins and lipids, whereas it presents a high diversity of genes involved in the transport and catabolism of sugars [10,11] as well as in the bacterial surface-expressed molecules [1,3,12], which determines the ability of each strain to interact with the host.
Considering that the complete genomes of L. plantarum MPL16, CRL1506, CRL681 and TL2766 has been sequenced, the aim of this work was to carry out comparative and functional genomics studies in order to identify the bacterial gene(s) responsible for the distinct immunomodulatory properties of each strain. For this purpose, special emphasis was given to the study of genes with potential immunomodulatory function, including surface and secreted proteins, adhesion factors, and genes involved in the biosynthesis of exopolysaccharides (EPS), lipoproteins, and teichoic acids.
2. Results
2.1. General Genomic Characteristics of L. plantarum
The complete genomes of L. plantarum MPL16, CRL1506 and CRL681 were sequenced with the Illumina MiSeq platform and uploaded to the NCBI repository. Genomes are available under the accession numbers indicated in Table 1. For the comparative genomic analysis, L. plantarum WCFS1, with recognized probiotic activity [13], and L. plantarum TL2966, which has not possess immunomodulatory properties [14,15] were also incorporated. The general genomic characteristics of the L. plantarum evaluated in this work are shown in Table 1.

Table 1.
General genomic characteristics of the L. plantarum strains studied.
The mean whole genome size and GC content of the strains were 3.26 Mb and 44.2%, respectively. This is in agreement with recent results obtained from the genomic analysis of 127 strains of L. plantarum which revealed a size and GC content of 3.32 Mb and 44.5%, respectively [1]. These data are also in line with the study reported by Mao et al. [11], describing that within the complete genomes of 114 strains of L. plantarum, genomes have a size between 2.94 and 3.90 Mb and a GC content of 44.1 to 46.5%. Among the five strains selected for this work, L. plantarum CRL681 presented the highest number of protein-coding genes (3081) whereas the lowest number (2918) was found for the CRL1506 strain.
Using the BlastKOALA tool, the functional characterization of L. plantarum genomes was carried out according to the KEGG database. As observed in Figure 1, no remarkable differences were observed in the number of genes associated with the metabolism pathways of carbohydrates (216 ± 3), amino acids (136 ± 2), lipids (40 ± 3), nucleotides (66 ± 2), glycans (136 ± 2), cofactors and vitamins (70 ± 3) or energy metabolism (68 ± 2) among the MPL16, CRL1506, CRL681, WCFS1 and TL2766 strains. L. plantarum CRL1506 presented a lower number of genes associated with the metabolism of terpenoids and polyketides as well as for the biosynthesis of other secondary metabolites compared with the other strains (Figure 1).
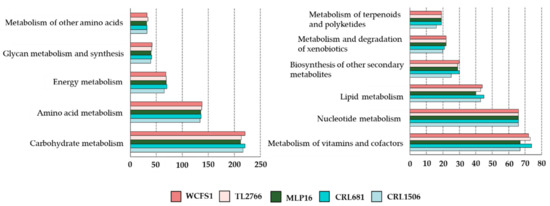
Figure 1.
Study of the number of genes in the different functional categories associated with metabolic pathways in the genomes of Lactiplantibacillus plantarum strains WCFS1, TL2766, MPL16, CRL681 and CRL1506. Functional characterization of genes was performed with the BlastKOALA tool according to the KEGG database.
There were also no significant differences in the numbers of genes associated with “genetic information processing”, “environmental information processing” or “cellular processes” when the five strains of L. plantarum were compared (Figure 2). The only exceptions were the numbers of genes related to “folding, sorting and degradation” and “transduction signal” for the CRL1506 strain which were lower than those found in the other lactobacilli.
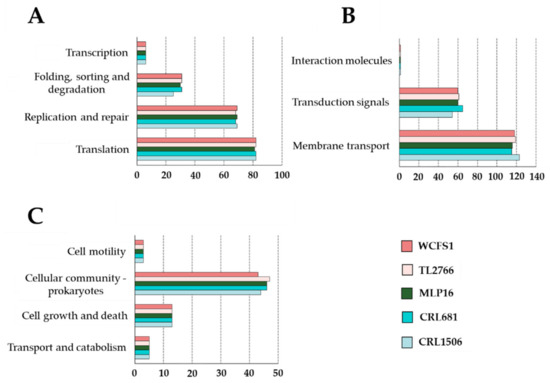
Figure 2.
Study of the number of genes in the different functional categories associated with genetic information processing (A), environmental information processing (B) and cellular processes (C) pathways of the genomes of Lactiplantibacillus plantarum strains WCFS1, TL2766, MPL16, CRL681 and CRL1506. Functional characterization of genes was performed with the BlastKOALA tool according to the KEGG database.
The MPL16, CRL1506, CRL681, WCFS1 and TL2766 strains were then compared by a Venn diagram using the orthologous genes (Figure 3). The analysis revealed that the five strains of L. plantarum share 2282 genes, whereas 194, 53, 106, 69 and 62 unique genes were detected for MPL16, CRL1506, CRL681, WCFS1 and TL2766, respectively. Among the unique genes for L. plantarum MPL16, two bacterial proteins with immunoglobulin-like (Ig-like) domains (OG0003093 and OG0002455), a PTS sugar transporter (OG0003210), an N-acetyl transferase of the GNAT family (OG0003311), two glycosyltransferases (GT) (OG0003314, OG0003357), two glycosylhydrolases (GH) (OG0003274, OG0003356), one lysophospholipase (OG0003265), the stress response membrane protein GlsB/YeaQ/YmgE (OG0003255), and several proteins associated with the type I-E CRISPR system (OG0003362, OG0003363, OG0003364, OG0003365, OG0003366, OG0003367, OG0003368, OG0003369) were detected (Table S1).
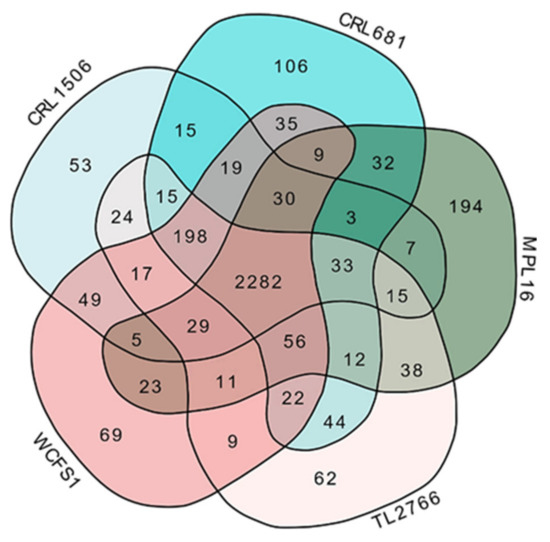
Figure 3.
Venn diagram of the number of orthologous genes shared by Lactiplantibacillus plantarum strains WCFS1, TL2766, MPL16, CRL681 and CRL1506. Amino acid sequences encoded in genomes were compared for ortholog group inference with OrthoFinder v2.5.2.
Unique genes for CRL1506 included an N-acetyl-muramyl-L-alanine amidase (OG0003105), a GNAT family N-acetyl transferase (OG0003102), a glycosyltransferase (OG0003069), and several phage-associated proteins (OG0003095, OG0003080, OG0003076, OG0003072). The genes for a YjzC family protein (OG0003135), a YjdF family protein (OG0003144), two glycosyltransferases (OG0003180, OG0003182), and other phage-associated proteins (OG0003167, OG0003168, OG0003169, OG0003174, OG0003176, OG0003201) were detected only in the genome of L. plantarum CRL681 (Table S1).
Among the seven genes shared by the MPL16 and CRL1506 strains, a polysaccharide pyruvyl transferase (OG0002887) was found, whereas among the 15 genes shared by the CRL1506 and CRL681 strains, a family 2 glycosyltransferase (OG0002860) was noticed (Figure 3, Table S1).
Genes shared by the probiotic bacterium L. plantarum WCSF1 and the immunomodulatory bacteria MPL16 and CRL1506 were also analyzed. Among the 49 genes shared by WCSF1 and CRL1506 we found a cell wall protein with an LPxTG domain (OG0002819), two α/β hydrolases (OG0002845, OG0002846), an amidohydrolase (OG0002874), an N-acetyl-neuraminatolyase (OG0002840), a sugar epimerase (OG0002842), and four subunits of an N-acetyl-galactosamine transporter PTS complex (OG0002876, OG0002877, OG0002878, OG0002879). Among the 23 genes shared by WCSF1 and MPL16, a glucose transporter (OG0002666), an aldose-1-epimerase (OG0002993), a deoxyribose-phosphate aldolase (OG0002996), a ceramidase (OG0003015) and an amidohydrolase (OG0003016) were detected.
Considering that the Venn diagram analysis revealed differences in the content of GH and GT, the difference between the strains with respect to the number of genes for GH and GT was further investigated in detail. Previous genomic analyzes found that L. plantarum has six main families of enzymes involved in carbohydrate metabolism: GH, GT, carbohydrate esterases, carbohydrate-binding enzymes, auxiliary active enzymes, and polysaccharide lyases [11]. The study also found that the most abundant sugar metabolism genes in the L. plantarum strains belonged to the GH family, followed by the GT family. Bearing in mind that these enzymes affect the ability of the strains to adapt to their environments, we then proceeded to study the number of genes that code for these enzymes. As observed in Figure 4, genes for several GT and GH families were detected in the genomes of L. plantarum MPL16, CRL1506, CRL681, WCFS1 and TL2766.
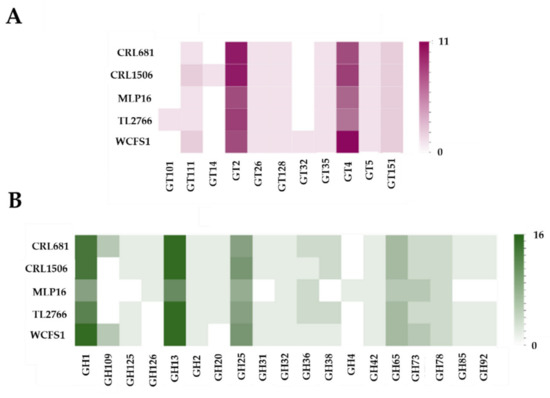
Figure 4.
Study of the number of genes of the different families of glycosyltransferases (GT) (A) and glycosylhydrolases (GH) (B) among Lactiplantibacillus plantarum strains WCFS1, TL2766, MPL16, CRL681 and CRL1506. The analysis of GH and GT was performed with the dbCAN2 server.
The most abundant GT families in L. plantarum were GT2 and GT4. The highest number of genes for GT2 was found in the CRL1506 and CRL681 strains, whereas L. plantarum WCSF1 had the highest number of genes for GT4. The WCSF1 strain was also the one with the highest number of genes for GT111 and the only one with GT32 (Figure 4). On the other hand, the analysis of GH gene numbers showed that GH1 and GH13 were the most abundant in the L. plantarum strains studied. The highest number for GH1 was found in the WCSF1 strain, whereas L. plantarum MPL16 had the lowest number of genes for GH13 (Figure 4). The MPL16 strain also presented a lower number of genes for the GH1, GH25, and GH65 families compared with the other strains evaluated and it did not contain genes for GH31 or GH38 families. These results show a variability between the strains when considering genes associated with carbohydrate metabolism, in line with previous genomic studies [10,11].
Unique genes or genes shared between specific strains detected in our comparative genomic analysis are involved in bacterial cell division, membrane biosynthesis, cell wall peptidoglycan biosynthesis and catabolism, EPS biosynthesis, or they encode for proteins that are secreted or expressed on the cell surface. These results suggest that the cell wall and surface molecules expressed in the different strains of L. plantarum could be involved in their differential ability to modulate the innate antiviral immune response.
2.2. Study of “Probiotic Markers” Genes
Some studies have proposed a set of genes involved in stress resistance, active metabolism in the host, adhesion, and immunomodulation as genes with probiotic functions [13,16,17]. Based on such studies, Carpi et al. [1] recently created an updated list of “probiotic marker” genes including genes responsible for resistance to stress (acid, osmotic, oxidative, temperature), bile salt hydrolase activity, adhesion capacity, and intestinal persistence. A total of 75 probiotic marker genes have been reported, of which approximately 70% correspond to genes located in the core/soft core genome (Table S2), whereas 12 genes are located in the shell/cloud genome (Table 2).

Table 2.
“Probiotic marker” genes of the shell/cloud genome of Lactiplantibacillus plantarum.
We next analyzed the presence/absence of these probiotic marker genes in the genomes of the five strains evaluated in this work. As expected, most of the core genome probiotic marker genes were found in L. plantarum MPL16, CRL1506, CRL681, TL2766 and WCFS1 (Figure 5).
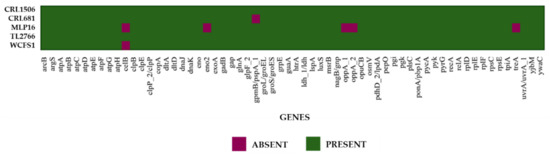
Figure 5.
Analysis of the presence/absence of probiotic marker genes from the core genome proposed by Carpi et al. [1] for the species Lactiplantibacillus plantarum. The genomes of L. plantarum strains WCFS1, TL2766, MPL16, CRL681 and CRL1506 were studied.
The celB (cellobiose-specific PTS system component) and gpmB (histidine phosphatase family protein) genes were not detected in the WCFS1 and CRL681 genomes, respectively. The analysis also revealed that L. plantarum MPL16 does not possess the celB, eno2 (enolase 2), oppA1 (OppA1 oligopeptide-binding protein), oppA2 (OppA2 oligopeptide-binding protein), nor treA (trehalose-6-phosphate hydrolase) genes belonging to the core genome of probiotic genes proposed for this bacterial species (Figure 5). It should be noted that the treA and celB genes have been associated with persistence in the gastrointestinal tract, whereas eno2, oppA1 and oppA2 are involved in resistance to bile salts [1].
In addition, when the analysis was carried out considering the probiotic marker genes of the cloud genome, an absence of several of them was found in the L. plantarum CRL1506, CRL681, TL2766 and WCFS1 genomes (Figure 6). The genes bshA (bile salt hydrolase), clpP (Clp-ATP-dependent protease), gbuB (glycine/carnitine transport protein GbuB), gla2 (aquaporin), oppA4 (oligopeptide-binding protein OppA4), and xylA (xylose isomerase) were not detected in any of the four genomes. The study also showed that L. plantarum MPL16 does not have the bshA, clpP, gla2, oppA4 and xylA genes. The absence of cbh (coliglycin hydrolase) and oppA3 (OppA3 oligopeptide binding protein) was also observed in the MPL16 strain, whereas it presented the gbuB gene, which is absent in L. plantarum CRL1506, CRL681, TL2766 and WCFS1 (Figure 6). The clpP gene was associated with resistance to acid stress, whereas gla2 was associated with resistance to osmotic stress [1]. On the other hand, bshA, cbh, oppA3 and oppA4 are involved in resistance to bile salts [1].
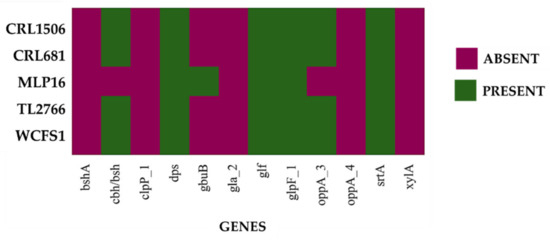
Figure 6.
Analysis of the presence/absence of probiotic marker genes from the cloud genome proposed by Carpi et al. [1] for the species Lactiplantibacillus plantarum. The genomes of L. plantarum strains WCFS1, TL2766, MPL16, CRL681 and CRL1506 were studied.
The study of the presence/absence of the probiotic marker genes proposed for L. plantarum [1] revealed a notable difference between the MPL16 strain and the other strains evaluated. However, the analysis was not able to clearly discriminate the other immunomodulatory strains from the non-immunomodulatory strains evaluated in this work. Considering these results, a comparative study focused on the molecules of the bacterial surface that could be responsible for the interaction of L. plantarum strains with the host’s immunological receptors was carried out.
2.3. Study of Genes Associated with the Synthesis of Exopolysaccharides
It was reported that L. plantarum might have four clusters for EPS biosynthesis [12]. To analyze whether the MPL16, CRL1506, CRL681 and TL2766 strains had these EPS biosynthesis-related genes in their genomes, a sequence comparison was made using the genes described in L. plantarum WCFS1 as a reference (Figure 7). The genes corresponding to the eps1 cluster (or cps1) were not found in the genomes of any of the L. plantarum studied, except for the eps1C and eps1D genes that were found in the CRL1506 strain and the eps1H gene that was observed in L. plantarum CRL681.
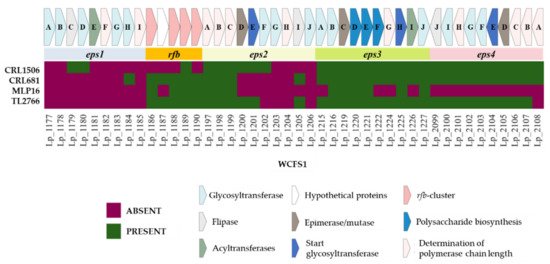
Figure 7.
Analysis of the presence/absence of genes involved in the biosynthesis of exopolysaccharides (EPS) in Lactiplantibacillus plantarum. The genomes of L. plantarum strains TL2766, MPL16, CRL681 and CRL1506 were studied and compared with the probiotic strain WCFS1 as reference. Four clusters for EPS biosynthesis (eps1, eps2, eps3 and eps4) and the rfb cluster involved in the incorporation of rhamnose to EPS are shown.
The first five genes of the eps2 (or cps2) cluster have been reported to be highly conserved among L. plantarum strains, including the eps2A, esp2B, and eps2C genes (also called wzd, wze, and wzh, respectively) that are involved in regulation of EPS biosynthesis dependent on tyrosine phosphoregulation [12,20]. In line with these previous studies, it was observed that L. plantarum MPL16, CRL1506, CRL681 and TL2766 possess the three genes eps2A, esp2B and eps2C (Figure 7). The other two conserved genes are eps2D, which encodes an epimerase enzyme that catalyzes the epimerization of UDP-acetylglucosamine to UDP-N-acetylgalactosamine, common precursors for EPS biosynthesis, and eps2E, which encodes a glycosyltransferase that mediates the initiation of synthesis of the repeated unit of the EPS [21]. Interestingly, these two genes were detected in L. plantarum CRL1506 and TL2766 but not in strains MPL16 or CRL681 (Figure 7). The other components of the eps2 cluster comprising eps2F-J have been shown to possess very limited sequence homology between the corresponding regions from different strains [12]. The eps2G, eps2H and eps2J genes of L. plantarum WCFS1 were not detected in any of the strains studied whereas eps2F was only observed in CRL1506 and eps2I in CRL681 and TL2766 (Figure 7).
Sequence and homology analysis revealed that eps3 is relatively conserved among different strains of L. plantarum, but that its genetic arrangement was different from that of eps2 [12]. It was observed that L. plantarum CRL1506, CRL681 and TL2766 possess the complete eps3 cluster, whereas in strain MPL16 the EPS biosynthesis membrane proteins cps3F (lp_1222) and cps3G (lp_1224) were not detected, nor was the O-acetyltransferase cps3I (lp_1226) (Figure 7). The eps3A gene (lp_2115) was also not observed in the MPL16 genome. On the other hand, it has been reported that the organization of the eps4 (or cps4) cluster in L. plantarum WCFS1 and NUC116 is similar to that of eps2 with esp4A-J genes [12]. The bioinformatic analysis carried out in this work showed that L. plantarum CRL1506, CRL681 and TL2766 have the complete cluster, except for the cps4A gene that codes for the EPS chain length regulatory protein (lp_2108), which is absent in the TL2766 strain. None of the eps4 cluster genes were detected in MPL16 (Figure 7).
It has been described that some strains of L. plantarum such as ST-III possess a group of conserved rmlACBD genes also called rfb (Figure 7). These genes produce a rhamnose precursor that is essential for the biosynthesis of some EPS [22]. Consistently, rhamnose has been detected as a component of surface EPS in L. plantarum WCFS1 [20]. Five of the four genes of this cluster were not detected in the genome of L. plantarum CRL1506, whereas the lp_1187 gene was not detected in the MPL16 strain. Both L. plantarum CRL681 and TL2766 displayed the complete rfb cluster in their genomes (Figure 7).
2.4. Study of Extracellular Proteins
The extracellular proteins, which together constitute the secretome, are involved in variable processes such as adherence to the host, recognition, degradation and absorption of nutrients, as well as signal transduction. It was suggested that extracellular and surface-exposed proteins play important roles in the various interactions that lactobacilli establish with their environments [23,24]. Therefore, we studied the genes that code for extracellular proteins as well as for their transport systems in L. plantarum MPL16, CRL1506, CRL681 and TL2766.
Several mechanisms of protein secretion have been characterized in Gram-positive bacteria, including the Sec (secretion), Tat (twin arginine translocation), FEA (flagella export apparatus), FPE (fimbrilin protein exporter), holins, and Wss (WXG100 secretion system) [25]. Of these extracellular protein transport systems, Sec, holins, and FPE are the most common in L. plantarum genomes [12,26].
The Sec system is the main modulator of protein transport across the cell membrane in Gram-positive bacteria [27]. Most of the genes of the Sec system were identified in the genomes of L. plantarum MPL16, CRL1506, CRL681 and TL2766 when the comparative analysis was performed with the WCFS1 strain (Figure 8). All the genomes analyzed presented the genes for secA (ATP-dependent motor protein), secYEG (membrane channel-forming complex) and two yidC genes (membrane protein insertases). The Sec system also has two proteins encoded by the ftsY and ffh genes, which bind to the cell membrane and interact directly with the secYEG channel, forming a binding pocket for the signal sequence that allows the preprotein to be recognized and presented to the secYEG channel [27]. Both genes, ftsY and ffh, were detected in the genomes of MPL16, CRL1506, CRL681 and TL2766 (Figure 8). It was established that this system also has three genes (secD, secF and YajC) that make up the trimeric complex secDF-YajC, which directs the motor protein SecA [27]. The secD or secF genes were not detected in the genomes of the L. plantarum in this study, which is in line with previous reports for the WCFS1 strain [26] as well as for other probiotic strains such as L. plantarum NCU116 [12]. On the other hand, it has been shown that YajC can bind to YidC and thus control the movement of the polypeptide chain through the secYEG channel, suggesting that YajC fulfills the function of preprotein translocation in L. plantarum NCU116 and WCFS1 [12] as well as in the strains of this work.
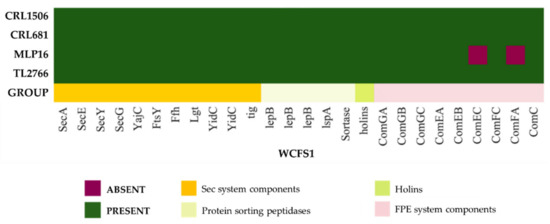
Figure 8.
Analysis of the presence/absence of genes involved in extracellular protein transport systems in Lactiplantibacillus plantarum. The genomes of L. plantarum strains TL2766, MPL16, CRL681 and CRL1506 were studied and compared with the probiotic strain WCFS1 as reference. The Sec (secretion), protein sorting peptidases, holins, and FPE (fimbrilin protein exporter) secretion systems are shown.
The FPE pathway allows uptake of exogenous DNA across the plasma membrane [28] whereas holins are small integral membrane proteins associated with permeabilizing lytic enzymes found in most lactobacilli [29]. Recently it was suggested that these two protein translocation systems could complement the Sec system in protein secretion function in L. plantarum [12]. Studies described that L. plantarum WCFS1 [26] and NCU116 [12] possess genes for the main components of the FPE system including the comGA-GC operon and the comC homologue, but to lack the comGD and comGF genes. Similarly, the L. plantarum strains analyzed in this study had the comGA-GC and comC genes (Figure 8) but not the comGD and comGF genes. Single copies of the comE and comF genes were identified in other species of lactobacilli, which are also involved in the DNA uptake process [30]. Similar to the WCFS1 strain, L. plantarum CRL1506, CRL681 and TL2766 presented the comEA, comEB, comEC, comFC and comFA genes (Figure 8). However, comEC and comFA were not detected in the genome of L. plantarum MPL16. The comEC gene codes for a channel protein involved in DNA binding and uptake whereas the comFA gene codes for a membrane-associated DNA-dependent ATPase. The genes encoding holins were found in the genomes of MPL16, CRL1506, CRL681 and TL2766 (Figure 8). One holin gene copy was found in each strain (Table S3) as described for L. plantarum WCFS1 [26]. It should be noted that this bacterial species may have more than one copy of this gene, as has been reported for L. plantarum NCU116, which has three copies [12].
The extracellular proteins in the L. plantarum strains evaluated in this work were identified by bioinformatic analysis taking into account their subcellular location (secreted or expressed on the surface), their type of anchorage and the secretory mechanism (proteins with signal peptides), following the guidelines previously described for L. plantarum WCFS1 [26] and NCU116 [12]. The WCFS1 genome was used as reference (Figure 9).
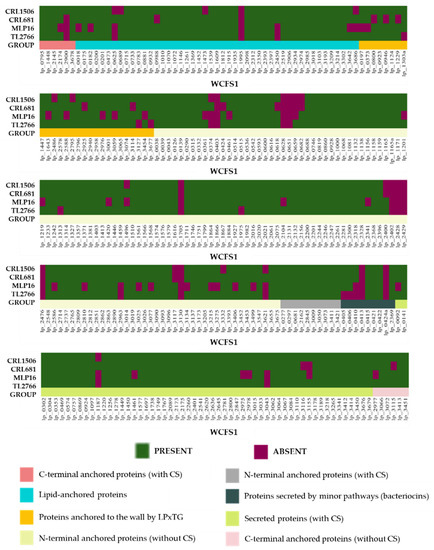
Figure 9.
Analysis of the presence/absence of extracellular proteins in the Lactiplantibacillus plantarum. The genomes of L. plantarum strains TL2766, MPL16, CRL681 and CRL1506 were studied and compared with the probiotic strain WCFS1 as reference. Extracellular proteins were grouped in C-terminal anchored proteins with and without cleavage sites (CS), N-terminal anchored proteins with and without CS, lipid-anchored proteins, proteins anchored to the wall by the LPxTG domain, secreted proteins with CS and proteins secreted by minor pathways.
We studied 304 proteins which were classified into eight groups, including: C-terminal anchored proteins (with and without cleavage sites), N-terminal anchored proteins (with and without cleavage sites), lipid-anchored proteins, proteins anchored to the wall by the LPxTG domain, secreted proteins (with cleavage sites) and proteins secreted by minor pathways (bacteriocins). Contrary to what was observed for the genes of protein secretion systems (Figure 8), notable differences were detected between the strains studied when analyzing the genes for extracellular proteins (Figure 9).
In L. plantarum MPL16, genes for OppA proteins (lp_0018, lp_3686, lp_0092), a mannosyl-beta-N-acetylglucosmidase (lp_0182), an ABC sugar transporter (lp_3642), two membrane-bound surface hydrolases (lp_0461, lp_2586), a lysine (lp_2810), a YxeA family protein (lp_3134), the shikimate 5-dehydrogenase aroD4 (lp_3499), the cell division protein FtsK (lp_1461), and various surface protein precursors (lp_2174, lp_0197, lp_0373, lp_1643, lp_02588, lp_2976, lp_3059, lp_2977) were not detected. The initiation glycosyltransferase cps2E (lp_1201) and a GH (lp_1187) were also not detected in the MPL16 strain (Figure 9) in agreement with the results obtained previously (Figure 7). On the other hand, the genes for the iron ABD transporter fecB (lp_1473), for a surface protein of the GY family (lp_2486), and for the transposase ISP2-C (lp_3332) were not detected in L. plantarum CRL1506 (Figure 9). It was also observed that both MPL16 and CRL1506 do not possess the gene for the MoeB protein (lp_1496) involved in molybdopterin biosynthesis. L. plantarum CRL681 was the only strain in which the activator of the sorbitol operon srlM1 (lp_3621) and a GH (lp_1187) were detected. In addition, genes for GY-family surface protein (lp_0946) and cell surface-bound hydrolase (lp_0618) were not found in the genome of L. plantarum CRL681.
These results reveal that the surface of each of the strains of L. plantarum studied in this work owns a great variation in the surface/secreted proteins.
2.5. Study of Adhesion Factors
Adhesion factors are believed to help probiotic bacteria persist in the host’s gastrointestinal tract and compete against pathogens as well as stimulate the immune system. Therefore, the property of adhesion to cells and molecules of the intestinal mucosa has been associated with health-promoting effects. We next investigated the presence of genes involved in the adhesion to the gastrointestinal tract in the genomes of L. plantarum MPL16, CRL1506, CRL681, TL2766 and WCFS1. Proteins with multiple copies of the MucBP or Mub_B2 mucin-binding domains have been identified in L. plantarum strains [12]. Genomic analysis revealed that the five strains evaluated in this work presented several proteins with multiple MucBP domains (PF06458) (Figure 10). All the strains presented the protein MucBP1 protein. L. plantarum CRL1506, CRL681, TL2766 and WCFS1 but not MPL16 had the genes for MucBP2 protein, whereas L. plantarum CRL681, TL2766, MPL16 and WCFS1 but not CRL1506 contained the MucBP3 protein. MucBP4 protein was detected in all strains except L. plantarum CRL681 (Figure 10). The MucBP5 protein, which in addition to contain MucBP domains has LPxTG cell wall anchor domains (IPR019931), was found in WCFS1, MPL16, CRL1506 and TL2766 strains.
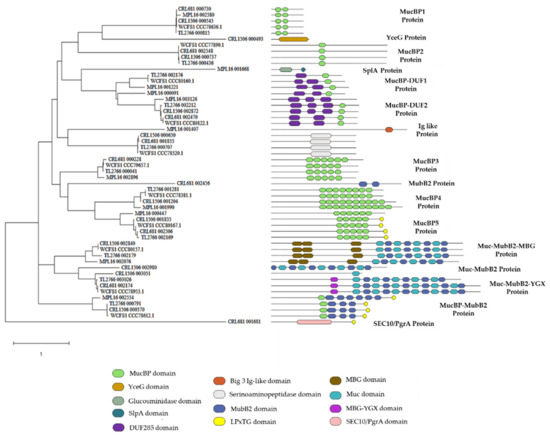
Figure 10.
Phylogeny analysis of mucin-binding domain proteins in Lactiplantibacillus plantarum. The genomes of L. plantarum strains TL2766, MPL16, CRL681 and CRL1506 were studied and compared with the probiotic strain WCFS1.
Proteins simultaneously containing MucBP and DUF285 domains (PF03382) were also found. One copy of the gene encoding the MucBP-DUF1 protein was detected in the genomes of TL2766 and WCFS1, whereas L. plantarum MPL16 had two genes for this putative adhesion molecule (Figure 10). Analysis of their sequences showed that they are not copies of each other (data not shown). On the other hand, the five strains presented the MucBP-DUF2 protein. Two proteins with alternating mucin-binding domains (PF17965) and MubB2-like domains (PF17966) were also observed. One of these proteins also had MGB domains (PF17883) and was designated here as Muc-MubB2-MBG protein. This protein was found in the genomes of L. plantarum CRL1506, TL2766, MPL16 and WCFS1 but not in the CRL681 strain (Figure 10). The other protein also possessed YGX-like MBG domains and was designated as Muc-MubB2-YGX protein. It was found in the genomes of L. plantarum CRL681, TL2766, MPL16 and WCFS1 and partially in the genome of the CRL1506 strain (Figure 10). A protein with a MucBP domain and several MubB2 domains was also detected in all strains except in L. plantarum CRL681.
Interestingly, some of the proteins with known adhesion-mediating domains were found exclusively in one of the L. plantarum strains studied. A protein with alternating MubB2 and mucin-binding domains but without MBG or YGX domains (Muc-MubB2 protein) was detected only in L. plantarum CRL1506 (Figure 10). It was also observed that only the CRL1506 strain had a protein with a YceG-type domain (PF02618), which is an endolytic murein transglycosylase (also known as MltG, YrrL or YceG) and functions as a peptidoglycan terminase by endolytically cleaving strands of nascent peptidoglycan to finish their elongation. It has been proposed that this protein is involved in the adhesion of Lacticaseibacillus paracasei under conditions of heat stress [31]. Only L. plantarum CRL681 had a protein with two MubB2 domains and a protein with a SEC10/PgrA surface exclusion domain (Figure 10). The SEC10/PgrA domain has been described in adhesins from Gram-positive bacteria such as SpyAD from group A streptococci and has been implicated in adhesion to the cell surface [32]. Proteins containing this domain have also been described in strains of Limosilactobacillus reuteri and it has been postulated that they mediate adhesion to the intestinal mucosa [33].
On the other hand, it was observed that only L. plantarum MPL16 has a protein with a SplA domain (PF03217) that also had a domain with mannosyl-endo-beta-N-acetylglucosaminidase activity (PF01832) (Figure 10). SplA domains have been associated with the ability of Lactobacillus acidophilus and Levilactobacillus brevis to adhere to intestinal epithelial cells (IECs) [34]. Additionally, only the MPL16 strain had a protein with a bacterial domain similar to immunoglobulin (Ig-like Big_3, PF07523).
In addition, the analysis of the secretome in L. plantarum WCFS1 identified other adhesion factors and three of them contained domains to adhere to collagen, chitin, and fibronectin [24]. These include lp_1229, which encodes the Msa protein. The deletion of the msa gene results in the loss of the ability of L. plantarum WCFS1 to agglutinate with yeast [35]. Recent studies comparing the mannose-specific binding ability of L. plantarum WCFS1 and 299v showed that L. plantarum 299v is superior to WCFS1 [36]. Variations in the promoter region of the msa gene produced higher levels of expression of the Msa adhesin in L. plantarum 299v than in WCFS1. On the other hand, it was reported that the adhesion capacity of the highly adhesive strains L. plantarum AR326 and AR269 depends on a protein that is 100% homologous with the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) of L. plantarum WCFS1 [37]. The results indicated that GAPDH mediates the adhesion of these lactobacilli to IECs. Studies carried out with L. plantarum KLDS1.0391 showed that the luxS gene plays an important role in the bacteria’s resistance to the gastrointestinal environment as well as in its adhesion capacity [38]. Therefore, the presence of the genes msa, gapdh and luxS as well as collagen binding proteins (cbp) and fibronectin (fbp) in the genomes of L. plantarum MPL16, CRL1506, CRL681, TL2766 and WCFS1 were also investigated (Figure 11).
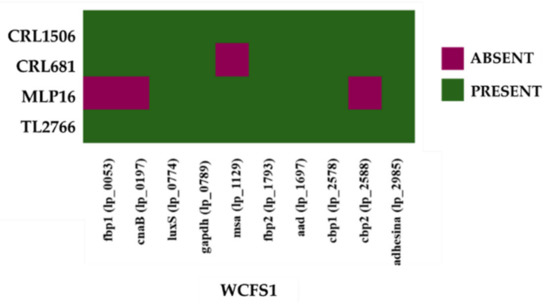
Figure 11.
Analysis of the presence/absence of adhesion factor genes in Lactiplantibacillus plantarum. The genomes of L. plantarum strains TL2766, MPL16, CRL681 and CRL1506 were studied and compared with the probiotic strain WCFS1 as reference.
Of the two fibronectin-binding proteins detected in the L. plantarum WCSF1 genome, only fbp2 was present in all the strains, whereas the gene for fbp1 was absent in the genome of the MPL16 strain. Two collagen-binding proteins were also observed. Of these, cbp1 was present in all the studied strains, whereas the cbp2 gene was not detected in L. plantarum MPL16 (Figure 11). In addition, the luxS, gapdh, aad (alpha-acetolactate decarboxylase) genes and a putative cell surface adhesin were detected in all L. plantarum strains (Figure 11). The gene for Cna protein type B (cnaB) was detected in L. plantarum CRL1506, CRL681 and TL2766 but not in MPL16 whereas msa was absent in the genome of L. plantarum CRL681.
2.6. Study of Lipoproteins and Teichoic Acids
Some studies have established that the composition of the lipoproteins expressed on bacterial surfaces conditions their interaction with the host’s immune system, since they are considered important microbial-associated molecular patterns (MAMPs) capable of being recognized by receptors such as TLR2. In this sense, it has been shown that the lgt gene (lp_0755), which encodes prolipoprotein diacylglyceryl transferase, an enzyme responsible for the lipidation of lipoprotein precursors, is important for the immunomodulatory properties of the probiotic strain L. plantarum WCFS1 [39]. As can be seen in Figure 12, the lgt gene was detected in all the L. plantarum strains evaluated in this work. Species belonging to the Lactobacillus group have been reported to be producers of diacylated lipoproteins [40]. However, recent studies with L. plantarum WCFS1 have suggested that this species could also contain triacylated lipoproteins [39]. It has been reported that the genome of L. plantarum WCFS1 has three acyltransferases (lp_0856, lp_0925 and lp_1181) that could be involved in lipoprotein triacylation. Bioinformatic analysis revealed that the acyltransferases lp_0856 and lp_0925 are present in the genomes of MPL16, CRL1506, CRL681, TL2766 and WCFS1 strains (Figure 12). The acyltransferase lp_1181 is also present in all strains except in L. plantarum MPL16.
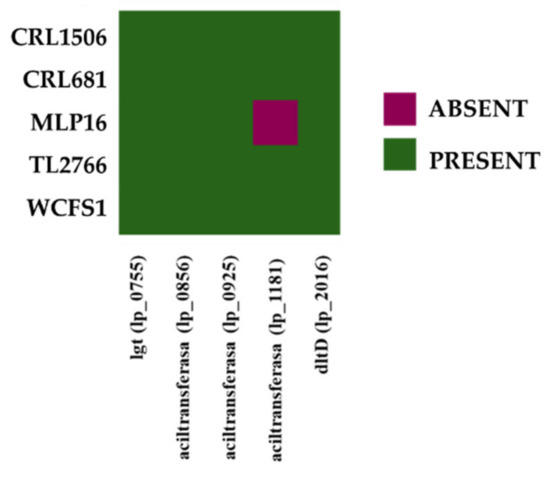
Figure 12.
Analysis of the presence/absence of genes involved in the biosynthesis of lipoproteins and teichoic acids in Lactiplantibacillus plantarum. The genomes of L. plantarum strains TL2766, MPL16, CRL681 and CRL1506 were studied and compared with the probiotic strain WCFS1 as reference.
Teichoic acids (TA), and in particular, lipoteichoic acids (LTA), have also been shown to play an important role in the immunomodulatory activity of L. plantarum WCFS1 both in vitro [41,42] and in vivo [43]. These studies demonstrated that the deletion of the dltX-D (or dltD) gene, involved in the incorporation of D-alanine into LTA, alters the immunomodulatory properties of L. plantarum WCFS1. Our own studies using a dltD mutant of L. plantarum CRL1506 revealed the importance of LTA in the immunomodulatory activity of the strain in the context of intestinal inflammation mediated by TLR3 activation [44]. As expected, the dltD gene was found in all five L. plantarum genomes studied (Figure 12).
3. Discussion
In this study, the complete genomes of L. plantarum MPL16, CRL1506, CRL681, TL2766 and WCFS1 were compared with the aim of identifying the genes involved in their differential immunomodulatory properties. The comparative analysis of the genes involved in the metabolic pathways of the five strains under study did not reveal differences in the metabolism of amino acids, lipids, nucleotides, cofactors and vitamins or in the genes associated with energy metabolism. However, differences were found between MPL16, CRL1506, CRL681, WCFS1 and TL2766 strains when considering carbohydrate metabolism pathways, particularly in the presence/absence of GH and GT. These findings are in line with previous studies carried out for other L. plantarum strains. Molenaar et al. [45] investigated 20 L. plantarum strains and reported that genes involved in protein and lipid synthesis or degradation were largely conserved, whereas genes involved in sugar transport and catabolism were highly variable among the strains. In another study, 24 different phenotypes were found when 185 strains of L. plantarum were studied to determine their fermentation and growth characteristics [3], indicating differences among the strains in their ability to metabolize sugars. These studies were complemented and extended by subsequent genomic studies in which 108 [10], 127 [1] and 133 [11] different strains of L. plantarum were used. They confirmed the wide variety of phenotypes that can be found in this species when carbohydrate metabolism is considered. Genomic studies focused on the carbohydrate-active enzymes of L. plantarum found no significant differences in the number of genes in the families of carbohydrate esterases, carbohydrate-binding enzymes, auxiliary active enzymes, and polysaccharide lyases for 114 L. plantarum isolated from different niches [11]. The same study found variations in the numbers of GH and GT genes in L. plantarum strains. The GH family includes the most abundant enzymes, which are necessary for the degradation of glycosidic bonds between carbohydrates. The GH encoded in all L. plantarum strains were members of the GH1, GH109, GH13, GH31 and GH25 families. On the other hand, within the GT family, GT2 (necessary for cellulose synthesis) and GT4 (necessary for sucrose synthesis) were identified in all L. plantarum strains.
One study showed a lack of association between gene content and habitat when various strains of L. plantarum were studied [46]. However, more recent works identified that certain origins were weakly associated with two phylogenetic groups based on SNP analysis: groups that were designated as G2 and G3 [10]. G2 contained L. plantarum strains isolated mainly from meat samples whereas G3 contained strains from plants. In groups G1, G4 and G5, strains from plants, milk and dairy products as well as intestine isolates were located. For the five defined phylogenetic groups, no critical differences in genome size, number of coding sequences, or G + C content was found. However, the study found variations in the presence of some genes in the genomes of the five groups [10]. Strains in G1 and G2 had a higher capacity to metabolize various carbohydrates such as glucose, fructose, galactose, and lactose, but genes related to these metabolic processes were rare in groups G4 and G5. Thus, the differences between L. plantarum MPL16, CRL1506, CRL681, TL2766 and WCFS1 in relation to carbohydrate metabolism could be associated with the different origin of each of the strains (Table 1). It should be noted that the differences in the metabolic capacities of the MPL16, CRL1506, CRL681 and TL2766 strains, and therefore their ability to persist more efficiently in the gastrointestinal tract, could not be associated with their different immunomodulatory capacities.
In an attempt to explain the differences observed between the L. plantarum strains in this study, a group of genes proposed as “probiotic markers” in a recent study published by Carpi et al. [1] was first analyzed. The study performed a comprehensive pan-genomic analysis of L. plantarum considering only closed complete genomes available in open access repositories. The ultimate goal was to generate information that would serve as a reference to help in the characterization and identification of genes involved in the beneficial properties of potential probiotic strains. The comparative genomics study using the complete genomes of 127 L. plantarum strains identified 1436 genes in the core genome, 414 in the soft core, 1858 genes in the shell, and 13,203 genes in the cloud genome, respectively, out of a total of 16,911 genes. Interestingly, this work detected 75 “probiotic marker” genes in the genomes of L. plantarum [1] based on previous works that proposed that genes associated with stress resistance, active metabolism in the host, adhesion and intestinal persistence are involved in the beneficial properties of lactobacilli [13,16,17]. Of the 75 probiotic marker genes, 70% corresponded to genes located in the core and soft-core genome. Most of these genes were present in all the L. plantarum genomes used for the study [1]. On the other hand, 12 genes (clpP1, bshA, oppA3, oppA4, srtA, xylA, gbuB, gla2, glf, dps, glpF1 and cbh/bsh) were located in the shell and cloud genome, indicating that they were only present in some strains. In fact, genes such as bshA and oppA4 were found in only one strain of L. plantarum each. Interestingly, the study did not correlate a gene or group of genes with documented probiotic properties for the L. plantarum strains. For example, the list of strains that possessed any of these 12 genes did not include L. plantarum WCFS1 and strains with documented probiotic activity such as L. plantarum Zhang-LL [47] or NCU116 [48,49,50,51] possessed 2 and 3 of those 12 genes, respectively. As a first attempt to find genes that could explain the differential immunomodulatory activity of the MPL16, CRL1506, CRL681, TL2766 and WCFS1 strains, the study of the presence/absence of the probiotic marker genes proposed for L. plantarum was carried out [1]. Although the results revealed a marked difference between L. plantarum MPL16 and the other strains, it was not possible to clearly discriminate the immunomodulatory strains from the non-immunomodulatory ones. Considering these results, a comparative study focused on the molecules of the bacterial surface that could be responsible for interacting with the host’s immunological receptors: EPS, adhesion factors, and LTA was carried out.
Studies performed by our research group demonstrated that the EPS produced by some immunobiotic bacteria have important roles in the immunomodulatory effect in the context of TLR3 activation in PIE cells. We showed that EPS produced by Lactobacillus delbrueckii TUA4408L [52], L. delbrueckii OLL1073R-1 [53], and Streptococcus thermophilus ST538 [44] participate in the ability of immunobiotic strains to increase the production of type I interferons (IFNs) and antiviral factors in PIE cells after poly(I:C) challenge. It was reported that L. delbrueckii TUA4408L and its EPS are capable of increasing the activation of the transcription factors IRF-3 and NF-κB, leading to an increase in the expression of IFN-β, MxA and RNAseL and a decrease in the replication of the rotavirus in PIE cells [52]. It was also described that the EPS of the strain OLL1073R-1 increases the expression of IFN-α, IFN-β, MxA and RNAseL in PIE cells challenged with poly(I:C) and that this effect depends on its ability to regulate the expression of the A20 protein [53]. The capacity of EPS-producing S. thermophilus ST538 to modulate the TLR3-mediated innate antiviral response in PIE cells was also demonstrated [44], by a successful development of two mutant strains lacking the epsB or epsC genes, which were unable to produce the macromolecule. Studies in PIE cells demonstrated that EPS from S. thermophilus ST538 was able to significantly enhance IFN-β, IL-6 and CXCL10 expression in response to TLR3 stimulation, whereas in S. thermophilus ΔepsB and ΔepsC mutant strains this effect was significantly decreased [44]. On the other hand, some studies have shown that EPS produced by L. plantarum strains are involved in their immunomodulatory properties. L. plantarum NCU116, isolated from a traditional Chinese sauerkraut called “paocai”, has been shown to possess probiotic functions including immunomodulation ability [54,55]. This strain produces a heteropolysaccharide called EPS116, which has been shown to attenuate intestinal inflammation through the STAT3 signaling pathway and inhibit cancer cell proliferation through the TLR2-dependent mechanism [56]. EPS isolated from L. plantarum JLK0142 and designated EPS0142 has also been described as possessing immunomodulatory properties [57]. In vivo studies in mice immunocompromised by chemotherapy showed that oral administration of EPS0142 increases intestinal IgA levels, serum TNF-α and IL-2 levels, and the proliferative capacity of spleen lymphocytes [57].
Considering this background, studies were carried out to compare the EPS clusters in the genomes of the MPL16, CRL1506, CRL681, TL2766 and WCFS1 strains. Interestingly, L. plantarum MPL16, which is the strain with the most remarkable immunomodulatory activity among the lactobacilli evaluated here, was the strain that did not present any complete EPS cluster of the 4 described for the species. In line with these findings, it has been shown that the MPL16 strain does not have the ability to produce EPS [58]. No differences could be recognized between the EPS clusters that could explain the different immunomodulatory properties of the CRL1506, CRL681 and TL2766 strains.
Adhesion factors are considered key genes in the probiotic activity of many lactobacilli [12,16]. Particularly, in L. plantarum WCFS1, various proteins with adhesion function have been described, including mucin binding proteins, collagen binding proteins, chitin binding proteins, fibronectin binding proteins [24], agglutination and adhesion protein Msa [35,36] as well as GAPDH [57], which are essential for the strain to optimally fulfill its probiotic functions. L. plantarum NCU116 is another strain with probiotic properties for which exhaustive studies of its adhesion factors have been carried out. In its genome, 17 proteins that can be directly associated with adherence were identified through analysis of the composition of their domains. Five of these proteins possess a bacterial immunoglobulin-like domain (Ig-like) including PF17961, PF02368 and PF07523 according to the Pfam database [12]. These proteins are involved in extracellular binding and adhesion processes [59]. Two collagen-binding domain proteins were also found in strain NCU116, which can mediate adherence to host IECs. In addition, 8 proteins with multiple copies of the MucBP (PF06458) or Mub_B2 (PF17966) mucin-binding domains were identified. They play an important role in the adhesion of lactobacilli to the gastrointestinal tract [12]. The presence of a 1343 amino acid protein in L. plantarum NCU116 with MucBP and Mub B2 domains (PF17966) was also reported, which was identical (98.91% homology) to the adhesion protein Msa of L. plantarum WCFS1, which has been shown to participate in bacterial adherence [13,35].
When proteins with adhesion functions in L. plantarum MPL16, CRL1506, CRL681, TL2766 and WCFS1 were evaluated, a great variability between strains was observed (Figure 13). L. plantarum MPL16 differed from the other strains under study due to the absence of the fbp1, cbp2 and cnaB genes; and for being the only one to present genes for a protein with a SplA domain (PF03217) and for a protein with a bacterial domain similar to immunoglobulin (Ig-like Big_3, PF07523).
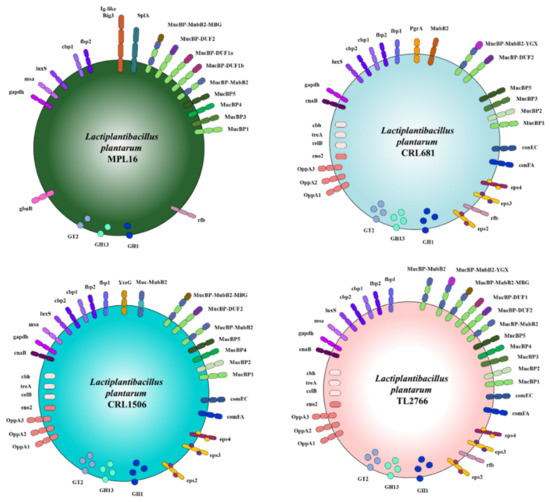
Figure 13.
Schematic representation of glycosyltransferases, glycosylhydrolases, proteins involved in resistance to passage through the gastrointestinal tract and bacterial surface molecules detected in the Lactiplantibacillus plantarum genomes. The genomes of L. plantarum strains TL2766, MPL16, CRL681 and CRL1506 were studied and compared with the probiotic strain WCFS1 as reference.
As mentioned above, the SplA domains have been associated with the ability of L. acidophilus and L. brevis to adhere to IECs [34]. On the other hand, bacterial Ig-like domains, which are frequently observed in cell surface proteins, have been shown to mediate cell-to-cell recognition via surface receptors [60,61]. L. acidophilus NCFM possesses a bacterial immunoglobulin-like domain-containing protein SlaP, which has been designated IgdA [60]. This protein is considered to be conserved among lactobacilli that form the S layer [62]. In silico analyzes revealed that IgdA and the corresponding orthologs were unique to host-adapted lactobacilli, whereas the immunoglobulin domain was specific to vertebrate-adapted species [60]. It was further observed that IgdA mutants of L. acidophilus possessed a visibly altered cell surface, which contributed to their increased salt sensitivity, altered immunogenicity profile, and severely reduced adhesion to intestinal Caco-2 cells, extracellular matrices, and mucin in vitro [60]. L. plantarum MPL16 was the only strain in this study in which the SlaP protein and a protein with bacterial Ig-like domains were detected; however, the data obtained in this work do not allow us to predict whether these proteins would be involved in its differential immunomodulatory activity. The development of mutants of the MPL16 strain that do not express these genes could contribute to revealing their specific role in the capacity of the strain to modulate the intestinal and respiratory antiviral immune response.
Bacterial lipoproteins are widely recognized MAMPs, which interact with TLR2. Lipoproteins are conjugated with two or three acyl (di- or tri-acyl) chains, which are essential for their proper anchoring in the cell membrane, as well as for interaction with TLR2. After export across the cell membrane, lipoprotein precursors (pre-prolipoproteins) undergo lipid modification, which is catalyzed by three conserved enzymes. Prolipoprotein diacylglyceryl transferase (Lgt) transfers a diacylglyceryl moiety to the cysteine residue, which is then directly cleaved by lipoprotein signal peptidase (Lsp) from the N-terminal signal sequence of the lipid-modified cysteine residue. Finally, lipoprotein N-acyl transferase (Lnt) adds a third acyl chain to the free amino group of the lipidated cysteine. Di- and tri-acyl lipoproteins produced by bacteria are differentially recognized by different TLR2 heterodimers: TLR2/6 and TLR1/2, respectively [39].
Information on the role of lipoproteins in the interaction of probiotic bacteria with the host is scarce. By deleting the lgt gene (lp_0755), which encodes prolipoprotein diacylglyceryl transferase, an enzyme responsible for the lipidation of lipoprotein precursors, the functions of the set of lipoproteins in the physiology of the probiotic strain L. plantarum WCFS1 was investigated [39]. These authors compared the ability of Δlgt mutant and wild type strain to stimulate TLR2 signaling and regulate the inflammatory response. The deletion of the lgt gene in the WCFS1 strain significantly reduced its ability to stimulate TLR1/2, whereas its ability to stimulate TLR2/6 signaling was not affected. Using human blood mononuclear cells, it was also shown that the Δlgt mutant induced a proinflammatory profile compared with the wild-type strain, characterized by increased production of IL-12, TNF-α, IL-1β and IL-8 and lower levels of the anti-inflammatory cytokine IL-10 [39]. It was then suggested that the loss of lipoproteins on the surface of the probiotic bacteria WCFS1, which were found in higher concentration in the culture medium, reduced its anti-inflammatory capacity. These results are in contrast to what occurs in pathogens such as Streptococcus agalactiae [63] and Staphylococcus aureus [64], in which the deletion of the lgt gene significantly increased its proinflammatory capacity.
Results of TLR1/2 and TLR2/6 activation showed that triacylated lipoproteins are dominant in L. plantarum WCFS1 [39]. This contrasts with previous reports describing the absence of orthologous genes for lnt, which is responsible for transferring a third acyl chain to the N-terminal cysteine of lipoproteins. Therefore, Gram-positive bacteria with low GC content such as species belonging to the Lactobacillus group have been considered as producers of diacylated lipoproteins [40]. The L. plantarum WCFS1 genome has been found to possess three acyltransferases (lp_0856, lp_0925, and lp_1181) that could be involved in a similar role to lnt in lipoprotein triacylation [39]. In this work, we revealed that the three acyltransferases are present in the genomes of CRL1506, CRL681 and TL2766 strains but that L. plantarum MPL16 does not have the acyltransferase lp_1181. Whether this difference impacts the composition of the surface-expressed lipoproteins of the MPL16 strain and consequently on its interaction with the host’s immune system is an interesting point for future research.
LTA are the most proinflammatory components of the Gram-positive bacterial envelope and this effect has been shown to be highly dependent on D-alanine substitution of the TA backbone [65]. Although the potency of immunomodulatory capacity differs among bacterial strains [66], it has been shown that LTA purified from L. plantarum WCFS1 can induce a potent pro-inflammatory response in immune cells in vitro [41,42], and that this response was dependent on D-alanine substitution in the LTA backbone. Indeed, the absence of D-alanine in LTA changed the ability of L. plantarum WCFS1 and its purified LTA to modulate immune responses in vitro towards a more anti-inflammatory cytokine profile [42]. In vivo studies compared the effects of L. plantarum WCFS1 and its negative mutant for D-Ala in LTA (ΔdltX-D) [43] on the distribution of populations of intestinal and systemic T cells and dendritic cells (DCs) in mice. It was shown that the distribution of not only pro-, but also anti-inflammatory T cell and DCs populations depends on the functionality of the dltX-D gene and LTA D-alanine in the cell envelope of L. plantarum WCFS1 [65]. Not only the proinflammatory immune responses were reduced in the absence of D-Ala substitution, but also anti-inflammatory responses, such as the generation of regulatory T cells. In line with these studies, our group demonstrated that a mutant strain of L. plantarum CRL1506 (Δdlt) has altered immunomodulatory capacity in the context of intestinal inflammation mediated by TLR3 activation compared with the wild-type strain [44]. L. plantarum CRL1506 (Δdlt), similar to the wild strain, was able to increase IFN-β expression levels in PIE cells stimulated with poly(I:C), but did not induce changes in IL-6 levels or MCP-1. The CRL1506 (Δdlt) strain was also unable to increase IL-10 levels or reduce CD3+NK1.1+CD8αα+ intraepithelial lymphocytes or TNF-α, IL-6, or IL-15 levels in the intestine of mice challenged with the TLR3 agonist. As expected, the dltD gene was found in the five genomes of L. plantarum studied (MPL16, CRL1506, CRL681 and TL2766), and therefore could not be associated with their different immunomodulatory capacities.
Interestingly, L. plantarum MPL16 was the strain that presented the least number of genes associated with resistance to passage through the gastrointestinal tract. Compared with CRL1506, CRL681, and TL2766 strains, the genome of L. plantarum MPL16 did not have the cbh, treA, celB, eno2, oppA1, oppA2, and oppA3 genes and was the only one to have the gbuB gene (Figure 13). These results allow us to speculate that the MPL16 strain would be less resistant to passage through the gastrointestinal tract. These findings also allow us to speculate that the action of gastric juice and digestive enzymes could more easily expose surface molecules of L. plantarum MPL16 that could interact with the immunological receptors of the intestinal mucosa.
In summary, the comparison of genes potentially involved in the in vitro and in vivo differential effects of the L. plantarum strains in this work revealed no obvious differences between the bacteria evaluated. The comparative and functional genomic studies carried out do not allow proposing bacterial genes involved in the immunomodulatory effect, since it was not possible to correlate a group of specific genes with the differential immunomodulatory capacity of the L. plantarum strains. Proteomic studies are ongoing to evaluate the possible role of protein expression patterns with the immunomodulatory differences among strains.
4. Materials and Methods
4.1. Microorganisms
L. plantarum CRL1506, L. plantarum CRL681 and L. plantarum MPL16 belong to CERELA Culture Collection. L. plantarum TL2766 was kindly provided by the Culture Collection of the Nutritional Immunology Group of the International Center for Agricultural and Nutritional Immunology of Tohoku University (Sendai, Japan). The published genomic sequence of L. plantarum WCFS1 was obtained from the GenBank database (ID number AL935263.2) [67].
4.2. Assembly and Annotation of Nucleotide Sequences
The quality of each sequence was controlled with FASTQC [68]. Sequences with low quality or with contaminations were eliminated with PrinSeq lite v0.20.4 [69]. The resulting readings were assembled using Bruijn graphs through the SPAdes v3.11.1 tool [70]. The obtained assemblies were uploaded to public access repositories (DDBJ/ENA/GenBank).
Genes were predicted with the Prokaryotic Genome Annotation Pipeline (PGAP) v4.8 using the stand-alone configuration. A virtual machine service in the cloud, provided by Microsoft Azure®, was used because the scorer requires a high computing resource. The configuration used was D8v3 consisting of 32 GB of RAM, 8 Intel Xeon® E5-2673 v3 (Haswell) 2.4 GHz processors and 200 GB of storage.
4.3. Bioinformatic Tools for Comparative Genomics Studies
The functional characterization of individual genes from the genomes of L. plantarum CRL1506, CRL681, MPL16, TL2766 and WCFS1 was performed with the BlastKOALA tool [71].
Amino acid sequences encoded in L. plantarum genomes were compared for ortholog group inference with OrthoFinder v2.5.2. The Venn Diagram was made with the Venn Diagram tool (https://bioinformatics.psb.ugent.be/webtools/Venn/; accessed on 15 November 2021). The dbCAN2 server, which allows automated notation of active carbohydrate enzymes, was used for the analysis of glycosylhydrolases and glycosyltransferases [72].
The GenBank database was used to obtain the amino acid sequences of the genes belonging to the potential probiotic markers, including: proteins involved in the biosynthesis of EPS, extracellular proteins, adhesion factors, lipoproteins and teichoic acids. These genes were searched in the genomes of L. plantarum with BLAST [73] in the stand-alone mode.
The phylogenetic tree was constructed from protein sequences possessing a MucBP domain. Using a multiple alignment program (MUSCLE) [74], the sequences were aligned and the tree was built from the maximum likelihood estimation statistical test [75], both tools are available in MEGAX [76].
Gene presence/absence graphs were performed with Python 3.7 using the Pandas and Seaborn libraries.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/bacteria1030012/s1, Table S1: Orthologous genes obtained by OrthoFinder analysis, with their corresponding PGAP annotation; Table S2: “Probiotic markers” genes of the core/soft core genome; Table S3: Mechanisms of protein secretion in L. plantarum CRL1506, CRL681, MPL16, TL2766 and WCFS1, including Sec (secretion), FPE (fimbrilin protein exporter), holins and peptidases associated with protein sorting.
Author Contributions
Conceptualization, L.A., H.K. and J.V.; methodology, L.A., F.R.T., K.F., Y.S., B.Z., A.A.B. and L.S.; software, L.A. and E.M.H.; validation, L.A., F.R.T., K.F., Y.S., B.Z., A.A.B. and L.S.; formal analysis, L.A. and J.V.; resources, Y.S., S.K., S.F., E.M.H., H.K. and J.V.; data curation, L.A.; writing—original draft preparation, L.A., F.R.T. and J.V.; writing—review and editing, L.S., S.K., S.F., E.M.H. and H.K.; visualization, L.A.; supervision, J.V.; project administration, H.K. and J.V.; funding acquisition, H.K. and J.V. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by ANPCyT-FONCyT grants PICT-2016-0410 and PICT-2018-03264 to Julio Villena, PICT-2018-01378 and PICT-2020-0599 to Elvira Hebert, and PICT-2018-02249 to Silvina Fadda. This study was also supported by a Grant-in-Aid for Scientific Research (A) (19H00965) and Open Partnership Joint Projects of JSPS Bilateral Joint Research Projects from the Japan Society for the Promotion of Science (JSPS), and grants from the project of NARO Bio-oriented Technology Research Advancement Institution (research program on the development of innovative technology, No. 01002AB2) to Haruki Kitazawa, and by JSPS Core-to-Core Program, A. Advanced Research Networks entitled Establishment of international agricultural immunology research-core for a quantum improvement in food safety, and by Tohoku University Research Program “Frontier Research in Duo” (FRiD), and by AMED (Moonshot R&D—MILLENNIA Program) Grant Number JP21zf0127001.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Carpi, F.M.; Coman, M.M.; Silvi, S.; Picciolini, M.; Verdenelli, M.C.; Napolioni, V. Comprehensive pan-genome analysis of Lactiplantibacillus plantarum complete genomes. J. Appl. Microbiol. 2022, 132, 592–604. [Google Scholar] [CrossRef] [PubMed]
- De Vries, M.C.; Vaughan, E.E.; Kleerebezem, M.; de Vos, W.M. Lactobacillus plantarum—Survival, functional and potential probiotic properties in the human intestinal tract. Int. Dairy J. 2006, 16, 1018–1028. [Google Scholar] [CrossRef]
- Siezen, R.J.; Tzeneva, V.A.; Castioni, A.; Wels, M.; Phan, H.T.K.; Rademaker, J.L.W.; Starrenburg, M.J.C.; Kleerebezem, M.; Molenaar, D.; Vlieg, J.E.V.H. Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environ. Microbiol. 2010, 12, 758–773. [Google Scholar] [CrossRef] [PubMed]
- Villena, J.; Li, C.; Vizoso-Pinto, M.; Sacur, J.; Ren, L.; Kitazawa, H. Lactiplantibacillus plantarum as a potential adjuvant and delivery system for the development of SARS-CoV-2 oral vaccines. Microorganisms 2021, 9, 683. [Google Scholar] [CrossRef] [PubMed]
- Behera, S.S.; Ray, R.C.; Zdolec, N. Lactobacillus plantarum with functional properties: An approach to increase safety and shelf-life of fermented foods. BioMed Res. Int. 2018, 2018, 9361614. [Google Scholar] [CrossRef] [Green Version]
- Albarracin, L.; Garcia-Castillo, V.; Masumizu, Y.; Indo, Y.; Islam, M.A.; Suda, Y.; Garcia-Cancino, A.; Aso, H.; Takahashi, H.; Kitazawa, H.; et al. Efficient selection of new immunobiotic strains with antiviral effects in local and distal mucosal sites by using porcine intestinal epitheliocytes. Front. Immunol. 2020, 11, 543. [Google Scholar] [CrossRef]
- Axelsson, L.; Rud, I.; Naterstad, K.; Blom, H.; Renckens, B.; Boekhorst, J.; Kleerebezem, M.; van Hijum, S.; Siezen, R.J. Genome sequence of the naturally plasmid-free Lactobacillus plantarum strain NC8 (CCUG 61730). J. Bacteriol. 2012, 194, 2391–2392. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.-J.; Wang, R.; Gong, F.-M.; Liu, X.-F.; Zheng, H.-J.; Luo, Y.-Y.; Li, X.-R. Complete genome sequences and comparative genome analysis of Lactobacillus plantarum strain 5-2 isolated from fermented soybean. Genomics 2015, 106, 404–411. [Google Scholar] [CrossRef]
- Kwak, W.; Kim, K.; Lee, C.; Lee, C.; Kang, J.; Cho, K.; Yoon, S.H.; Kang, D.-K.; Kim, H.; Heo, J.; et al. Comparative analysis of the complete genome of Lactobacillus plantarum GB-LP2 and potential candidate genes for host immune system enhancement. J. Microbiol. Biotechnol. 2016, 26, 684–692. [Google Scholar] [CrossRef]
- Choi, S.; Jin, G.-D.; Park, J.; You, I.; Kim, E.B. Pan-genomics of Lactobacillus plantarum revealed group-specific genomic profiles without habitat association. J. Microbiol. Biotechnol. 2018, 28, 1352–1359. [Google Scholar] [CrossRef]
- Mao, B.; Yin, R.; Li, X.; Cui, S.; Zhang, H.; Zhao, J.; Chen, W. Comparative genomic analysis of Lactiplantibacillus plantarum isolated from different niches. Genes 2021, 12, 241. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Peng, Z.; Hu, M.; Xiao, Y.-S.; Liu, Z.-G.; Guan, Q.-Q.; Xie, M.-Y.; Xiong, T. Interactions between Lactobacillus plantarum NCU116 and its environments based on extracellular proteins and polysaccharides prediction by comparative analysis. Genomics 2020, 112, 3579–3587. [Google Scholar] [CrossRef] [PubMed]
- Van den Nieuwboer, M.; van Hemert, S.; Claassen, E.; de Vos, W.M. Lactobacillus plantarum WCFS1 and its host interaction: A dozen years after the genome. Microb. Biotechnol. 2016, 9, 452–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimazu, T.; Villena, J.; Tohno, M.; Fujie, H.; Hosoya, S.; Shimosato, T.; Aso, H.; Suda, Y.; Kawai, Y.; Saito, T.; et al. Immunobiotic Lactobacillus jensenii elicits anti-inflammatory activity in porcine intestinal epithelial cells by modulating negative regulators of the toll-like receptor signaling pathway. Infect. Immun. 2012, 80, 276–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villena, J.; Suzuki, R.; Fujie, H.; Chiba, E.; Takahashi, T.; Tomosada, Y.; Shimazu, T.; Aso, H.; Ohwada, S.; Suda, Y.; et al. Immunobiotic Lactobacillus jensenii modulates the toll-like receptor 4-induced inflammatory response via negative regulation in porcine antigen-presenting cells. Clin. Vaccine Immunol. 2012, 19, 1038–1053. [Google Scholar] [CrossRef] [Green Version]
- Lebeer, S.; Vanderleyden, J.; De Keersmaecker, S.C.J. Genes and molecules of lactobacilli supporting probiotic action. Microbiol. Mol. Biol. Rev. 2008, 72, 728–764. [Google Scholar] [CrossRef] [Green Version]
- Muscariello, L.; De Siena, B.; Marasco, R. Lactobacillus cell surface proteins involved in interaction with mucus and extracellular matrix components. Curr. Microbiol. 2020, 77, 3831–3841. [Google Scholar] [CrossRef]
- Nguyen, T.L.; Kim, D.-H. Genome-wide comparison reveals a probiotic strain Lactococcus lactis WFLU12 isolated from the gastrointestinal tract of olive flounder (Paralichthys olivaceus) harboring genes supporting probiotic action. Mar. Drugs 2018, 16, 140. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, L.C.; Saraiva, T.D.; Silva, W.M.; Pereira, U.P.; Campos, B.C.; Benevides, L.J.; Rocha, F.S.; Figueiredo, H.C.; Azevedo, V.; Soares, S.C. Analyses of the probiotic property and stress resistance-related genes of Lactococcus lactis subsp. lactis NCDO 2118 through comparative genomics and in vitro assays. PLoS ONE 2017, 12, e0175116. [Google Scholar] [CrossRef]
- Remus, D.M.; van Kranenburg, R.; van Swam, I.I.; Taverne, N.; Bongers, R.S.; Wels, M.; Wells, J.M.; Bron, P.A.; Kleerebezem, M. Impact of 4 Lactobacillus plantarum capsular polysaccharide clusters on surface glycan composition and host cell signaling. Microb. Cell Fact. 2012, 11, 149. [Google Scholar] [CrossRef] [Green Version]
- Yother, J. Capsules of Streptococcus pneumoniae and other bacteria: Paradigms for polysaccharide biosynthesis and regulation. Annu. Rev. Microbiol. 2011, 65, 563–581. [Google Scholar] [CrossRef] [PubMed]
- Péant, B.; LaPointe, G.; Gilbert, C.; Atlan, D.; Ward, P.; Roy, D. Comparative analysis of the exopolysaccharide biosynthesis gene clusters from four strains of Lactobacillus rhamnosus. Microbiology 2005, 151, 1839–1851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramírez, M.D.F.; Groot, M.N.N.; Smid, E.J.; Hols, P.; Kleerebezem, M.; Abee, T. Role of cell surface composition and lysis in static biofilm formation by Lactobacillus plantarum WCFS1. Int. J. Food Microbiol. 2018, 271, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Boekhorst, J.; Wels, M.; Kleerebezem, M.; Siezen, R.J. The predicted secretome of Lactobacillus plantarum WCFS1 sheds light on interactions with its environment. Microbiology 2006, 152, 3175–3183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desvaux, M.; Hébraud, M.; Talon, R.; Henderson, I.R. Secretion and subcellular localizations of bacterial proteins: A semantic awareness issue. Trends Microbiol. 2009, 17, 139–145. [Google Scholar] [CrossRef]
- Kleerebezem, M.; Hols, P.; Bernard, E.; Rolain, T.; Zhou, M.; Siezen, R.J.; Bron, P.A. The extracellular biology of the lactobacilli. FEMS Microbiol. Rev. 2010, 34, 199–230. [Google Scholar] [CrossRef]
- Driessen, A.J.; Nouwen, N. Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 2008, 77, 643–667. [Google Scholar] [CrossRef] [Green Version]
- Tjalsma, H.; Bolhuis, A.; Jongbloed, J.D.; Bron, S.; van Dijl, J.M. Signal peptide-dependent protein transport in Bacillus subtilis: A genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 2000, 64, 515–547. [Google Scholar] [CrossRef] [Green Version]
- Wang, I.-N.; Smith, D.L.; Young, R. Holins: The protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 2000, 54, 799–825. [Google Scholar] [CrossRef]
- Chen, I.; Dubnau, D. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2004, 2, 241–249. [Google Scholar] [CrossRef]
- Adu, K.T.; Wilson, R.; Baker, A.L.; Bowman, J.; Britz, M.L. Prolonged heat stress of Lactobacillus paracasei GCRL163 improves binding to human colorectal adenocarcinoma HT-29 cells and modulates the relative abundance of secreted and cell surface-located proteins. J. Proteome Res. 2020, 19, 1824–1846. [Google Scholar] [CrossRef] [PubMed]
- Gallotta, M.; Gancitano, G.; Pietrocola, G.; Mora, M.; Pezzicoli, A.; Tuscano, G.; Chiarot, E.; Nardi-Dei, V.; Taddei, A.R.; Rindi, S.; et al. SpyAD, a moonlighting protein of group A streptococcus contributing to bacterial division and host cell adhesion. Infect. Immun. 2014, 82, 2890–2901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wegmann, U.; MacKenzie, D.A.; Zheng, J.; Goesmann, A.; Roos, S.; Swarbreck, D.; Walter, J.; Crossman, L.C.; Juge, N. The pan-genome of Lactobacillus reuteri strains originating from the pig gastrointestinal tract. BMC Genom. 2015, 16, 1023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vélez, M.P.; De Keersmaecker, S.C.; Vanderleyden, J. Adherence factors of Lactobacillus in the human gastrointestinal tract. FEMS Microbiol. Lett. 2007, 276, 140–148. [Google Scholar] [CrossRef] [Green Version]
- Pretzer, G.; Snel, J.; Molenaar, D.; Wiersma, A.; Bron, P.A.; Lambert, J.; de Vos, W.M.; van der Meer, R.; Smits, M.A.; Kleerebezem, M. Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J. Bacteriol. 2005, 187, 6128–6136. [Google Scholar] [CrossRef] [Green Version]
- Holst, B.; Glenting, J.; Holmstrøm, K.; Israelsen, H.; Vrang, A.; Antonsson, M.; Ahrné, S.; Madsen, S.M. Molecular switch controlling expression of the mannose-specific adhesin, Msa, in Lactobacillus plantarum. Appl. Environ. Microbiol. 2019, 85, e02954-18. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Zhang, M.; Zhao, J.; Xia, Y.; Lai, P.F.; Ai, L. A surface protein from Lactobacillus plantarum increases the adhesion of Lactobacillus strains to human epithelial cells. Front. Microbiol. 2018, 9, 2858. [Google Scholar] [CrossRef] [Green Version]
- Jia, F.-F.; Zheng, H.-Q.; Sun, S.-R.; Pang, X.-H.; Liang, Y.; Shang, J.-C.; Zhu, Z.-T.; Meng, X.-C. Role of luxS in stress tolerance and adhesion ability in Lactobacillus plantarum KLDS1.0391. BioMed Res. Int. 2018, 2018, 4506829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, I.-C.; Van Swam, I.I.; Boeren, S.; Vervoort, J.; Meijerink, M.; Taverne, N.; Starrenburg, M.; Bron, P.A.; Kleerebezem, M. Lipoproteins contribute to the anti-inflammatory capacity of Lactobacillus plantarum WCFS1. Front. Microbiol. 2020, 11, 1822. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, H.; Kurokawa, K.; Lee, B.L. Lipoproteins in bacteria: Structures and biosynthetic pathways. FEBS J. 2012, 279, 4247–4268. [Google Scholar] [CrossRef]
- Grangette, C.; Nutten, S.; Palumbo, E.; Morath, S.; Hermann, C.; Dewulf, J.; Pot, B.; Hartung, T.; Hols, P.; Mercenier, A. Enhanced antiinflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc. Natl. Acad. Sci. USA 2005, 102, 10321–10326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirose, Y.; Murosaki, S.; Fujiki, T.; Yamamoto, Y.; Yoshikai, Y.; Yamashita, M. Lipoteichoic acids on Lactobacillus plantarum cell surfaces correlate with induction of interleukin-12p40 production. Microbiol. Immunol. 2010, 54, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Bron, P.A.; Tomita, S.; van Swam, I.I.; Remus, D.M.; Meijerink, M.; Wels, M.; Okada, S.; Wells, J.M.; Kleerebezem, M. Lactobacillus plantarum possesses the capability for wall teichoic acid backbone alditol switching. Microb. Cell Factories 2012, 11, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizuno, H.; Arce, L.; Tomotsune, K.; Albarracin, L.; Funabashi, R.; Vera, D.; Islam, M.A.; Vizoso-Pinto, M.G.; Takahashi, H.; Sasaki, Y.; et al. Lipoteichoic acid is involved in the ability of the immunobiotic strain Lactobacillus plantarum CRL1506 to modulate the intestinal antiviral innate immunity triggered by TLR3 activation. Front. Immunol. 2020, 11, 571. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, D.; Bringel, F.; Schuren, F.H.; de Vos, W.M.; Siezen, R.J.; Kleerebezem, M. Exploring Lactobacillus plantarum genome diversity by using microarrays. J. Bacteriol. 2005, 187, 6119–6127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martino, M.E.; Bayjanov, J.R.; Caffrey, B.E.; Wels, M.; Joncour, P.; Hughes, S.; Gillet, B.; Kleerebezem, M.; van Hijum, S.A.; Leulier, F. Nomadic lifestyle of Lactobacillus plantarum revealed by comparative genomics of 54 strains isolated from different habitats. Environ. Microbiol. 2016, 18, 4974–4989. [Google Scholar] [CrossRef]
- Chen, C.; Li, J.; Zhang, H.; Xie, Y.; Xiong, L.; Liu, H.; Wang, F. Effects of a probiotic on the growth performance, intestinal flora, and immune function of chicks infected with Salmonella pullorum. Poult. Sci. 2020, 99, 5316–5323. [Google Scholar] [CrossRef]
- Gao, H.; Wen, J.-J.; Hu, J.-L.; Nie, Q.-X.; Chen, H.-H.; Xiong, T.; Nie, S.-P.; Xie, M.-Y. Polysaccharide from fermented Momordica charantia L. with Lactobacillus plantarum NCU116 ameliorates type 2 diabetes in rats. Carbohydr. Polym. 2018, 201, 624–633. [Google Scholar] [CrossRef]
- Li, C.; Nie, S.-P.; Zhu, K.-X.; Xiong, T.; Li, C.; Gong, J.; Xie, M.-Y. Effect of Lactobacillus plantarum NCU116 on loperamide-induced constipation in mice. Int. J. Food Sci. Nutr. 2015, 66, 533–538. [Google Scholar] [CrossRef]
- Wen, J.-J.; Li, M.-Z.; Gao, H.; Hu, J.-L.; Nie, Q.-X.; Chen, H.-H.; Zhang, Y.-L.; Xie, M.-Y.; Nie, S.-P. Polysaccharides from fermented Momordica charantia L. with Lactobacillus plantarum NCU116 ameliorate metabolic disorders and gut microbiota change in obese rats. Food Funct. 2021, 12, 2617–2630. [Google Scholar] [CrossRef]
- Xie, J.; Yu, Q.; Nie, S.; Fan, S.; Xiong, T.; Xie, M. Effects of Lactobacillus plantarum NCU116 on intestine mucosal immunity in immunosuppressed mice. J. Agric. Food Chem. 2015, 63, 10914–10920. [Google Scholar] [CrossRef] [PubMed]
- Kanmani, P.; Albarracin, L.; Kobayashi, H.; Hebert, E.M.; Saavedra, L.; Komatsu, R.; Gatica, B.; Miyazaki, A.; Ikeda-Ohtsubo, W.; Suda, Y.; et al. Genomic characterization of Lactobacillus delbrueckii TUA4408L and evaluation of the antiviral activities of its extracellular polysaccharides in porcine intestinal epithelial cells. Front. Immunol. 2018, 9, 2178. [Google Scholar] [CrossRef] [PubMed]
- Kanmani, P.; Albarracin, L.; Kobayashi, H.; Iida, H.; Komatsu, R.; Kober, A.K.M.H.; Ikeda-Ohtsubo, W.; Suda, Y.; Aso, H.; Makino, S.; et al. Exopolysaccharides from Lactobacillus delbrueckii OLL1073R-1 modulate innate antiviral immune response in porcine intestinal epithelial cells. Mol. Immunol. 2018, 93, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.-H.; Fan, S.-T.; Nie, S.-P.; Yu, Q.; Xiong, T.; Gong, D.; Xie, M.-Y. Lactobacillus plantarum NCU116 attenuates cyclophosphamide-induced intestinal mucosal injury, metabolism and intestinal microbiota disorders in mice. Food Funct. 2016, 7, 1584–1592. [Google Scholar] [CrossRef]
- Xie, J.; Nie, S.; Yu, Q.; Yin, J.; Xiong, T.; Gong, D.; Xie, M. Lactobacillus plantarum NCU116 attenuates cyclophosphamide-induced immunosuppression and regulates Th17/Treg cell immune responses in mice. J. Agric. Food Chem. 2016, 64, 1291–1297. [Google Scholar] [CrossRef]
- Zhou, X.; Hong, T.; Yu, Q.; Nie, S.; Gong, D.; Xiong, T.; Xie, M. Exopolysaccharides from Lactobacillus plantarum NCU116 induce c-Jun dependent Fas/Fasl-mediated apoptosis via TLR2 in mouse intestinal epithelial cancer cells. Sci. Rep. 2017, 7, 14247. [Google Scholar] [CrossRef]
- Wang, J.; Wu, T.; Fang, X.; Min, W.; Yang, Z. Characterization and immunomodulatory activity of an exopolysaccharide produced by Lactobacillus plantarum JLK0142 isolated from fermented dairy tofu. Int. J. Biol. Macromol. 2018, 115, 985–993. [Google Scholar] [CrossRef]
- Masumizu, Y.; Zhou, B.; Kober, A.; Islam, M.A.; Iida, H.; Ikeda-Ohtsubo, W.; Suda, Y.; Albarracin, L.; Nochi, T.; Aso, H.; et al. Isolation and immunocharacterization of Lactobacillus salivarius from the intestine of wakame-fed pigs to develop novel “immunosynbiotics”. Microorganisms 2019, 7, 167. [Google Scholar] [CrossRef] [Green Version]
- Gagic, D.; Wen, W.; Collett, M.A.; Rakonjac, J. Unique secreted–surface protein complex of Lactobacillus rhamnosus, identified by phage display. MicrobiologyOpen 2013, 2, 1–17. [Google Scholar] [CrossRef]
- Klotz, C.; Goh, Y.J.; O’Flaherty, S.; Johnson, B.; Barrangou, R. Deletion of S-layer associated Ig-like domain protein disrupts the Lactobacillus acidophilus cell surface. Front. Microbiol. 2020, 11, 345. [Google Scholar] [CrossRef] [Green Version]
- Pavkov, T.; Egelseer, E.M.; Tesarz, M.; Svergun, D.I.; Sleytr, U.B.; Keller, W. The structure and binding behavior of the bacterial cell surface layer protein SbsC. Structure 2008, 16, 1226–1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klotz, C.; O’Flaherty, S.; Goh, Y.J.; Barrangou, R. Investigating the effect of growth phase on the surface-layer associated proteome of Lactobacillus acidophilus using quantitative proteomics. Front. Microbiol. 2017, 8, 2174. [Google Scholar] [CrossRef] [PubMed]
- Henneke, P.; Dramsi, S.; Mancuso, G.; Chraibi, K.; Pellegrini, E.; Theilacker, C.; Hübner, J.; Santos-Sierra, S.; Teti, G.; Golenbock, D.T.; et al. Lipoproteins are critical TLR2 activating toxins in group B streptococcal sepsis. J. Immunol. 2008, 180, 6149–6158. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, M.; Nguyen, M.-T.; Engdahl, C.; Na, M.; Jarneborn, A.; Hu, Z.; Karlsson, A.; Pullerits, R.; Ali, A.; Götz, F.; et al. The YIN and YANG of lipoproteins in developing and preventing infectious arthritis by Staphylococcus aureus. PLoS Pathog. 2019, 15, e1007877. [Google Scholar] [CrossRef] [PubMed]
- Smelt, M.J.; De Haan, B.J.; Bron, P.A.; Van Swam, I.; Meijerink, M.; Wells, J.M.; Kleerebezem, M.; Faas, M.M.; De Vos, P. The impact of Lactobacillus plantarum WCFS1 teichoic acid D-alanylation on the generation of effector and regulatory T-cells in healthy mice. PLoS ONE 2013, 8, e63099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, Y.H.; Baik, J.E.; Yang, J.S.; Kang, S.-S.; Im, J.; Yun, C.-H.; Kim, D.W.; Lee, K.; Chung, D.K.; Ju, H.R.; et al. Differential immunostimulatory effects of Gram-positive bacteria due to their lipoteichoic acids. Int. Immunopharmacol. 2009, 9, 127–133. [Google Scholar] [CrossRef]
- Kleerebezem, M.; Boekhorst, J.; Van Kranenburg, R.; Molenaar, D.; Kuipers, O.P.; Leer, R.; Tarchini, R.; Peters, S.A.; Sandbrink, H.M.; Fiers, M.W.; et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 2003, 100, 1990–1995. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S. Babraham Bioinformatics—FastQC a Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 15 November 2021).
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Cam, L. Maximum likelihood: An introduction. Int. Stat. Rev. 1990, 58, 153–171. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).