Hatchery-Imposed Selection Does Not Impact the Genetic Diversity of Australian Farmed Blue Mussels (Mytilus spp.)
Abstract
1. Introduction
2. Methodology
2.1. Sample Collection
2.2. DNA Extraction and Microsatellite Genotyping
2.3. Statistical Analysis
3. Results
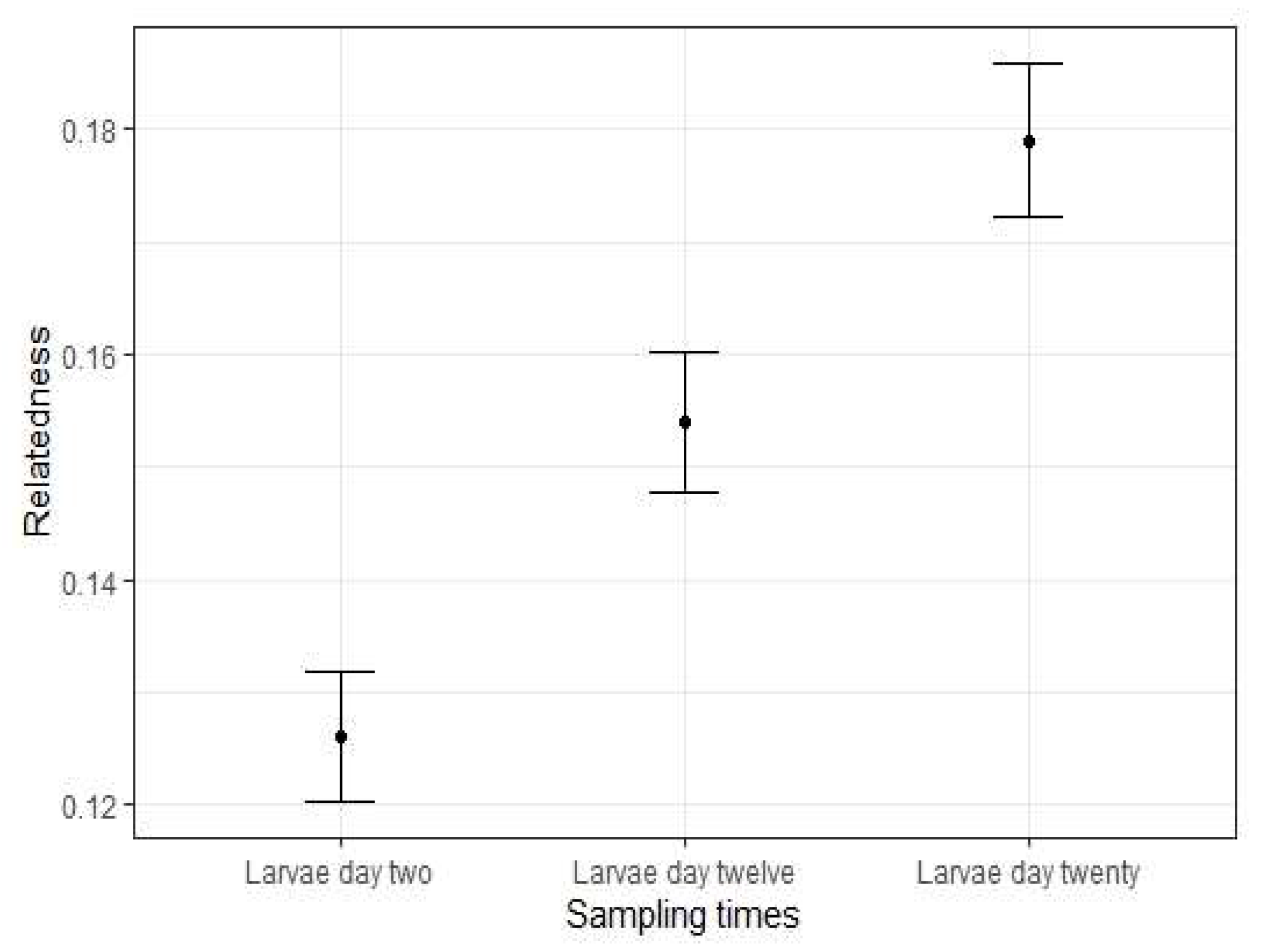
3.1. Genetic Diversity of Hatchery-Raised Larvae
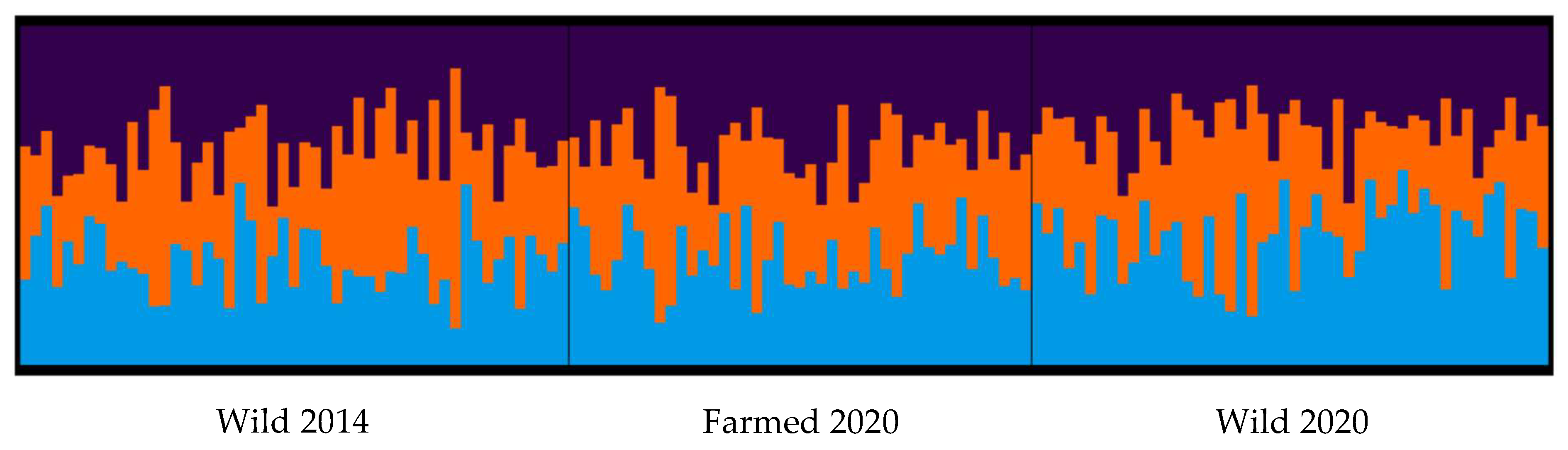
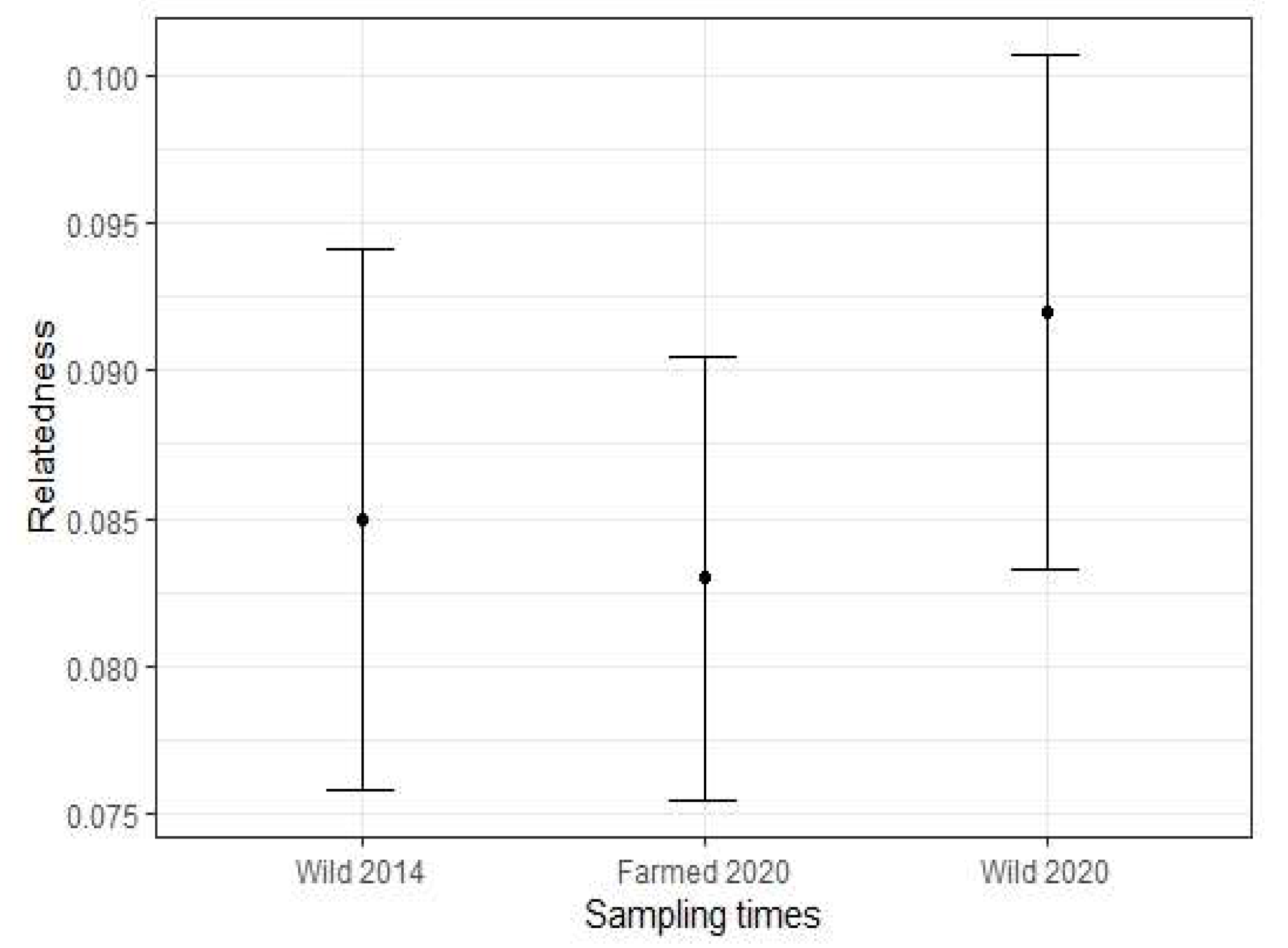
3.2. Genetic Diversity of Wild and Farmed Adult Mussel Populations
3.3. Genetic Diversity of Hatchery-Raised Larvae Compared to Adult Farmed and Wild Mussel Populations
4. Discussion
5. Management Implications and Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lehane, S. Fish for the Future: Aquaculture and Food Security; Future Directions International Pty Ltd.: Nedlands, Australia, 2013; Available online: www.futuredirections.org.au/publication/fish-for-the-future-aquaculture-andfoodsecurity (accessed on 1 March 2024).
- Fisheries, F. The State of World Fisheries and Aquaculture—Towards Blue Transformation; FAO: Rome, Italy, 2022. [Google Scholar]
- Wijsman, J.W.M.; Troost, K.; Fang, J.; Roncarati, A. Global Production of Marine Bivalves. Trends and Challenges. In Goods and Services of Marine Bivalves; Smaal, A.C., Ferreira, J.G., Grant, J., Petersen, J.K., Strand, Ø., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 7–26. [Google Scholar]
- Costelloe, T. Regional Report for Oceania; CMS: Bonn, Germany, 2022; Volume 682, p. 181. [Google Scholar]
- Boyd, C.E.; D’Abramo, L.R.; Glencross, B.D.; Huyben, D.C.; Juarez, L.M.; Lockwood, G.S.; McNevin, A.A.; Tacon, A.G.J.; Teletchea, F.; Tomasso, J.R., Jr.; et al. Achieving sustainable aquaculture: Historical and current perspectives and future needs and challenges. J. World Aquac. Soc. 2020, 51, 578–633. [Google Scholar] [CrossRef]
- Waples, R.S.; Hindar, K.; Hard, J.J. Genetic Risks Associated with Marine Aquaculture; U.S. Department of Commerce: Washington, DC, USA, 2012. [Google Scholar]
- Popovic, I.; Matias, A.M.A.; Bierne, N.; Riginos, C. Twin introductions by independent invader mussel lineages are both associated with recent admixture with a native congener in Australia. Evol. Appl. 2020, 13, 515–532. [Google Scholar] [CrossRef]
- Roodt-Wilding, R. Abalone ranching: A review on genetic considerations. Aquac. Res. 2007, 38, 1229–1241. [Google Scholar] [CrossRef]
- Zbawicka, M.; Wenne, R.; Dias, P.J.; Gardner, J.P.A. Combined threats to native smooth-shelled mussels (genus Mytilus) in Australia: Bioinvasions and hybridization. Zool. J. Linn. Soc. 2021, 194, 1194–1211. [Google Scholar] [CrossRef]
- Gjedrem, T.; Robinson, N.; Rye, M. The importance of selective breeding in aquaculture to meet future demands for animal protein: A review. Aquaculture 2012, 350–353, 117–129. [Google Scholar] [CrossRef]
- Argue, B.J.; Arce, S.M.; Lotz, J.M.; Moss, S.M. Selective breeding of Pacific white shrimp (Litopenaeus vannamei) for growth and resistance to Taura Syndrome Virus. Aquaculture 2002, 204, 447–460. [Google Scholar] [CrossRef]
- Moss, S.M.; Moss, D.R.; Arce, S.M.; Lightner, D.V.; Lotz, J.M. The role of selective breeding and biosecurity in the prevention of disease in penaeid shrimp aquaculture. J. Invertebr. Pathol. 2012, 110, 247–250. [Google Scholar] [CrossRef]
- Jaris, H.; Brown, D.S.; Proestou, D.A. Assessing the contribution of aquaculture and restoration to wild oyster populations in a Rhode Island coastal lagoon. Conserv. Genet. 2019, 20, 503–516. [Google Scholar] [CrossRef]
- Šegvić-Bubić, T.; Žužul, I.; Talijančić, I.; Ugrin, N.; Lepen Pleić, I.; Žuvić, L.; Stagličić, N.; Grubišić, L. Translocation and aquaculture impact on genetic diversity and composition of wild self-sustainable Ostrea edulis populations in the Adriatic sea. Front. Mar. Sci. 2020, 7, 84. [Google Scholar] [CrossRef]
- Taylor, E.; Fraser, D.J.; Minto, C.; Calvert, A.M.; Eddington, J.D.; Hutchings, J.A. Potential for domesticated–wild interbreeding to induce maladaptive phenology across multiple populations of wild Atlantic salmon (Salmo salar). Can. J. Fish. Aquat. Sci. 2010, 67, 1768–1775. [Google Scholar] [CrossRef]
- Heath, D.D.; Heath, J.W.; Bryden, C.A.; Johnson, R.M.; Fox, C.W. Rapid Evolution of Egg Size in Captive Salmon. Science 2003, 299, 1738. [Google Scholar] [CrossRef] [PubMed]
- Bramwell, G.; Schultz, A.G.; Jennings, G.; Nini, U.N.; Vanbeek, C.; Biro, P.A.; Beckmann, C.; Dujon, A.M.; Thomas, F.; Sherman, C.D. The effect of mitochondrial recombination on fertilization success in blue mussels. Sci. Total Environ. 2024, 913, 169491. [Google Scholar] [CrossRef] [PubMed]
- Larraín, M.A.; Díaz, N.F.; Lamas, C.; Uribe, C.; Jilberto, F.; Araneda, C. Heterologous microsatellite-based genetic diversity in blue mussel (Mytilus chilensis) and differentiation among localities in southern Chile. Lat. Am. J. Aquat. Res. 2015, 43, 998–1010. [Google Scholar] [CrossRef]
- Zbawicka, M.; Trucco, M.I.; Wenne, R. Single nucleotide polymorphisms in native South American Atlantic coast populations of smooth shelled mussels: Hybridization with invasive European Mytilus galloprovincialis. Genet. Sel. Evol. 2018, 50, 5. [Google Scholar] [CrossRef]
- ABARE. Australian Fisheries and Aquaculture Statistics 2018; ABARE: Canberra, Australia, 2019. [Google Scholar]
- Jahangard, S.; Williams, M.; Mercer, J.; Ab Rahim, E.; Ingram, B. A technical report on hatchery production of blue mussel Mytilus galloprovincialis at the Victoria Shellfish Hatchery (VSH), Queenscliff-2008. Fish. Vic. Tech. Rep. 2010, 112, 42. [Google Scholar]
- Beaumont, A.R. Genetic considerations in hatchery culture of bivalve shellfish. Recent Adv. Mar. Biotechnol. 2000, 4, 87–109. [Google Scholar]
- Pettersen, A.K.; Turchini, G.M.; Jahangard, S.; Ingram, B.A.; Sherman, C.D. Effects of different dietary microalgae on survival, growth, settlement and fatty acid composition of blue mussel (Mytilus galloprovincialis) larvae. Aquaculture 2010, 309, 115–124. [Google Scholar] [CrossRef]
- Sherman, C.D.; Ab Rahim, E.S.; Olsson, M.; Careau, V. The more pieces, the better the puzzle: Sperm concentration increases gametic compatibility. Ecol. Evol. 2015, 5, 4354–4364. [Google Scholar] [CrossRef]
- Nguyen, T.; Hayes, B.; Guthridge, K.; Ab Rahim, E.; Ingram, B. Use of a microsatellite-based pedigree in estimation of heritabilities for economic traits in Australian blue mussel, Mytilus galloprovincialis. J. Anim. Breed. Genet. 2011, 128, 482–490. [Google Scholar] [CrossRef]
- Lallias, D.; Stockdale, R.; Boudry, P.; Lapègue, S.; Beaumont, A.R. Characterization of ten microsatellite loci in the blue mussel Mytilus edulis. J. Shellfish Res. 2009, 28, 547–551. [Google Scholar] [CrossRef]
- Presa, P.; Pérez, M.; Pérez Diz, Á. Polymorphic microsatellite markers for blue mussels (Mytilus spp.). Conserv. Genet. 2002, 3, 441–443. [Google Scholar] [CrossRef]
- Li, J.; Wang, G.; Bai, Z. Genetic variability in four wild and two farmed stocks of the Chinese freshwater pearl mussel (Hyriopsis cumingii) estimated by microsatellite DNA markers. Aquaculture 2009, 287, 286–291. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Rousset, F. genepop’007: A complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. 2005, 1, 117693430500100003. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Morvezen, R.; Boudry, P.; Laroche, J.; Charrier, G. Stock enhancement or sea ranching? Insights from monitoring the genetic diversity, relatedness and effective population size in a seeded great scallop population (Pecten maximus). Heredity 2016, 117, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, M.L. Statistical Computing with R, 2nd, ed.; Chapman and Hall/CRC: New York, NY, USA, 2019. [Google Scholar]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; vonHoldt, B.M. Structure harvester: A website and program for visualizing structure output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Wang, J. COANCESTRY: A program for simulating, estimating and analysing relatedness and inbreeding coefficients. Mol. Ecol. Resour. 2011, 11, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. Triadic IBD coefficients and applications to estimating pairwise relatedness. Genet. Res. 2007, 89, 135–153. [Google Scholar] [CrossRef]
- Hughes, A.R.; Inouye, B.D.; Johnson, M.T.J.; Underwood, N.; Vellend, M. Ecological consequences of genetic diversity. Ecol. Lett. 2008, 11, 609–623. [Google Scholar] [CrossRef]
- Ujvari, B.; Klaassen, M.; Raven, N.; Russell, T.; Vittecoq, M.; Hamede, R.; Thomas, F.; Madsen, T. Genetic diversity, inbreeding and cancer. Proc. R. Soc. B Biol. Sci. 2018, 285, 20172589. [Google Scholar] [CrossRef]
- de Boer, R.A.; Eens, M.; Müller, W. Sex-specific effects of inbreeding on reproductive senescence. Proc. R. Soc. B Biol. Sci. 2018, 285, 20180231. [Google Scholar] [CrossRef]
- Sumida, T.S.; Hafler, D.A. Population genetics meets single-cell sequencing. Science 2022, 376, 134–135. [Google Scholar] [CrossRef]
- Gwak, W.; Nakayama, K. Genetic variation of hatchery and wild stocks of the pearl oyster Pinctada fucata martensii (Dunker, 1872), assessed by mitochondrial DNA analysis. Aquac. Int. 2011, 19, 585–591. [Google Scholar] [CrossRef]
- Fang, J.; Han, Z.; Li, Q. Effect of inbreeding on performance and genetic parameters of growth and survival traits in the Pacific oyster Crassostrea gigas at larval stage. Aquac. Rep. 2021, 19, 100590. [Google Scholar] [CrossRef]
- Turelli, M.; Ginzburg, L.R. Should individual fitness increase with heterozygosity? Genetics 1983, 104, 191–209. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, J.; Morrison, C.L.; Reece, K.S. Wild and aquaculture populations of the eastern oyster compared using microsatellites. J. Hered. 2006, 97, 595–598. [Google Scholar] [CrossRef] [PubMed]
- Gurney-Smith, H.J.; Wade, A.J.; Abbott, C.L. Species composition and genetic diversity of farmed mussels in British Columbia, Canada. Aquaculture 2017, 466, 33–40. [Google Scholar] [CrossRef]
- Kochmann, J.; Carlsson, J.; Crowe, T.P.; Mariani, S. Genetic evidence for the uncoupling of local aquaculture activities and a population of an invasive species—A case study of Pacific oysters (Crassostrea gigas). J. Hered. 2012, 103, 661–671. [Google Scholar] [CrossRef]
- Lind, C.E.; Evans, B.S.; Knauer, J.; Taylor, J.J.; Jerry, D.R. Decreased genetic diversity and a reduced effective population size in cultured silver-lipped pearl oysters (Pinctada maxima). Aquaculture 2009, 286, 12–19. [Google Scholar] [CrossRef]
- Geist, J.; Bayerl, H.; Stoeckle, B.C.; Kuehn, R. Securing genetic integrity in freshwater pearl mussel propagation and captive breeding. Sci. Rep. 2021, 11, 16019. [Google Scholar] [CrossRef] [PubMed]
- Araki, H.; Schmid, C. Is hatchery stocking a help or harm?: Evidence, limitations and future directions in ecological and genetic surveys. Aquaculture 2010, 308, S2–S11. [Google Scholar] [CrossRef]
- Jansen, P.A.; Kristoffersen, A.B.; Viljugrein, H.; Jimenez, D.; Aldrin, M.; Stien, A. Sea lice as a density-dependent constraint to salmonid farming. Proc. R. Soc. B Biol. Sci. 2012, 279, 2330–2338. [Google Scholar] [CrossRef]
- Ben-Horin, T.; Burge, C.A.; Bushek, D.; Groner, M.L.; Proestou, D.A.; Huey, L.I.; Bidegain, G.; Carnegie, R.B. Intensive oyster aquaculture can reduce disease impacts on sympatric wild oysters. Aquac. Environ. Interact. 2018, 10, 557–567. [Google Scholar] [CrossRef]
- Michalek, K.; Ventura, A.; Sanders, T. Mytilus hybridisation and impact on aquaculture: A minireview. Mar. Genom. 2016, 27, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, E.; Sadler-Riggleman, I.; Beck, D.; Skinner, M.K. Differential DNA methylation in somatic and sperm cells of hatchery vs wild (natural-origin) steelhead trout populations. Environ. Epigenetics 2021, 7, dvab002. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez Barreto, D.; Garcia de Leaniz, C.; Verspoor, E.; Sobolewska, H.; Coulson, M.; Consuegra, S. DNA methylation changes in the sperm of captive-reared fish: A route to epigenetic introgression in wild populations. Mol. Biol. Evol. 2019, 36, 2205–2211. [Google Scholar] [CrossRef] [PubMed]
- Schenekar, T.; Weiss, S. Selection and genetic drift in captive versus wild populations: An assessment of neutral and adaptive (MHC-linked) genetic variation in wild and hatchery brown trout (Salmo trutta) populations. Conserv. Genet. 2017, 18, 1011–1022. [Google Scholar] [CrossRef]
- Maine, U.O. Maine Seafood Guide—Mussels. Available online: https://seagrant.umaine.edu/maine-seafood-guide/mussels/ (accessed on 1 March 2024).
- Carroll, A. The Great Mussel and Clam Cookbook; R&R Publications Marketing Pty. Ltd.: Brunswick, Australia, 2004. [Google Scholar]
- Schagerström, E.; Christophersen, G.; Sunde, J.; Bakke, S.; Matusse, N.R.; Dupont, S.; Sundell, K.S. Controlled spawning and rearing of the sea cucumber, Parastichopus tremulus. J. World Aquac. Soc. 2022, 53, 224–240. [Google Scholar] [CrossRef]
- Smith, T.I.; Denson, M.R. Controlled spawning of Southern flounder Paralichthys lethostigma: Issues and progress. In Spawning and Maturation of Aquaculture Species, Proceedings of the 28th US-Japan Natural Resources Aquaculture Panel, Kihei, HI, USA, 10–12 November 1999; South Carolina Department of Natural Resources: Charleston, SC, USA, 10–12 November 2000. [Google Scholar]

| Larvae Day 2 | Larvae Day 12 | Larvae Day 20 | Wild 2014 | Farmed 2020 | Wild 2020 | |
|---|---|---|---|---|---|---|
| Larvae Day 2 | 0.000 | |||||
| Larvae Day 12 | 0.053 *** | 0.000 | ||||
| Larvae Day 20 | 0.055 *** | 0.000 | 0.000 | |||
| Wild 2014 | 0.100 *** | 0.111 *** | 0.108 *** | 0.000 | ||
| Farmed 2020 | 0.123 *** | 0.112 *** | 0.122 *** | 0.023 *** | 0.000 | |
| Wild 2020 | 0.130 *** | 0.105 *** | 0.095 *** | 0.040 *** | 0.026 ** | 0.000 |
| N | HO | HE | FIS | Relatedness | ||
|---|---|---|---|---|---|---|
| Larvae Day 2 | 96 | 6.500 | 0.398 | 0.613 | 0.350 | 0.126 * |
| Larvae Day 12 | 96 | 5.833 | 0.607 | 0.597 | −0.035 | 0.155 * |
| Larvae Day 20 | 96 | 5.500 | 0.550 | 0.581 | 0.013 | 0.179 * |
| Wild 2014 | 52 | 9.500 | 0.609 | 0.708 | 0.146 | 0.085 |
| Farmed 2020 | 42 | 9.667 | 0.657 | 0.761 | 0.124 | 0.083 |
| Wild 2020 | 47 | 10.800 | 0.489 | 0.754 | 0.346 | 0.092 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bramwell, G.; Ab Rahim, E.S.; Schultz, A.G.; Dujon, A.M.; Weston, K.; Ingram, B.A.; Jahangard, S.; Thomas, F.; Ujvari, B.; Sherman, C.D.H. Hatchery-Imposed Selection Does Not Impact the Genetic Diversity of Australian Farmed Blue Mussels (Mytilus spp.). Hydrobiology 2024, 3, 51-62. https://doi.org/10.3390/hydrobiology3010004
Bramwell G, Ab Rahim ES, Schultz AG, Dujon AM, Weston K, Ingram BA, Jahangard S, Thomas F, Ujvari B, Sherman CDH. Hatchery-Imposed Selection Does Not Impact the Genetic Diversity of Australian Farmed Blue Mussels (Mytilus spp.). Hydrobiology. 2024; 3(1):51-62. https://doi.org/10.3390/hydrobiology3010004
Chicago/Turabian StyleBramwell, Georgina, Emi S. Ab Rahim, Aaron G. Schultz, Antoine M. Dujon, Kim Weston, Brett A. Ingram, Sam Jahangard, Frédéric Thomas, Beata Ujvari, and Craig D. H. Sherman. 2024. "Hatchery-Imposed Selection Does Not Impact the Genetic Diversity of Australian Farmed Blue Mussels (Mytilus spp.)" Hydrobiology 3, no. 1: 51-62. https://doi.org/10.3390/hydrobiology3010004
APA StyleBramwell, G., Ab Rahim, E. S., Schultz, A. G., Dujon, A. M., Weston, K., Ingram, B. A., Jahangard, S., Thomas, F., Ujvari, B., & Sherman, C. D. H. (2024). Hatchery-Imposed Selection Does Not Impact the Genetic Diversity of Australian Farmed Blue Mussels (Mytilus spp.). Hydrobiology, 3(1), 51-62. https://doi.org/10.3390/hydrobiology3010004









