RUNX1-Regulated Pathways and Biomarkers in Acute Myeloid Leukaemia †
Abstract
:1. Introduction
2. Methods
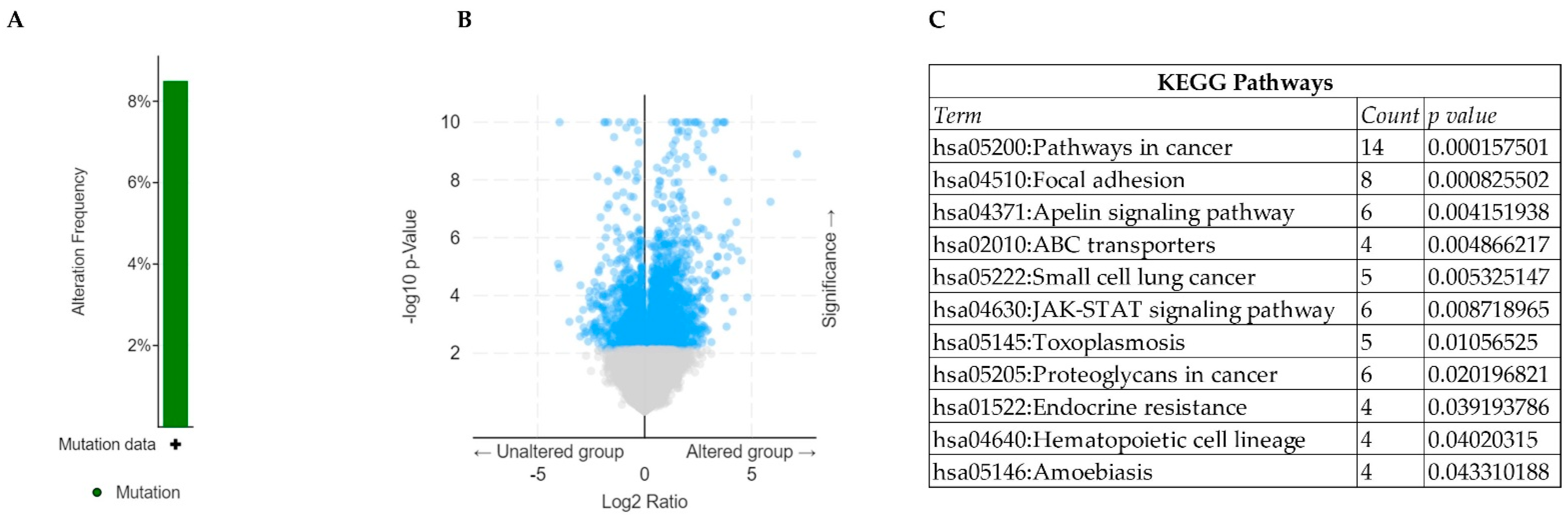
2.1. Identification of Mutational Landscape of RUNX1 in TCGA-AML
2.2. Analysis of Differentially Expressed Genes (DEGs) in RUNX1-Mutated TCGA-AML
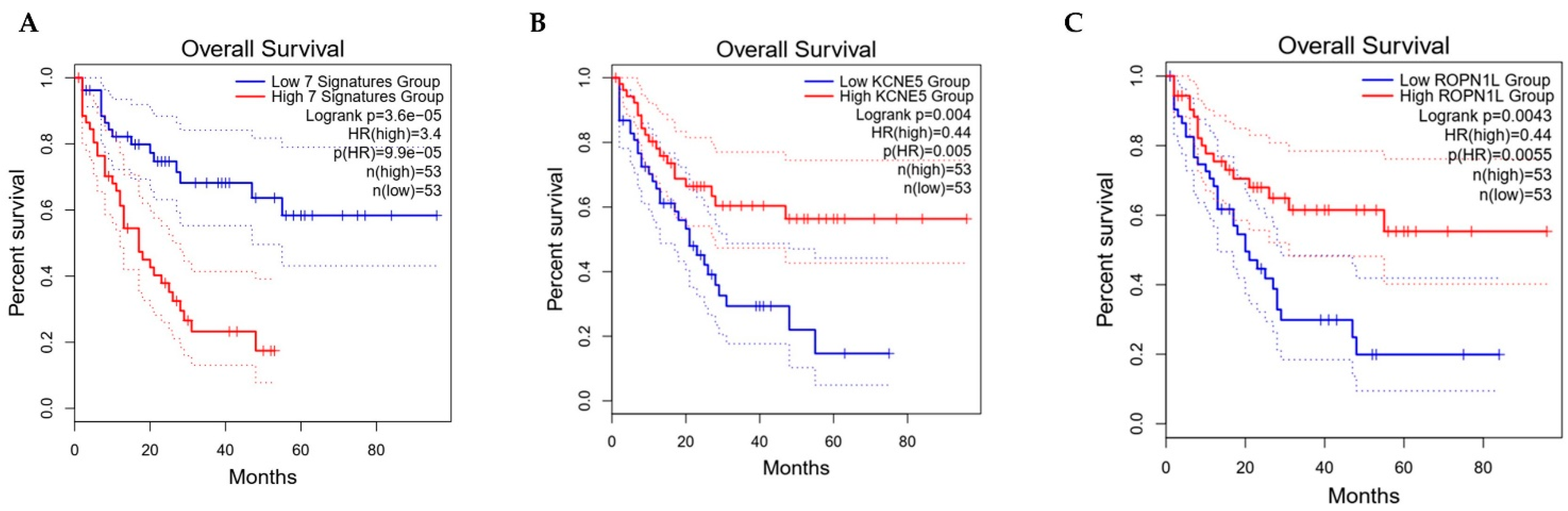
2.3. Functional Annotation and Survival Analysis
3. Results and Discussions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mendler, J.H.; Maharry, K.; Radmacher, M.D.; Mrozek, K.; Becker, H.; Metzeler, K.H.; Schwind, S.; Whitman, S.P.; Khalife, J.; Kohlschmidt, J.; et al. RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and MicroRNA expression signatures. J. Clin. Oncol. 2012, 30, 3109–3118. [Google Scholar] [CrossRef] [PubMed]
- Bullinger, L.; Dohner, K.; Dohner, H. Genomics of Acute Myeloid Leukemia Diagnosis and Pathways. J. Clin. Oncol. 2017, 35, 934–946. [Google Scholar] [CrossRef] [PubMed]
- Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, A.; Hoadley, K.; Triche, T.J., Jr.; Laird, P.W.; Baty, J.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Brlek, P.; Kafka, A.; Bukovac, A.; Pecina Slaus, N. Integrative cBioPortal Analysis Revealed Molecular Mechanisms That Regulate EGFR PI 3 K AKT mTOR Pathway in Diffuse Gliomas of the Brain. Cancers 2021, 13, 3247. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verma, D.K.; Chauhan, H.S.; Namani, A. RUNX1-Regulated Pathways and Biomarkers in Acute Myeloid Leukaemia. Med. Sci. Forum 2023, 20, 2. https://doi.org/10.3390/IECC2023-14279
Verma DK, Chauhan HS, Namani A. RUNX1-Regulated Pathways and Biomarkers in Acute Myeloid Leukaemia. Medical Sciences Forum. 2023; 20(1):2. https://doi.org/10.3390/IECC2023-14279
Chicago/Turabian StyleVerma, Deepesh Kumar, Hrishika Singh Chauhan, and Akhileshwar Namani. 2023. "RUNX1-Regulated Pathways and Biomarkers in Acute Myeloid Leukaemia" Medical Sciences Forum 20, no. 1: 2. https://doi.org/10.3390/IECC2023-14279
APA StyleVerma, D. K., Chauhan, H. S., & Namani, A. (2023). RUNX1-Regulated Pathways and Biomarkers in Acute Myeloid Leukaemia. Medical Sciences Forum, 20(1), 2. https://doi.org/10.3390/IECC2023-14279





