Abstract
Breast cancer (BC) remains one of the leading causes of cancer deaths among women worldwide. Recently, studies of long non-coding RNAs (lncRNAs) involved in the regulation of various signaling pathways in cells have become increasingly important, since they demonstrate great prognostic potential in cancer. The aim of our work was to identify new aberrantly expressed lncRNAs in BC. We identified 30 aberrantly expressed lncRNAs in BC. For most lncRNAs, a decrease in the expression level by 2.34–13.2 times (p < 0.05) was noted, and only for lncRNA TERC, an increase in the expression level by 2.24 times (p = 0.034) was noted. Of greatest interest are the data obtained for the lncRNAs ADAMTS9-AS2, EMX2OS, HOTAIRM1 and MEG3, as they are consistent with the data of the bioinformatic analysis.
1. Introduction
According to the data of the International Agency for Research on Cancer (IARC), breast cancer (BC) is recognized as the most frequently diagnosed type of cancer (more than 2.26 million new cases) and was the cause of death of almost 685,000 people in 2020. Thus, in 2020, breast cancer overtook lung cancer in terms of the number of diagnosed cases and became the most common cause of cancer death among women and the fifth most common cause of cancer death overall. IARC estimates the incidence of breast cancer will increase to 3 million new cases per year, and mortality from breast cancer will increase to 1 million deaths per year by 2040 [1].
BC is characterized by significant variability in cellular composition, as well as histological, expressional, genotypic and epigenetic heterogeneity. Based on immunohistochemistry data, namely the expression of estrogen and progesterone receptors in cancer cells, BC is divided into four molecular subtypes: luminal A, luminal B, Her-2 positive and triple negative breast cancer. The molecular subtype of breast cancer determines the response of patients to various therapies, including targeted anticancer therapies [2,3,4].
Epigenetic deregulation in cells has a decisive influence on the development and progression of cancer [5,6]. To date, such ways of epigenetic regulation as DNA methylation, histone modifications [7], abnormal expression of miRNAs, long non-coding RNAs (lncRNAs) and small nucleolar RNAs are known. lncRNAs, their genes accounting for up to 80% of the mammalian genome, play a key role in the regulation of gene expression and in cancer biology through various mechanisms, including chromosome remodeling and transcriptional and post-transcriptional modifications [4,8,9,10].
Despite the increasing effectiveness of diagnostic and therapeutic strategies, the recovery for BC remains limited [11]. The lack of validated diagnostic and prognostic biomarkers is one of the reasons for failures in the early detection and treatment of breast cancer worldwide [12,13]. We conducted an experimental search for long non-coding RNAs that change expression in breast cancer, which can be used in the future as markers of this disease or targets/agents of targeted therapy, and compared our data with available online databases.
2. Materials and Methods
Sample collection. In total, 24 paired tissue samples from the tumor and adjacent histologically normal tissue were collected and characterized at the National Medical Research Center of Oncology named after N.N. Blokhin. When performing a planned surgical operation, samples were collected, and tumor areas with a tumor cell content of at least 70% and adjacent tissue areas in which no tumor cells were found during a histological analysis were taken for the study.
RNA Isolation. Total RNA was isolated by a modified guanidine thiocyanate–phenol–chloroform extraction method in an acidic acetate buffer (previously described in [14]). The concentration of total RNA was determined spectrophotometrically using the NanoDrop 1000 (Thermo Scientific, Waltham, MA, USA). RIN was determined from the results of total-RNA-denaturing electrophoresis in 1% agarose gel [14].
Reverse transcription and PCR. In total, 1 μg of total RNA was taken into the reverse transcription reaction; the reaction was carried out using the RT2 First Strand Kit (QIAGEN Sciences, Frederick, MD, USA) according to the manufacturer’s protocol. PCR was performed using the RT2 SYBR® Green qPCR Mastermix and RT2 lncRNA PCR Array #LAHS-002Z (QIAGEN Sciences, USA) according to the manufacturer’s protocol. Controls for the genomic DNA contamination, reverse transcription reaction and PCR, and reference genes, were included in the RT2 lncRNA PCR Array panel.
Statistical processing of the results was carried out using QIAGEN’s GeneGlobe Data Analysis Center (https://geneglobe.qiagen.com/us/analyze/, accessed on 4 September 2022) [15]; Cq was 39 because some lncRNAs are relatively weakly expressed in breast tissue. RNA isolated from histologically normal tissue was used in the Control Group, and RNA from tumor tissue was used in the Experimental Group (Test Group 1).
Bioinformatics. In parallel with the analysis of the expression of lncRNAs in clinical samples, a search for differentially expressed lncRNAs in BC was carried out using the GEPIA database (http://gepia.cancer-pku.cn/, accessed on 6 November 2022) [16].
3. Results and Discussion
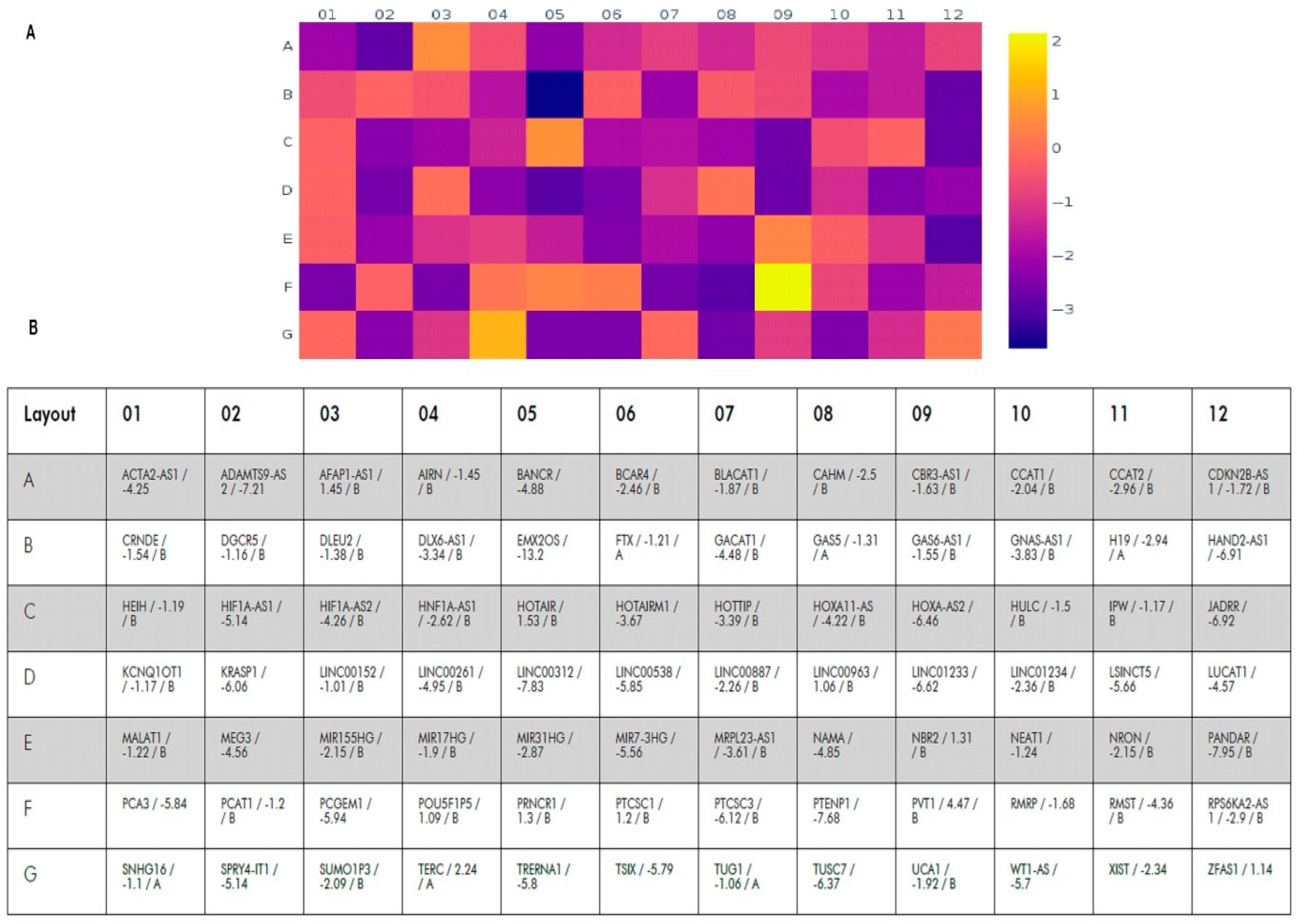
We analyzed the expression of 84 lncRNAs validated by QIAGEN Sciences for BC (Figure 1). Expression of 30 of them significantly changed in tumor tissues compared to the histological norm (p < 0.05); a change in expression by two or more times was considered significant. Moreover, for 29 lncRNAs, a decrease of 2.34–13.2 times was revealed, and only for TERC, an increase in expression by 2.24 times was found (Table 1).
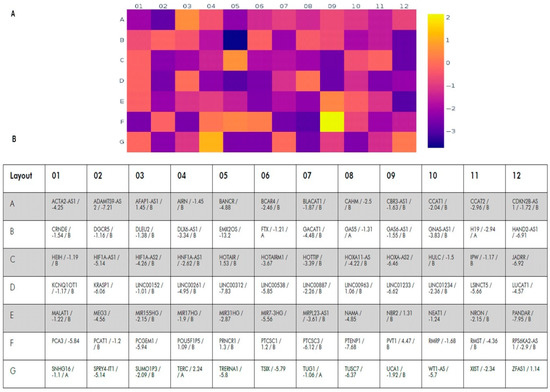
Figure 1.
Heat map of changes in the lncRNA expression levels in tumor tissues compared to histologically normal tissues. (A)—heat map; color scale of changes (log2) is given on the right. (B)—the table indicating the symbol of the lncRNA, the expression changes (fold regulation) and statistical significance ((A)—expression varies greatly within the group, which makes statistical analysis difficult and (B)—the value may be insignificant, p ≥ 0.05).

Table 1.
The most significant lncRNA expression changes in tumor tissue compared to the histological norm (mean values are shown).
In parallel with the experimental search for lncRNA aberrantly expressed in BC, we analyzed data from the Gene Expression Profiling Interactive Analysis database (GEPIA) [16] (Figure 2).
TERC (telomerase RNA component) serves as a template for telomere replication catalyzed by the telomerase enzyme, which contributes to the uncontrolled proliferation of cancer cells. Despite the fact that our experimental data were not confirmed by the data of the GEPIA database for breast cancer, an increase in TERC expression was shown for gastric cancer [17], and TERC was proposed as a diagnostic marker in cervical cancer [18] and lung cancer [19].
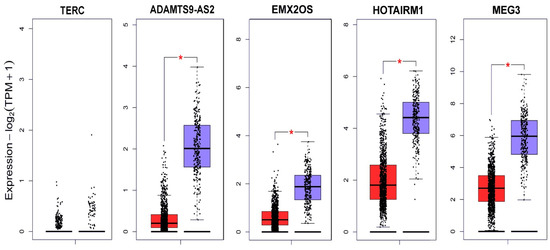
Figure 2.
lncRNA expression profile of TERC, ADAMTS9-AS2, EMX2OS, HOTAIRM1 and MEG3 in BC according to the GEPIA database [18]. Red indicates the expression level in the tumor tissue (1085 samples); blue indicates the expression level in normal tissue (291 samples); and * p < 0.01.
Figure 2.
lncRNA expression profile of TERC, ADAMTS9-AS2, EMX2OS, HOTAIRM1 and MEG3 in BC according to the GEPIA database [18]. Red indicates the expression level in the tumor tissue (1085 samples); blue indicates the expression level in normal tissue (291 samples); and * p < 0.01.
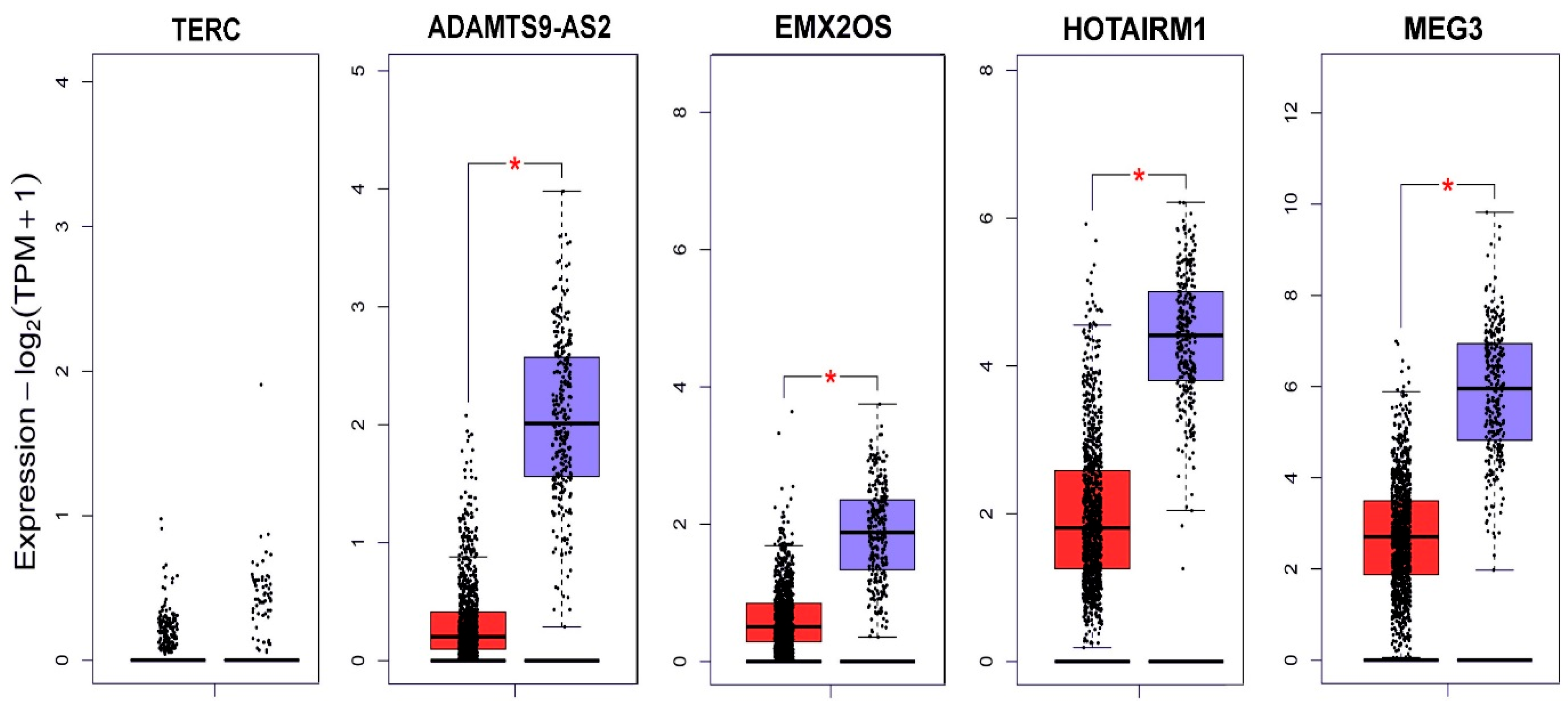
MEG3 (maternally expressed 3) is a relatively well-studied lncRNA demonstrating oncosuppressive properties. A systemic meta-analysis showed an association between the suppression of MEG3 expression and poor outcome in patients with cancer of different localizations [20]. MEG3 expression is reduced in cell lines and tissues of ovarian cancer [21]. An increase in MEG3 expression prevented cell proliferation and induced apoptosis in cell lines of ovarian, cervical, and other cancers [22]. Competitively interacting with miR-421, MEG3 enhances the action of E-cadherin, which blocks the epithelial–mesenchymal transition [23].
ADAMTS9-AS2 also exhibits suppressor properties in cancer. Various authors have studied ADAMTS9-AS2 in regulating the development of gastric cancer [24] and lung cancer [25]. EMX2OS is still undergoing relatively little study; however, competitive interaction of this lncRNA with some microRNAs in the Wilms tumor [26] and its prognostic value in gastric cancer have been shown [27].
The lncRNA HOTAIRM1 has already been proposed as a biomarker for oral squamous cell carcinoma [28], and its role in the proliferation and metastasis of breast cancer has also been proposed [29]; the development of resistance to tamoxifen therapy due to the regulation of HOXA1 protein expression is being studied [30].
4. Conclusions
lncRNAs are involved in the regulation of multiple cancer features. Their association with inhibition of apoptosis, initiation of invasion and metastasis and activation of angiogenesis has been shown (see review [31]).
During the experimental and bioinformatic analysis of changes in lncRNA expression in breast cancer, we have identified TERC, ADAMTS9-AS2, EMX2OS, HOTAIRM1 and MEG3 lncRNAs with biomarker potential. The further study of lncRNA expression and its regulation will also help in the development of targeted anticancer therapy.
Author Contributions
Conceptualization, I.P. and E.F.; methodology, I.P.; software, V.L., A.B. and E.F.; validation, I.P., S.L. and E.B.; formal analysis, E.B.; investigation, I.P. and E.F.; resources, T.K.; data curation, T.K. and E.B.; writing—original draft preparation, I.P.; writing—review and editing, I.P., V.L., E.B. and E.F.; visualization, I.P. and V.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Science and Higher Education of the Russian Federation, the state task number: FGFU-2022-0007 (the sample collection, RNA isolation and online database analysis), and the Russian Science Foundation, grant no.: 22-75-00132 (the experimental RT2 lncRNA PCR Array analysis).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on reasonable request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- International Agency for Research on Cancer. Available online: https://www.iarc.who.int/featured-news/breast-cancer-awareness-month-2021/ (accessed on 19 January 2023).
- Joseph, C.; Papadaki, A.; Althobiti, M.; Alsaleem, M.; Aleskandarany, M.A.; Rakha, E.A. Breast cancer intratumour heterogeneity: Current status and clinical implications. Histopathology 2018, 73, 717–731. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Kansara, S.; Pandey, V.; Lobie, P.E.; Sethi, G.; Garg, M.; Pandey, A.K. Mechanistic Involvement of Long Non-Coding RNAs in Oncotherapeutics Resistance in Triple-Negative Breast Cancer. Cells 2020, 9, 1511. [Google Scholar] [CrossRef]
- Kumar, R.; Paul, A.M.; Rameshwar, P.; Pillai, M.R. Epigenetic Dysregulation at the Crossroad of Women’s Cancer. Cancers 2019, 11, 1193. [Google Scholar] [CrossRef]
- Kagohara, L.T.; Stein-O’Brien, G.L.; Kelley, D.; Flam, E.; Wick, H.C.; Danilova, L.V.; Easwaran, H.; Favorov, A.V.; Qian, J.; A Gaykalova, D.; et al. Epigenetic regulation of gene expression in cancer: Techniques, resources and analysis. Briefings Funct. Genom. 2017, 17, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, M.K.; Arfuso, F.; Arumugam, S.; Chinnathambi, A.; Jinsong, B.; Warrier, S.; Wang, L.Z.; Kumar, A.P.; Ahn, K.S.; Sethi, G.; et al. Role of novel histone modifications in cancer. Oncotarget 2018, 9, 11414–11426. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Wei, G.-H. Genomic Insight into the Role of lncRNAs in Cancer Susceptibility. Int. J. Mol. Sci. 2017, 18, 1239. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Tamizkar, K.H.; Hussen, B.M.; Taheri, M. An update on the role of long non-coding RNAs in the pathogenesis of breast cancer. Pathol.-Res. Pract. 2021, 219, 153373. [Google Scholar] [CrossRef]
- Nounou, M.I.; ElAmrawy, F.; Ahmed, N.; Abdelraouf, K.; Goda, S.; Syed-Sha-Qhattal, H. Breast Cancer: Conventional Diagnosis and Treatment Modalities and Recent Patents and Technologies. Breast Cancer Basic Clin. Res. 2015, 9, 17–34. [Google Scholar] [CrossRef]
- Feldman, R.; Kim, E.S. Prognostic and predictive biomarkers post curative intent therapy. Ann. Transl. Med. 2017, 5, 374. [Google Scholar] [CrossRef]
- Kashyap, D.; Kaur, H. Cell-free miRNAs as non-invasive biomarkers in breast cancer: Significance in early diagnosis and metastasis prediction. Life Sci. 2020, 246, 117417. [Google Scholar] [CrossRef]
- Pronina, I.V.; Loginov, V.I.; Prasolov, V.S.; Klimov, E.A.; Khodyrev, D.S.; Kazubskaya, T.P.; Gar’Kavtseva, R.F.; Sulimova, G.E.; Braga, E.A. Altered expression of the SEMA3B gene in epithelial tumors. Mol. Biol. 2009, 43, 403–409. [Google Scholar] [CrossRef]
- QIAGEN’s GeneGlobe Data Analysis Center. Available online: https://geneglobe.qiagen.com/us/analyze (accessed on 30 October 2022).
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Wang, J.; Wu, M.; Chang, L.; Jin, Z.; Yang, X.; Li, D.; Wang, J.; Qu, J.; Hou, Q.; Huang, X.; et al. The lncRNA TERC promotes gastric cancer cell proliferation, migration, and invasion by sponging miR-423-5p to regulate SOX12 expression. Ann. Transl. Med. 2022, 10, 963. [Google Scholar] [CrossRef]
- Zhao, H.; He, Y.; Fan, B.; Wang, Y.; Wu, Y.-M. Human papillomavirus E6E7 mRNA and TERC lncRNA in situ detection in cervical scraped cells and cervical disease progression assessment. Virol. J. 2022, 19, 18. [Google Scholar] [CrossRef] [PubMed]
- Storti, C.B.; de Oliveira, R.A.; de Carvalho, M.; Hasimoto, E.N.; Cataneo, D.C.; Cataneo, A.J.M.; De Faveri, J.; Vasconcelos, E.J.R.; dos Reis, P.P.; Cano, M.I.N. Telomere-associated genes and telomeric lncRNAs are biomarker candidates in lung squamous cell carcinoma (LUSC). Exp. Mol. Pathol. 2020, 112, 104354. [Google Scholar] [CrossRef] [PubMed]
- Binabaj, M.M.; Bahreyni, A.; Khazaei, M.; Avan, A.; Hassanian, S.M. The prognostic value of long noncoding RNA MEG3 expression in the survival of patients with cancer: A meta-analysis—Response. J. Cell. Biochem. 2019, 120, 18599. [Google Scholar] [CrossRef]
- Tao, P.; Yang, B.; Zhang, H.; Sun, L.; Wang, Y.; Zheng, W. The overexpression of lncRNA MEG3 inhibits cell viability and invasion and promotes apoptosis in ovarian cancer by sponging miR-205-5p. Int. J. Clin. Exp. Pathol. 2020, 13, 869–879. [Google Scholar] [PubMed]
- He, Y.; Luo, Y.; Liang, B.; Ye, L.; Lu, G.; He, W. Potential applications of MEG3 in cancer diagnosis and prognosis. Oncotarget 2017, 8, 73282–73295. [Google Scholar] [CrossRef]
- Zhang, W.; Shi, S.; Jiang, J.; Li, X.; Lu, H.; Ren, F. LncRNA MEG3 inhibits cell epithelial-mesenchymal transition by sponging miR-421 targeting E-cadherin in breast cancer. Biomed. Pharmacother. 2017, 91, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, X.-C.; Zhu, J.-R.; Ma, Z.-J.; Ran, J.-T.; Zhou, Y.-N. Construction of a novel ceRNA network and identification of lncRNA ADAMTS9-AS2 and PVT1 as hub regulators of miRNA and coding gene expression in gastric cancer. Transl. Cancer Res. 2021, 10, 938–952. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Huang, W.; Yi, Y.; Li, D.; Xie, Z.; Li, Z.; Ye, M. LncRNA ADAMTS9-AS2 is a Prognostic Biomarker and Correlated with Immune Infiltrates in Lung Adenocarcinoma. Int. J. Gen. Med. 2021, 14, 8541–8555. [Google Scholar] [CrossRef]
- Chen, Z.H.; Cui, M.Y.; Zhang, H.M. EMX2OS Delays Wilms’ Tumor Progression via Targeting miR-654-3p. Ann. Clin. Lab. Sci. 2022, 52, 12–20. [Google Scholar] [PubMed]
- Liu, G.-X.; Tan, Y.-Z.; He, G.-C.; Zhang, Q.-L.; Liu, P. EMX2OS plays a prognosis-associated enhancer RNA role in gastric cancer. Medicine 2021, 100, e27535. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Niu, J.; Zhang, X.; Wang, X.; Song, H.; Liu, Y.; Jiao, X.; Chen, F. Identification and Validation of HOTAIRM1 as a Novel Biomarker for Oral Squamous Cell Carcinoma. Front. Bioeng. Biotechnol. 2022, 9, 798584. [Google Scholar] [CrossRef]
- Lai, G.-E.; Zhou, J.; Huang, C.-L.; Mai, C.-L.H.C.-J.; Lai, Y.-M.; Lin, Z.-Q.; Peng, T.; Luo, Y.; Liu, F.-E. A combination of transcriptome and methylation analyses reveals the role of lncRNA HOTAIRM1 in the proliferation and metastasis of breast cancer. Gland. Surg. 2022, 11, 826–836. [Google Scholar] [CrossRef]
- Kim, C.Y.; Oh, J.H.; Lee, J.-Y.; Kim, M.H. The LncRNA HOTAIRM1 Promotes Tamoxifen Resistance by Mediating HOXA1 Expression in ER+ Breast Cancer Cells. J. Cancer 2020, 11, 3416–3423. [Google Scholar] [CrossRef]
- Kashyap, D.; Sharma, R.; Goel, N.; Buttar, H.S.; Garg, V.K.; Pal, D.; Rajab, K.; Shaikh, A. Coding roles of long non-coding RNAs in breast cancer: Emerging molecular diagnostic biomarkers and potential therapeutic targets with special reference to chemotherapy resistance. Front. Genet. 2023, 13, 993687. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).