Abstract
Machine Learning (ML), a branch of Artificial Intelligence (AI), has been successfully applied in the healthcare domain to diagnosing diseases. The ML techniques have not only been able to diagnose common diseases but are also equally capable of diagnosing rare diseases. Although ML offers systematic and sophisticated algorithms for multi-dimensional clinical data, the accuracy of ML in diagnosing diseases is still a concern. As different ML approaches perform differently for different healthcare datasets, we need an approach to apply multiple state of art algorithms with optimal lines of codes, so that the search for the best ML method to diagnose a particular disease can be pursued efficiently. In our work, we show that, the use of libraries such as AutoGluon can be used to compare the performances of multiple ML approaches to diagnosing a disease for a given dataset with a couple of lines of codes. This will decrease the probability of inaccurate diagnosis, which is a significantly important consideration while dealing with the health of the people. We have tested the performance of 20 ML approaches such as Naïve Bayes, Support Vector Machine (SVD), K Nearest Neighbors (KNN), perceptron, and robust deep neural networks in AutoGluon such as LightGBM, XGBoost, MXNet, etc., based on the Pima Indian Diabetes Dataset.
1. Introduction
Machine Learning (ML), a branch of Artificial Intelligence (AI), learns from the data using various algorithms and is a self-improving process in terms of performance as making adjustments during the learning process [1]. ML has been successfully applied to practically every domain such as robotics, education, travel to health care [2]. In the healthcare domain, the ML approaches are mainly used for the purpose of disease diagnosis [3].
The machine learning approaches came into the health sector domain in the 1970s and an international AI journal Artificial Intelligence in Medicine was established in 1980 [4]. In the next two decades, disease diagnosis domain adopted the classical ML approaches such as Support Vector Machine, Naïve Bayes, and some artificial neural networks [5]. The introduction of AlexNet in 2012 initiated the current wave of deep learning in this field as neural networks demonstrated superior performance [6]. Also, in this past decade, the investment in AI in healthcare applications has increased significantly. The studies in [7,8,9,10,11] show that the use of AI and ML technologies in healthcare is leading to the development of software, platforms, automated systems and devices to check as well as improve the health condition of people.
The analysis of the clinical data can lead to the timely diagnosis of the disease which will help to start cure for the patient in time as well [3]. Traditional approach of diagnosing disease is generally costly and time-consuming. As well, the potential of time and cost-proficient machine learning-based disease diagnosis approaches are proven by the researchers [12]. ML techniques have not only been able to diagnose the common diseases but are also equally capable of diagnosing the rare diseases [2,13]. Authors in [14] demonstrate the significance and robustness of AI and ML techniques to solve health care problems.
In general, a dataset table used to build an ML model for diagnosing a disease has columns for different attributes and a column variable for the class variable. Here, class variable indicates whether the instance in the table indicated is positively diagnosed with the disease under consideration. Usually, class values of 1 means positively diagnosed and 0 means negatively diagnosed. Supervised and unsupervised ML [15] approaches have been in practice for analyzing the health care data. In general, disease diagnosis problems are based on supervised learning. We will present a detailed analysis of the used dataset and ML algorithms in Section 2.
Problem Statement
Although ML offers systematic and sophisticated algorithms of multi-dimensional clinical data, the accuracy of the ML in diagnosing the diseases is still a concern [16]. As well, the improvement in the performance of ML to diagnose disease is a hot topic in this domain. As different ML approaches perform differently for different healthcare dataset, we are also in need to find the way to apply many state-of-the-art algorithms to same dataset in reasonable time with minimal lines of codes, so that the search of best ML method can be pursued efficiently to diagnose a particular disease.
The use of libraries such as AutoGluon can help find the best performing ML approach out of many approaches in diagnosing the disease for a given dataset with optimal lines of codes. This will decrease the probability of inaccurate diagnosis, which is a significantly important consideration while dealing with the health of the people. We will test the performance of 20 ML approaches in diagnosing diabetes based on a public dataset discussed in Section 2.1.
2. Data, Algorithms, and Methods
2.1. Data
For this study, we have chosen a healthcare dataset related to diabetes. The dataset is the Pima Indian Diabetes Dataset which is frequently used to evaluate the performance of developed ML techniques [17,18]. We downloaded the dataset from [18]. This data set has 8 attributes and one class variable named Outcome. The Outcome variable has a possible value of 0 or 1, 1 being interpreted as tested positive for diabetes. The dataset has 768 instances, out of which 268 were those who tested positive for diabetes.
2.1.1. Data Exploration
Two of the attributes (BMI and Diabetes Pedigree Function) in the dataset are continuous numerical variables and the rest are discrete numerical integers. Also, no data is missing for each of the attributes. The detailed statistical description of each attribute is shown below in Table 1.

Table 1.
Statistical description of data based on attributes.
2.1.2. Data Exploratory Visualization
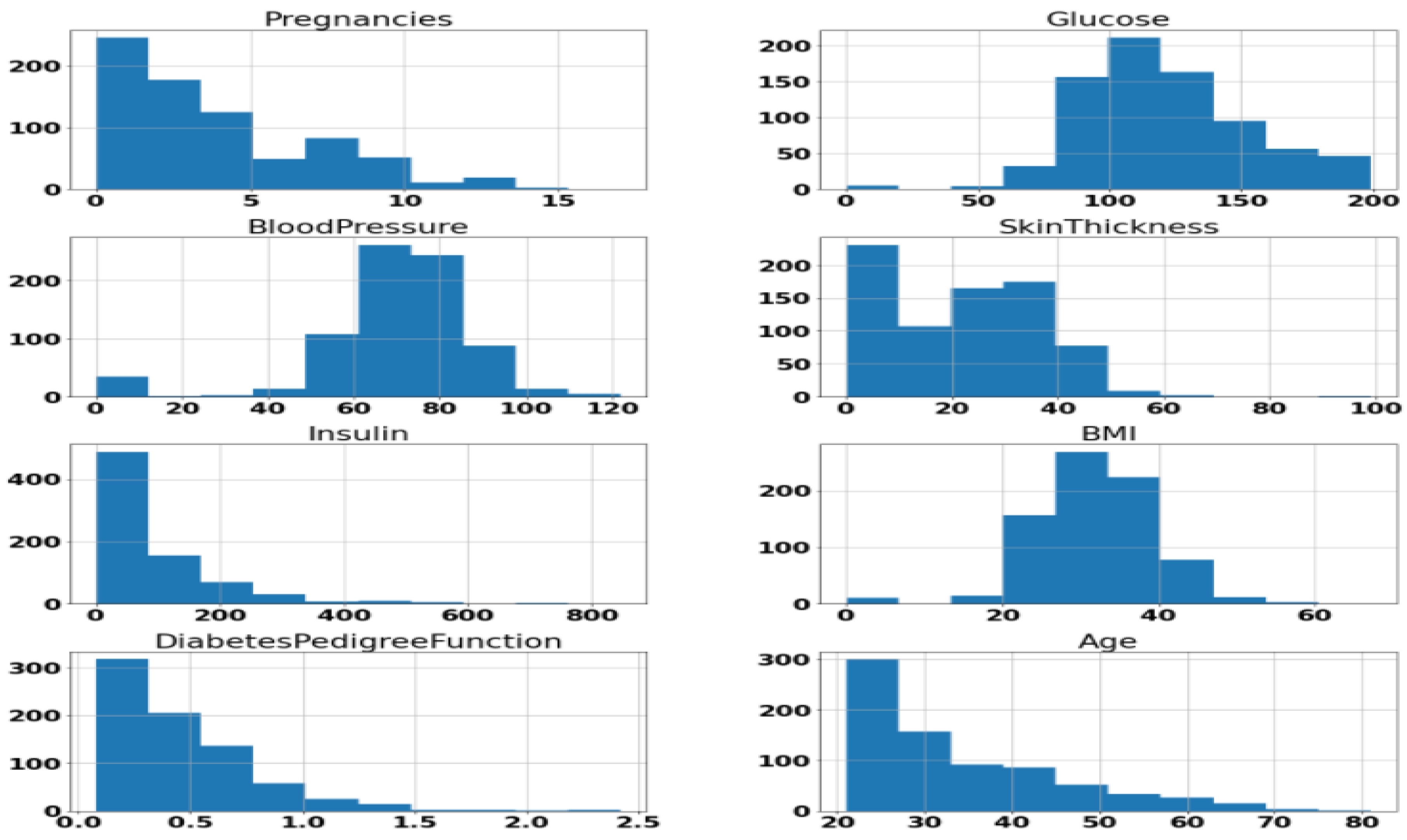
We performed exploratory visualization of the attributes with the histogram. The results are shown in Figure 1. The idea behind the exploratory visualization was to check whether some variables are constant over the range. Such variables can be avoided while building the modes. However, our exploratory visualization showed that every attribute can be important for disease diagnosis with Machine Learning. Also, Figure 1 shows that the mean BMI of the collected data is more than 30, however the dataset does have a significantly smaller proportion of instances diagnosed with diabetes, which is against the general assumption. Thus, the BMI cannot only account for a high probability of having diabetes.
Figure 1.
Histogram of attributes.
2.2. Machine Learning Algorithms and Techniques
Here, we will be applying classification algorithms from the scikit-learn library [19] and AutoGluon library [20] and checking the capacity of the algorithms to diagnose diabetes. Scikit-learn is the most successful and robust library for machine learning in Python. This library is primarily written in Python and is based on the modules such as NumPy [21], SciPy [22] and Matplotlib [23]. As well, the open source AutoML library AutoGluon-Tabular can train highly accurate different machine learning models with a single line of code [20]. The ML algorithms from the scikit-learn library and Auto-Gluon library are implemented with AWS SageMaker [24]. The Amazon SageMaker is capable of building, training, and deploying state of art Machine Learning models with full managed infrastructure tools and workflows [25]. Some of the classification ML used are Naïve Bayes, Support Vector Machine (SVD), K Nearest Neighbors (KNN), perceptron and robust deep neural networks in AutoGluon such as LightGBM, XGBoost, MXNet etc. The list of ML algorithms evaluated for diabetes diagnosis are shown in Table 2 [20,26]. The detail of the algorithm shadows the main goal of this study which is the implementation of ML for disease diagnosis. Please visit the reference [20,26], if the details of the Algorithms are of interest.

Table 2.
List of ML algorithms used.
2.3. Evaluation Metric
Disease diagnosis is a classification task. As well, Classification ML Algorithms are evaluated using Classification Accuracy Measures such as Accuracy, Precision, Recall and F1-score [27,28]. Let us consider a value of 1 (having diabetes) to be positive and a value of 0 in the class variable be negative in the considered dataset. Let True Positive (TP) be the correctly classified number of positive classes from an ML model. Similarly let False Positive (FP) be the number of incorrectly classified as positive classes, True Negative (TN) be the correctly classified number of negative classes and False Negative (FN) be the number of classes incorrectly classified as Negative classes. Various classification accuracy measures are computed based on TP, FP, TN and, FN [29]. The four classification evaluation metrics can be computed as: , , , and .
These four classification accuracy measures have been used to evaluate the performance of applied classifier algorithms. In general, only one (mostly accuracy) evaluation metric is used to evaluate the performance of the ML algorithms. However, in our study we are using four evaluation metrics primarily because of two reasons. The first reason is that in the used diabetes dataset Outcome class variables is highly imbalanced toward the value 0, and the accuracy measure from the imbalanced dataset can be misleading [30]. The next reason is that we are trying to avoid the case of the accuracy paradox by considering four evaluation metrics [31,32].
2.4. Overview of the Methodology
2.4.1. Data Preprocessing
The exploratory analysis and visualization of the data did not suggest any preprocessing of the data for learning the ML models, as no anomaly was detected. Therefore, the process of evaluating an ML for diagnosing the disease was performed with no data preprocessing.
2.4.2. Implementation of ML Algorithms
The implementation and evaluation of ML algorithms were performed in the notebook instance in Amazon SageMaker. The six ML techniques from Scikit-Learn module were applied by importing the module directly as it was already installed in the Cuda Python 3 Kernel. However, the AutoGluon library is not pre-installed in the kernel. There, it had to be downloaded before importing the ML algorithms from it. The detailed implementation process is presented in the notebook project.ipynb which is kept in the author’s GitHub respiratory [33]. The results can be reproduced using the project.ipynb notebook. A total of 14 ML algorithms from the AutoGluon library were trained with only a couple of lines of code as implemented in [33]. We made sure that same training and test set were used for each of the ML algorithms by defining the parameter seed = 42 during the random splitting of the original data into training and test set.
2.4.3. Refinement
We trained the 14 AutoGluon ML algorithms, first using the evaluation metric accuracy. As, the dataset we have an imbalanced dataset in terms of Outcome class, therefore we used the evaluation metric F1-score, which is a more favored evaluation metric while training with imbalanced data. We also tuned hyperparameters to check if better results are possible but the prediction accuracy with the tune hyperparameters came out to be lower than the untuned ones. Therefore, future research with the extensive tuning of different hyper parameters is recommended to check the existence of better models with a different set of hyper parameters.
3. Result and Discussion
The evaluation of different ML techniques in diagnosing diabetes based on the given dataset is shown in Table 3.

Table 3.
Evaluation of ML Algorithms.
Our study shows that most of the ML methods perform better than the benchmark of baseline accuracy of 65 percent, set by the authors in [18] for this dataset while diagnosing diabetes. About 77 percent of the accuracy seems to be the best case for the state of art ML algorithms for the dataset considered in this study. Considering the case of having imbalanced data, we can emphasize the capability of the Naïve Bayes method to perform better among the rest considering the combined analysis of all the evaluation metrics.
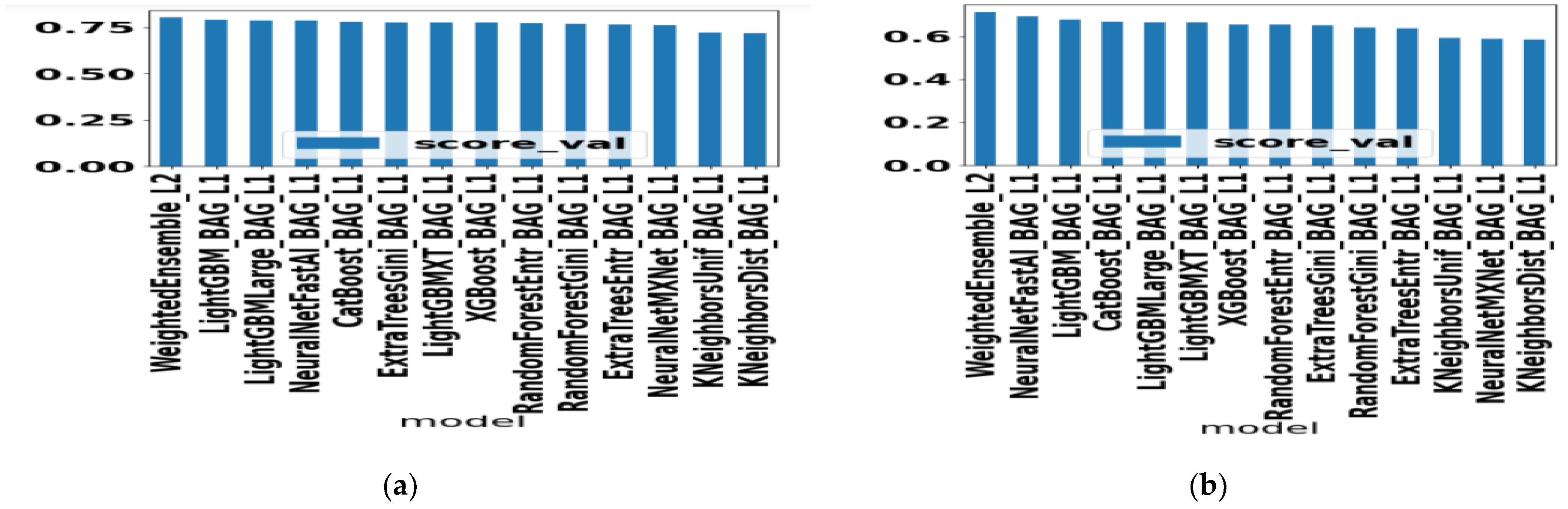
We present the accuracy performance of different AutoGluon ML algorithms when trained with accuracy as a validation metric in Figure 2a. Similarly, performance in terms of F1 scores is shown when trained with F1 scores as validation metric in Figure 2b. It is seen that the Weighted Ensemble ML technique performs better for both cases and KNN-based ML has the least performance.
Figure 2.
(a) Evaluation of AutoGluon ML algorithms when trained with accuracy as validation metric (b) Evaluation of AutoGluon ML algorithms when trained with F1-score as validation metric.
4. Conclusions and Future Work
Machine Learning (ML) algorithms have been successfully applied in the healthcare domain to diagnosing diseases. In our work we show that, the use of libraries such as AutoGluon can help to compare the performances of different ML approaches in diagnosing a disease for a given dataset with optimal lines of code. This helps in finding the best performing ML algorithm for a particular dataset or a particular type of disease as well. Furthermore, it decreases the probability of inaccurate diagnosis, which is a significantly important consideration while dealing with the health of the people. In this study we have tested the performance of 20 ML approaches in diagnosing diabetes based on the Pima Indian Diabetes Dataset. For the dataset considered in this study, the Naïve Bayes algorithm performed better among the other algorithms. This shows that using complex and computationally costly algorithms does not necessarily improve the accuracy of diagnosing a disease.
The possibility of the improvement in the performance of ML models in the future can be started by finding the correlation among each attribute and dropping the highly correlated attributes, because the highly correlated attributes can confuse a model in the learning phase. The evidence of applying multiple ML algorithms with optimal lines of codes in this study strongly suggests that such investigations are to be pursued.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
A freely available Pima Indian Diabetes Dataset which has been widely used in research articles and available in multiple public database platforms has been used.
Data Availability Statement
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mitchell, T.M. Machine Learning; McGraw-Hill: New York, NY, USA, 1997. [Google Scholar]
- Fatima, M.; Pasha, M. Survey of machine learning algorithms for disease diagnostic. J. Intell. Learn. Syst. Appl. 2017, 9, 73781. [Google Scholar] [CrossRef]
- Machine Learning Use Cases|Neural Designer. Available online: https://www.neuraldesigner.com/solutions (accessed on 13 January 2022).
- Demystifying AI in Healthcare: Historical Perspectives and Current Considerations. Available online: https://www.physicianleaders.org/news/demystifying-ai-in-healthcare-historical-perspectives-and-current-considerations (accessed on 13 January 2022).
- Kononenko, I. Machine learning for medical diagnosis: History, state of the art and perspective. Artif. Intell. Med. 2001, 23, 89–109. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Adv. Neural Inf. Process. Syst. 2012, 1097–1105. [Google Scholar] [CrossRef]
- Massaro, A.; Ricci, G.; Selicato, S.; Raminelli, S.; Galiano, A. Decisional Support System with Artificial Intelligence oriented on Health Prediction using a Wearable Device and Big Data. In Proceedings of the 2020 IEEE International Workshop on Metrology for Industry 4.0 & IoT, Rome, Italy, 3–5 June 2020; pp. 718–723. [Google Scholar] [CrossRef]
- Habib, M.; Faris, M.; Qaddoura, R.; Alomari, M.; Alomari, A.; Faris, H. Toward an automatic quality assessment of voice-based telemedicine consultations: A deep learning approach. Sensors 2021, 21, 3279. [Google Scholar] [CrossRef] [PubMed]
- Massaro, A.; Galiano, A.; Scarafile, D.; Vacca, A.; Frassanito, A.; Melaccio, A.; Solimando, A.; Ria, R.; Calamita, G.; Bonomo, M.; et al. Telemedicine DSS-AI Multi Level Platform for Monoclonal Gammopathy Assistance. In Proceedings of the 2020 IEEE International Symposium on Medical Measurements and Applications (MeMeA), Bari, Italy, 1 June–1 July 2020. [Google Scholar] [CrossRef]
- Niculescu, M.S.; Florescu, A.; Pasca, S. LabConcept—A new mobile healthcare platform for standardizing patient results in telemedicine. Appl. Sci. 2021, 11, 1935. [Google Scholar] [CrossRef]
- Massaro, A.; Maritati, V.; Savino, N.; Galiano, A. Neural Networks for Automated Smart Health Platforms oriented on Heart Predictive Diagnostic Big Data Systems. In Proceedings of the 2018 AEIT International Annual Conference, Bari, Italy, 3–5 October 2018. [Google Scholar] [CrossRef]
- Sajda, P. Machine learning for detection and diagnosis of disease. Annu. Rev. Biomed. Eng. 2006, 8, 537–565. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, J.; Lehne, M.; Schepers, J.; Prasser, F.; Thun, S. The use of machine learning in rare diseases: A scoping review. Orphanet J. Rare Dis. 2020, 15, 145. [Google Scholar] [CrossRef] [PubMed]
- Béjar, L.R.; Suleiman-Martos, N.; Mhlanga, D. The Role of Artificial Intelligence and Machine Learning Amid the COVID-19 Pandemic: What Lessons Are We Learning on 4IR and the Sustainable Development Goals. Int. J. Environ. Res. Public Health 2022, 19, 1879. [Google Scholar] [CrossRef]
- Schmidt, J.; Marques, M.R.G.; Botti, S.; Marques, M.A.L. Recent advances and applications of machine learning in solid-state materials science. npj Comput. Mater. 2019, 5, 83. [Google Scholar] [CrossRef]
- Deep Learning for Disease Diagnosis Confounded by Image Labels—Physics World. Available online: https://physicsworld.com/a/deep-learning-for-disease-diagnosis-confounded-by-image-labels/ (accessed on 13 January 2022).
- Smith, J.W.; Everhart, J.E.; Dickson, W.C.; Knowler, W.C.; Johannes, R.S. Using the ADAP Learning Algorithm to Forecast the Onset of Diabetes Mellitus. In Proceedings of the Annual Symposium on Computer Application in Medical Care, Washington, DC, USA, 6–9 November 1988; p. 261. [Google Scholar]
- 10 Standard Datasets for Practicing Applied Machine Learning. Available online: https://machinelearningmastery.com/standard-machine-learning-datasets/ (accessed on 12 January 2022).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python Gaël Varoquaux Bertrand Thirion Vincent Dubourg Alexandre Passos PEDREGOSA, VAROQUAUX, GRAMFORT ET AL. Matthieu Perrot. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Erickson, N.; Mueller, J.; Shirkov, A.; Zhang, H.; Larroy, P.; Li, M.; Smola, A. AutoGluon-Tabular: Robust and Accurate AutoML for Structured Data. arXiv 2020, arXiv:2003.06505. [Google Scholar]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Amazon SageMaker—Machine Learning—Amazon Web Services. Available online: https://aws.amazon.com/sagemaker/ (accessed on 13 January 2022).
- Amazon SageMaker: Amazon Sagemaker API Reference. Available online: https://docs.aws.amazon.com/sagemaker/latest/APIReference/API_Search.html (accessed on 13 January 2022).
- 1. Supervised Learning—Scikit-Learn 1.0.2 Documentation. Available online: https://scikit-learn.org/stable/supervised_learning.html (accessed on 14 January 2022).
- Poudel, S. Improving Collaborative Filtering Recommendation System via Optimal Sub-Sampling and Aspect-Based Interpretability. Ph.D. Thesis, North Carolina Agricultural and Technical State University, Greensboro, NC, USA, 2022. Available online: https://www.proquest.com/dissertations-theses/improving-collaborative-filtering-recommendation/docview/2680264335/se-2 (accessed on 14 January 2022).
- Poudel, S.; Bikdash, M. Optimal dependence of performance and efficiency of collaborative filtering on random stratified subsampling. Big Data Min. Anal. 2022, 5, 192–205. [Google Scholar] [CrossRef]
- Galdi, P.; Tagliaferri, R. Data Mining: Accuracy and Error Measures for Classification and Prediction Neonatal MRI View project Computational methods for omics data View project Data Mining: Accuracy and Error Measures for Classification and Prediction. Encycl. Bioinform. Comput. Biol. 2019, 1, 431–436. [Google Scholar] [CrossRef]
- Johnson, J.M.; Khoshgoftaar, T.M. Survey on deep learning with class imbalance. J. Big Data 2019, 6, 27. [Google Scholar] [CrossRef]
- Accuracy Paradox—Wikipedia. Available online: https://en.wikipedia.org/wiki/Accuracy_paradox (accessed on 14 January 2022).
- Valverde-Albacete, F.J.; Peláez-Moreno, C. 100% Classification Accuracy Considered Harmful: The Normalized Information Transfer Factor Explains the Accuracy Paradox. PLoS ONE 2014, 9, e84217. [Google Scholar] [CrossRef] [PubMed]
- Saminsm/Disease-Diagnosis-Using-Machine-Learning. Available online: https://github.com/saminsm/Disease-Diagnosis-using-Machine-Learning (accessed on 1 February 2022).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).