The Evolution of Life History Traits and Their Thermal Plasticity in Daphnia
Abstract
1. Introduction
2. Materials and Methods
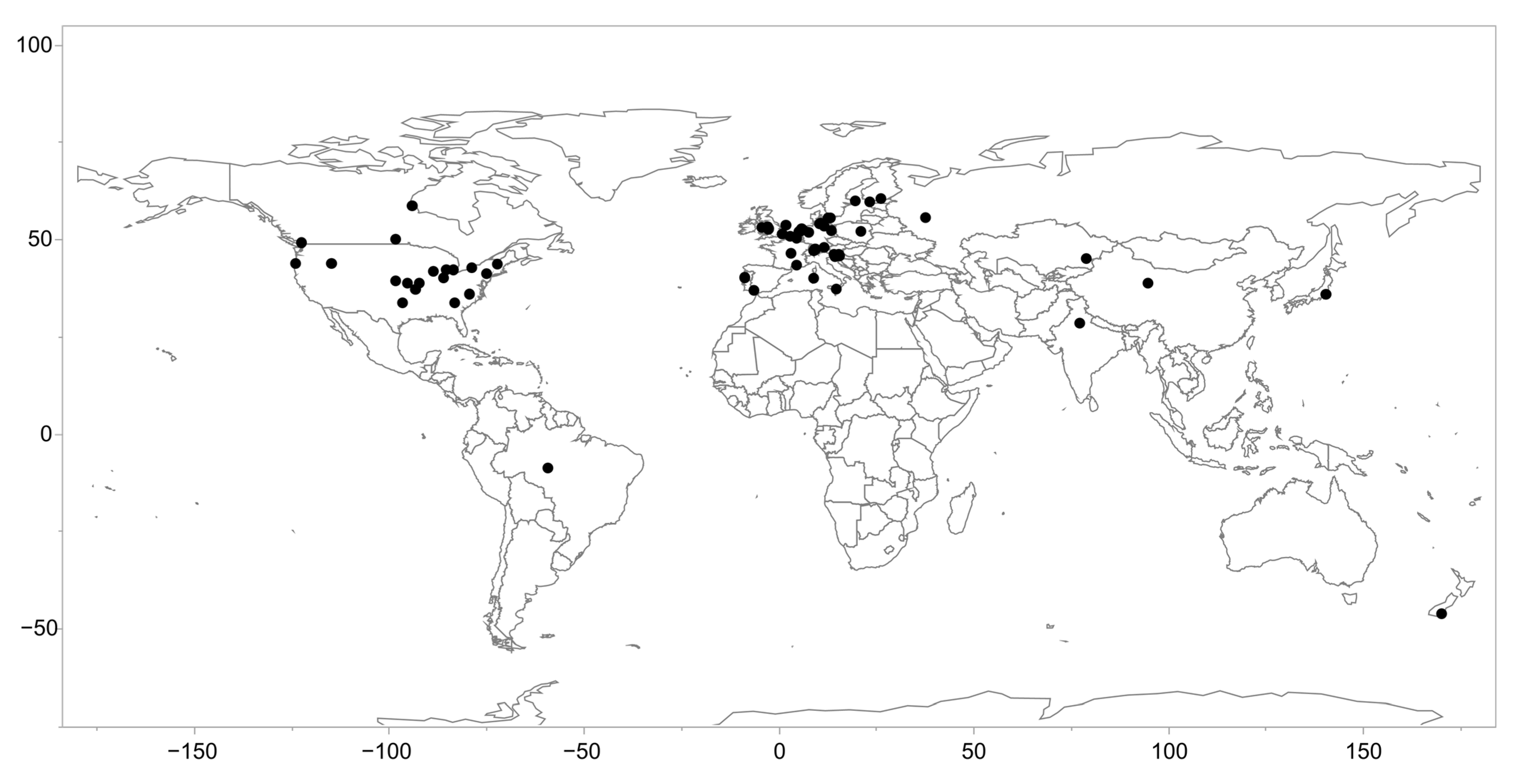
2.1. The Life History Dataset and Literature Review
2.2. Phylogeny
2.3. Statistical Analyses
2.3.1. Phylogenetic Signal
2.3.2. Climate and Temperature Variables
2.3.3. Life History Response Variables
2.3.4. Life History Traits and Climate Variables Regressions
- Species-level analyses—To test how climate affects life history traits, we used three different modeling structures. We used ordinary least squares (OLS) regressions to examine the predictive power of climate for life history traits. However, OLS models do not account for the known issues of phylogenetic relatedness among closely related lineages [33,34,35], more especially those that may have recent introgression events due to Daphnia’s elevated rate of hybridization. OLS models were conducted in JMP Pro 13.2 [36].
- Population-level analyses—We expected climate variables to be good predictor variables on a global scale or species-level, but when assessing individual populations, we were interested in whether climate, temperature, or latitude alone were enough, regardless of species, to predict variation in life history traits. We used a stepwise model selection approach for our population-level analysis. We built a full model for each life history trait that included the four climate variables (PC1temp, PC2temp, PC1precip, PC2precip), the reported experimental temperature, the latitude, and species identity as a fixed effect. Then, we added and reduced the model by individual variables and compared the Akaike information criterion corrected for small sample sizes (AICc) scores to choose the optimum model for each trait (Table 4). Individual species identity coefficients can be found in the Supplement (Table S3). All statistical analyses were performed in JMP Pro 13.2 [36] or R [46]; we used a 5% significance level to reject null hypotheses.
3. Results
3.1. Phylogeny
3.2. Life History Traits and Climate
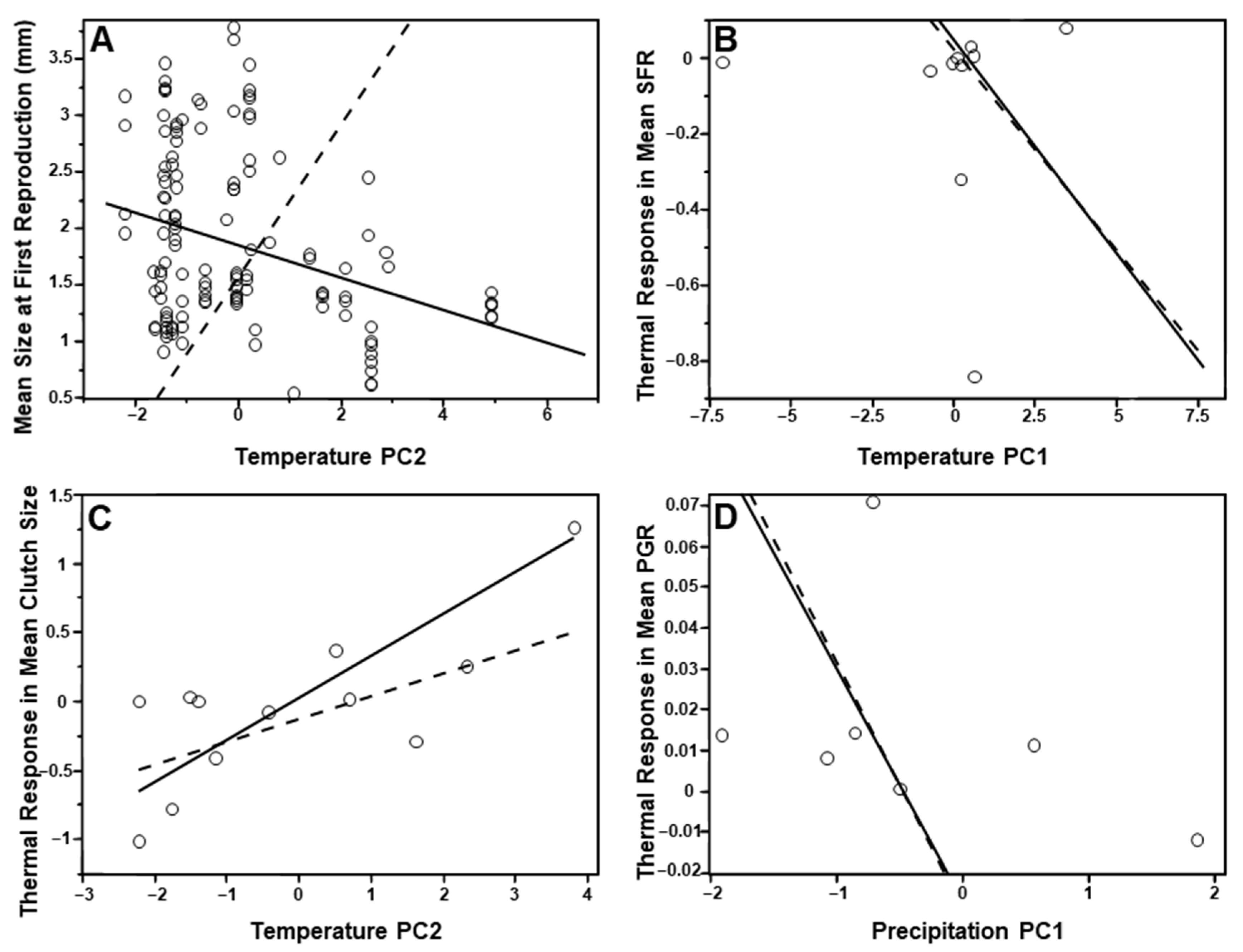
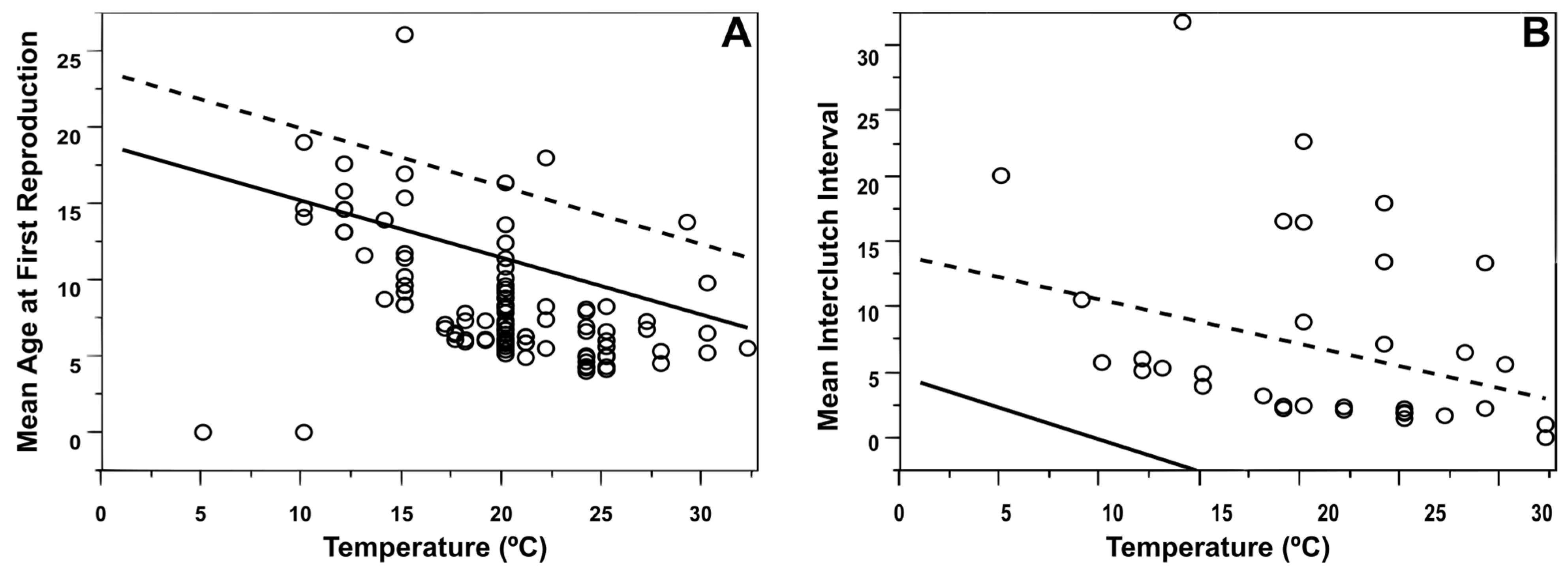
3.3. Species-Level Analyses
3.4. Population-Level Analyses
4. Discussion
4.1. Species-Level Analyses
4.2. Phylogeny
4.3. Life History Traits and Climate Dataset
4.4. Population-Level Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stearns, S.C. The Evolution of Life Histories; Oxford University Press: Oxford, UK, 1992. [Google Scholar]
- Price, T.; Schluter, D. On the low heritability of life-history traits. Evolution 1991, 45, 853–861. [Google Scholar] [CrossRef]
- Stearns, S.C. Life history evolution: Successes, limitations, and prospects. Naturwissenschaften 2000, 87, 476–486. [Google Scholar] [CrossRef]
- Van Noordwijk, A.J.; de Jong, G. Acquisition and allocation of resources: Their influence on variation in life history tactics. Am. Nat. 1986, 128, 137–142. [Google Scholar] [CrossRef]
- Metcalfe, N.B.; Monaghan, P. Compensation for a bad start: Grow now, pay later? Trends Ecol. Evol. 2001, 16, 254–260. [Google Scholar] [CrossRef]
- Pinsky, M.L.; Worm, B.; Fogarty, M.J.; Sarmiento, J.L.; Levin, S.A. Marine taxa track local climate velocities. Science 2013, 341, 1239–1242. [Google Scholar] [CrossRef] [PubMed]
- Nylin, S.; Gotthard, K. Plasticity in life-history traits. Annu. Rev. Entomol. 1998, 43, 63–83. [Google Scholar] [CrossRef] [PubMed]
- Partridge, L.; Barrie, B.; Barton, N.H.; Fowler, K.; French, V. Rapid laboratory evolution of adult life-history traits in Drosophila melanogaster in response to temperature. Evolution 1995, 49, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Sponaugle, S.; Grorud-Colvert, K.; Pinkard, D. Temperature-mediated variation in early life history traits and recruitment success of the coral reef fish Thalassoma bifasciatum in the Florida Keys. Mar. Ecol. Prog. Ser. 2006, 308, 1–15. [Google Scholar] [CrossRef]
- Bernot, R.J.; Dodds, W.K.; Quist, M.C.; Guy, C.S. Temperature and kairomone induced life history plasticity in coexisting Daphnia. Aquatic Ecology 2006, 40, 361–372. [Google Scholar] [CrossRef]
- Mitchell, S.E.; Lampert, W. Temperature adaptation in a geographically widespread zooplankter. Daphnia Magna. J. Evol. Biol. 2000, 13, 371–382. [Google Scholar] [CrossRef]
- Sakwińska, O. Plasticity of Daphnia magna life history traits in response to temperature and information about a predator. Freshw. Biol. 1998, 39, 681–687. [Google Scholar] [CrossRef]
- Arnett, A.E.; Gotelli, N.J. Geographic variation in life-history traits of the ant lion, Myrmeleon immaculatus: Evolutionary implications of Bergmann’s rule. Evolution 1999, 53, 1180–1188. [Google Scholar] [PubMed]
- Lombardi-Carlson, L.A.; Cortés, E.; Parsons, G.R.; Manire, C.A. Latitudinal variation in life-history traits of bonnethead sharks, Sphyrna tiburo,(Carcharhiniformes: Sphyrnidae) from the eastern Gulf of Mexico. Mar. Freshw. Res. 2003, 54, 875–883. [Google Scholar] [CrossRef]
- Geller, W.; Müller, H. Seasonal variability in the relationship between body length and individual dry weight as related to food abundance and clutch size in two coexisting Daphnia species. J. Plankton Res. 1985, 7, 1–18. [Google Scholar] [CrossRef]
- Gillooly, J.F.; Dodson, S.I. Latitudinal patterns in the size distribution and seasonal dynamics of new world, freshwater cladocerans. Limnol. Oceanogr. 2000, 45, 22–30. [Google Scholar] [CrossRef]
- Dodson, S. Predicting crustacean zooplankton species richness. Limnol. Oceanogr. 1992, 37, 848–856. [Google Scholar] [CrossRef]
- Hessen, D.O.; Faafeng, B.A.; Smith, V.H.; Bakkestuen, V.; Walseng, B. Extrinsic and intrinsic controls of zooplankton diversity in lakes. Ecology 2006, 87, 433–443. [Google Scholar] [CrossRef]
- Weber, A.; Declerck, S. Phenotypic plasticity of Daphnia life history traits in response to predator kairomones: Genetic variability and evolutionary potential. Hydrobiologia 1997, 360, 89–99. [Google Scholar] [CrossRef]
- Rohatgi, A. Webplotdigitizer: Version 4.4. 2020. Available online: https://automeris.io/WebPlotDigitizer (accessed on 1 December 2018).
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Mesquita, D.O.; Faria, R.G.; Colli, G.R.; Vitt, L.J.; Pianka, E.R. Lizard life-history strategies. Austral Ecol. 2016, 41, 1–5. [Google Scholar] [CrossRef]
- Revell, L.J. Phylogenetic signal and linear regression on species data. Methods Ecol. Evolution. 2010, 1, 319–329. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Colbourne, J.K.; Hebert, P.D. The systematics of North American Daphnia (Crustacea: Anomopoda): A molecular phylogenetic approach. Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 1996, 351, 349–360. [Google Scholar]
- Petrusek, A.; Hobæk, A.; Nilssen, J.P.; Skage, M.; ČErný, M.; Brede, N.; Schwenk, K. A taxonomic reappraisal of the European Daphnia longispina complex (Crustacea, Cladocera, Anomopoda). Zool. Scr. 2008, 37, 507–519. [Google Scholar] [CrossRef]
- Taylor, D.J.; Hebert, P.D. Genetic assessment of species boundaries in the North American Daphnia longispina complex (Crustacea: Daphniidae). Zool. J. Linn. Soc. 1994, 110, 27–40. [Google Scholar] [CrossRef]
- Pagel, M. Inferring evolutionary processes from phylogenies. Zool. Scr. 1997, 26, 331–348. [Google Scholar] [CrossRef]
- Pagel, M. Inferring the historical patterns of biological evolution. Nature 1999, 401, 877. [Google Scholar] [CrossRef]
- Blomberg, S.P.; Garland, T.; Ives, A.R. Testing for phylogenetic signal in comparative data: Behavioral traits are more labile. Evolution. 2003, 57, 717–745. [Google Scholar]
- Webb, C.O.; Ackerly, D.D.; McPeek, M.A.; Donoghue, M.J. Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 2002, 33, 475–505. [Google Scholar] [CrossRef]
- Ives, A.R.; Garland, T., Jr. Phylogenetic logistic regression for binary dependent variables. Syst. Biol. 2009, 59, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Ives, A.R.; Midford, P.E.; Garland, T., Jr. Within-species variation and measurement error in phylogenetic comparative methods. Syst. Biol. 2007, 56, 252–270. [Google Scholar] [CrossRef] [PubMed]
- SAS Institute Inc. JMP® Pro 13, 13.0.0 (64-bit); SAS Institute Inc.: Cary, NC, USA, 2016.
- Grafen, A. The phylogenetic regression. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1989, 326, 119–157. [Google Scholar]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D. R Core Team Nlme: Linear and Nonlinear Mixed Effects Models. R Package Version 3.1-117. 2014. Available online: https://CRAN.R-project.org/package=nlme. (accessed on 1 December 2018).
- Cavalli-Sforza, L.L.; Edwards, A.W. Phylogenetic analysis: Models and estimation procedures. Evolution 1967, 21, 550–570. [Google Scholar] [CrossRef]
- Lande, R. Natural selection and random genetic drift in phenotypic evolution. Evolution 1976, 30, 314–334. [Google Scholar] [CrossRef]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef]
- Felsenstein, J. Phylogenies and the comparative method. Am. Nat. 1985, 125, 1–15. [Google Scholar] [CrossRef]
- Martins, E.P.; Hansen, T.F. Phylogenies and the comparative method: A general approach to incorporating phylogenetic information into the analysis of interspecific data. Am. Nat. 1997, 149, 646–667. [Google Scholar] [CrossRef]
- Ripley, B.; Venables, B.; Bates, D.M.; Hornik, K.; Gebhardt, A.; Firth, D. R Package ‘mass’. 2013. Available online: https://CRAN.R-project.org/package=MASS (accessed on 1 December 2018).
- Harmon, L.J.; Weir, J.T.; Brock, C.D.; Glor, R.E.; Challenger, W. GEIGER: Investigating evolutionary radiations. Bioinformatics 2007, 24, 129–131. [Google Scholar] [CrossRef]
- R Development Core Team, R stats. RA Lang Environ. Stat Comput. 2013, 55, 275–286.
- Freckleton, R.P.; Harvey, P.H.; Pagel, M. Phylogenetic analysis and comparative data: A test and review of evidence. Am. Nat. 2002, 160, 712–726. [Google Scholar] [CrossRef]
- Colbourne, J.K.; Pfrender, M.E.; Gilbert, D.; Thomas, W.K.; Tucker, A.; Oakley, T.H.; Tokishita, S.; Aerts, A.; Arnold, G.J.; Boore, J.L.; et al. The ecoresponsive genome of Daphnia pulex. Science 2011, 331, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Gliwicz, Z.M.; Boavida, M.J. Clutch size and body size at first reproduction in Daphnia pulicaria at different levels of food and predation. J. Plankton Res. 1996, 18, 863–880. [Google Scholar] [CrossRef]
- Shine, R. “Costs” of reproduction in reptiles. Oecologia 1980, 46, 92–100. [Google Scholar] [CrossRef] [PubMed]
- McNab, B.K. On the ecological significance of Bergmann’s rule. Ecology 1971, 52, 845–854. [Google Scholar] [CrossRef]
- Blackburn, T.M.; Gaston, K.J.; Loder, N. Geographic gradients in body size: A clarification of Bergmann’s rule. Divers. Distrib. 1999, 5, 165–174. [Google Scholar] [CrossRef]
- Steiner, C.F. Temporal stability of pond zooplankton assemblages. Freshwat. Biol. 2005, 50, 105–112. [Google Scholar] [CrossRef]
- Vasseur, D.A.; Gaedke, U. Spectral analysis unmasks synchronous and compensatory dynamics in plankton communities. Ecology 2007, 88, 2058–2071. [Google Scholar]
- Drake, J.M.; Lodge, D.M. Effects of environmental variation on extinction and establishment. Ecol. Lett. 2004, 7, 26–30. [Google Scholar] [CrossRef]
- Kramer, A.M.; Drake, J.M. Experimental demonstration of population extinction due to a predator-driven Allee effect. J. Anim. Ecol. 2010, 79, 633–639. [Google Scholar] [CrossRef]
- Hanski, I.; Ranta, E. Coexistence in a patchy environment: Three species of Daphnia in rock pools. J. Anim. Ecol. 1983, 52, 263–279. [Google Scholar] [CrossRef]
- West, D.C.; Post, D.M. Impacts of warming revealed by linking resource growth rates with consumer functional responses. J. Anim. Ecol. 2016, 85, 671–680. [Google Scholar] [CrossRef]
- Luecke, C.; Vanni, M.J.; Magnuson, J.J.; Kitchell, J.F.; Jacobson, P.T. Seasonal regulation of Daphnia populations by planktivorous fish: Implications for the spring clear-water phase. Limnol. Oceanogr. 1990, 35, 1718–1733. [Google Scholar] [CrossRef]
- Scheffer, M.; Rinaldi, S.; Kuznetsov, Y.A.; van Nes, E.H. Seasonal dynamics of Daphnia and algae explained as a periodically forced predator-prey system. Oikos 1997, 80, 519–532. [Google Scholar] [CrossRef]
- Müller-Navarra, D.; Lampert, W. Seasonal patterns of food limitation in Daphnia galeata: Separating food quantity and food quality effects. J. Plankton Res. 1996, 18, 1137–1157. [Google Scholar] [CrossRef]
- Edmondson, W. The seasonal life history of Daphnia in an arctic lake. Ecology 1955, 36, 439–455. [Google Scholar] [CrossRef]
- Fairbairn, D.J. Allometry for sexual size dimorphism: Pattern and process in the coevolution of body size in males and females. Annu. Rev. Ecol. Syst. 1997, 28, 659–687. [Google Scholar] [CrossRef]
- Gould, S.J. Allometry and size in ontogeny and phylogeny. Biol. Rev. 1966, 41, 587–638. [Google Scholar] [CrossRef]
- Ryan, M.J.; Brenowitz, E.A. The role of body size, phylogeny, and ambient noise in the evolution of bird song. Am. Nat. 1985, 126, 87–100. [Google Scholar] [CrossRef]
- Scheiner, S.M.; Berrigan, D. The genetics of phenotypic plasticity. VIII. The cost of plasticity in Daphnia pulex. Evolution 1998, 52, 368–378. [Google Scholar] [CrossRef] [PubMed]
- Giesel, J.T. Reproductive strategies as adaptations to life in temporally heterogeneous environments. Annu. Rev. Ecol. Syst. 1976, 7, 57–79. [Google Scholar] [CrossRef]
- Zhang, L.; Baer, K. The influence of feeding, photoperiod and selected solvents on the reproductive strategies of the water flea, Daphnia magna. Environ. Pollut. 2000, 110, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Jönsson, K.I. Capital and income breeding as alternative tactics of resource use in reproduction. Oikos 1997, 78, 57–66. [Google Scholar] [CrossRef]
- Bonnet, X.; Bradshaw, D.; Shine, R. Capital versus income breeding: An ectothermic perspective. Oikos 1998, 83, 333–342. [Google Scholar] [CrossRef]
- Bowman, L.L.; Kondrateva, E.S.; Silow, E.A.; Wilburn, P.; Yampolsky, L.Y. A capital breeder in a heterogeneous environment: Lipid reserves and RNA: DNA ratio in Lake Baikal’s endemic Epischura. J. Great Lakes Res. 2017, 43, 280–288. [Google Scholar] [CrossRef]
- Craddock, D.R. Effects of increased water temperature on Daphnia pulex. Fish. Bull 1976, 74, 403–408. [Google Scholar]
- Burns, C.W. Relation between filtering rate, temperature, and body size in four species of Daphnia. Limnol. Oceanogr. 1969, 14, 693–700. [Google Scholar] [CrossRef]
- Tessier, A.J.; Henry, L.L.; Goulden, C.E.; Durand, M.W. Starvation in Daphnia: Energy reserves and reproductive allocation1. Limnol. Oceanogr. 1983, 28, 667–676. [Google Scholar] [CrossRef]
- Scheiner, S.M.; Yampolsky, L.Y. The evolution of Daphnia pulex in a temporally varying environment. Genet. Res. 1998, 72, 25–37. [Google Scholar] [CrossRef]
- Adamowicz, S.J.; Hebert, P.D.; Marinone, M.C. Species diversity and endemism in the Daphnia of Argentina: A genetic investigation. Zool. J. Linn. Soc. 2004, 140, 171–205. [Google Scholar] [CrossRef]
- Hebert, P.D.; Wilson, C.C. Provincialism in plankton: Endemism and allopatric speciation in Australian Daphnia. Evolution. 1994, 48, 1333–1349. [Google Scholar] [CrossRef]
- Beaulieu, J.M.; Jhwueng, D.; Boettiger, C.; O’Meara, B.C. Modeling stabilizing selection: Expanding the Ornstein–Uhlenbeck model of adaptive evolution. Evol. Int. J. Org. Evol. 2012, 66, 2369–2383. [Google Scholar] [CrossRef] [PubMed]
- Ingram, T.; Mahler, D.L. SURFACE: Detecting convergent evolution from comparative data by fitting Ornstein-Uhlenbeck models with stepwise Akaike Information Criterion. Methods Ecol. Evol. 2013, 4, 416–425. [Google Scholar] [CrossRef]
- Sinervo, B.; Licht, P. Hormonal and physiological control of clutch size, egg size, and egg shape in side-blotched lizards (Uta stansburiana): Constraints on the evolution of lizard life histories. J. Exp. Zool. 1991, 257, 252–264. [Google Scholar] [CrossRef]
- Monaghan, P.; Bolton, M.; Houston, D. Egg production constraints and the evolution of avian clutch size. Proc.R.Soc.Lond.B. 1995, 259, 189–191. [Google Scholar]
- Stoks, R.; De Block, M.; Van De Meutter, F.; Johansson, F. Predation cost of rapid growth: Behavioural coupling and physiological decoupling. J. Anim. Ecol. 2005, 74, 708–715. [Google Scholar] [CrossRef]
- Schulte, P.M.; Healy, T.M.; Fangue, N.A. Thermal performance curves, phenotypic plasticity, and the time scales of temperature exposure. Integr. Comp. Biol. 2011, 51, 691–702. [Google Scholar] [CrossRef] [PubMed]
- De Meester, L.; Boersma, M.; Spaak, P. Environmental stress and local adaptation in Daphnia magna. Limnol. Oceanogr 1999, 44, 393–402. [Google Scholar] [CrossRef]
- Tessier, A.J.; Young, A.; Leibold, M. Population dynamics and body-size selection in Daphnia. Limnol. Oceanogr. 1992, 37, 1–13. [Google Scholar] [CrossRef]
- Stibor, H.; Lampert, W. Components of additive variance in life-history traits of Daphnia hyalina: Seasonal differences in the response to predator signals. Oikos 2000, 88, 129–138. [Google Scholar] [CrossRef]
- Lennon, J.T.; Smith, V.H.; Williams, K. Influence of temperature on exotic Daphnia lumholtzi and implications for invasion success. J. Plankton Res. 2001, 23, 425–433. [Google Scholar] [CrossRef]
- Duffy, M.A.; Hall, S.R.; Cáceres, C.E.; Ives, A.R. Rapid evolution, seasonality, and the termination of parasite epidemics. Ecology 2009, 90, 1441–1448. [Google Scholar] [CrossRef] [PubMed]
- Borenstein, E.; Meilijson, I.; Ruppin, E. The effect of phenotypic plasticity on evolution in multipeaked fitness landscapes. J. Evol. Biol. 2006, 19, 1555–1570. [Google Scholar] [CrossRef]
- Duggan, I.C.; Robinson, K.V.; Burns, C.W.; Banks, J.C.; Hogg, I.D. Identifying invertebrate invasions using morphological and molecular analyses: North American Daphnia ‘pulex’ in New Zealand fresh waters. Aquat. Invasions 2012, 7, 585–590. [Google Scholar] [CrossRef]
- Adamowicz, S.J.; Gregory, T.R.; Marinone, M.C.; Hebert, P.D. New insights into the distribution of polyploid Daphnia: The Holarctic revisited and Argentina explored. Mol. Ecol. 2002, 11, 1209–1217. [Google Scholar] [CrossRef]
- Penton, E.; Hebert, P.; Crease, T. Mitochondrial DNA variation in North American populations of Daphnia obtusa: Continentalism or cryptic endemism? Mol. Ecol. 2004, 13, 97–107. [Google Scholar] [CrossRef]
- Gilbert, B.; Bennett, J.R. Partitioning variation in ecological communities: Do the numbers add up? J. Appl. Ecol. 2010, 47, 1071–1082. [Google Scholar] [CrossRef]
- Guisan, A.; Zimmermann, N.E. Predictive habitat distribution models in ecology. Ecol. Model. 2000, 135, 147–186. [Google Scholar] [CrossRef]
- Vasseur, D.A.; DeLong, J.P.; Gilbert, B.; Greig, H.S.; Harley, C.D.; McCann, K.S.; Savage, V.; Tunney, T.D.; O’Connor, M. Increased temperature variation poses a greater risk to species than climate warming. Proc. Biol. Sci. 2014, 281, 20132612. [Google Scholar] [CrossRef]
- Wieczynski, D.J.; Vasseur, D.A. Environmental fluctuations promote intraspecific diversity and population persistence via inflationary effects. Oikos 2016, 125, 1173–1181. [Google Scholar] [CrossRef]
- Lampert, W. Phenotypic plasticity of the size at first reproduction in Daphnia: The importance of maternal size. Ecology 1993, 74, 1455–1466. [Google Scholar] [CrossRef]
- Post, D.M.; Palkovacs, E.P.; Schielke, E.G.; Dodson, S.I. Intraspecific variation in a predator affects community structure and cascading trophic interactions. Ecology 2008, 89, 2019–2032. [Google Scholar] [CrossRef] [PubMed]
- Roff, D.A.; Heibo, E.; Vøllestad, L. The importance of growth and mortality costs in the evolution of the optimal life history. J. Evol. Biol. 2006, 19, 1920–1930. [Google Scholar] [CrossRef] [PubMed]
- Hampton, S.E.; Galloway, A.W.; Powers, S.M.; Ozersky, T.; Woo, K.H.; Batt, R.D.; Labou, S.G.; O’Reilly, C.M.; Sharma, S.; Lottig, N.R.; et al. Ecology under lake ice. Ecol. Lett. 2017, 20, 98–111. [Google Scholar] [CrossRef] [PubMed]
- Sainte-Marie, B. A review of the reproductive bionomics of aquatic gammaridean amphipods: Variation of life history traits with latitude, depth, salinity and superfamily. Hydrobiologia 1991, 223, 189–227. [Google Scholar] [CrossRef]
- Costa, D.P. Reproductive and foraging energetics of high latitude penguins, albatrosses and pinnipeds: Implications for life history patterns. Am. Zool. 1991, 31, 111–130. [Google Scholar] [CrossRef]

| Variable | N | Blomberg’s K | p | Pagel’s λ | p | Mode of Evolution |
|---|---|---|---|---|---|---|
| Life History Traits: | ||||||
| Age at First Reproduction | 111 | 0.773 | 0.09 | 1.00 | 0.08 | BM |
| Size at First Reproduction | 144 | 0.635 | 0.03 * | 1.00 | 0.06 | BM |
| Clutch Size | 116 | 0.304 | 0.56 | <0.001 | 1.0 | OU |
| Juvenile Growth Rate | 120 | 0.872 | 0.07 | 1.00 | 0.26 | OU |
| Population Growth Rate | 140 | 0.769 | 0.15 | 1.00 | 0.40 | OU |
| Size at Death | 28 | 10.14 | 0.08 | 1.00 | 0.26 | BM |
| Interclutch Interval | 37 | 0.718 | 0.58 | <0.001 | 1.0 | OU |
| Lifespan | 38 | 0.671 | 0.83 | <0.001 | 1.0 | BM |
| Temperature Response | ||||||
| Age at First Reproduction | 111 | 0.799 | 0.13 | 0.536 | 0.33 | BM |
| Size at First Reproduction | 144 | 0.382 | 0.32 | <0.001 | 1.0 | BM |
| Clutch Size | 116 | 0.257 | 0.61 | <0.001 | 1.0 | OU |
| Juvenile Growth Rate | 120 | 0.397 | 0.45 | <0.001 | 1.0 | BM |
| Population Growth Rate | 140 | 0.669 | 0.44 | <0.001 | 1.0 | BM |
| Size at Death | 28 | 10.27 | 0.23 | 0.975 | 0.29 | BM |
| Interclutch Interval | 37 | 0.735 | 0.56 | <0.001 | 1.0 | BM |
| Lifespan | 38 | 0.637 | 0.79 | <0.001 | 1.0 | BM |
| Climate: | ||||||
| Temp PC1 | 0.337 | 0.50 | 0.354 | 0.69 | ||
| Temp PC2 | 0.355 | 0.39 | <.001 | 1.0 | ||
| Precip PC1 | 0.526 | 0.17 | 0.809 | 0.56 | ||
| Precip PC2 | 0.297 | 0.59 | <0.001 | 1.0 | ||
| * p < 0.05 |
| Response Variable and Model | Regression Summaries | β (Explanatory Variables) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Adj r2 | F/χ2 | df | p | Intercept | PC1temp | PC2temp | PC1precip | PC2precip | AIC † | |
| Age at First Reproduction | ||||||||||
| OLS | 0.478 | 3.06 | 5, 10 | 0.126 | 8.60 *** | 1.16 * | 0.946 | −0.292 | −1.98 * | 639 |
| PGLSBM | 0.640 | 5.03 | 5 | 0.039 | 8.40 | 1.20 | 1.22 | −0.190 | −2.12 | 47.75 |
| PGLSOU | 0.930 | 13.2 | 5 | <0.001 | 8.47 ** | 1.26 | 1.12 | −0.219 | −2.24 | 50.50 |
| Size at First Reproduction | ||||||||||
| OLS | −0.133 | 0.678 | 4, 11 | 0.628 | 1.52 *** | 0.167 | 0.656 | 0.222 | 0.019 | 304 |
| PGLSBM | 0.335 | 1.81 | 4 | 0.460 | 1.52 ** | 0.058 | −0.223 | 0.202 | −0.046 | 37.67 |
| PGLSOU | 0.868 | 10.4 | 4 | <0.001 | 1.55 ** | 0.035 | −0.258 * | 0.173 | −0.050 | 39.97 |
| Clutch Size | ||||||||||
| OLS | 0.606 | 5.22 | 4, 11 | 0.246 | 14.9 *** | 0.089 | 1.70 * | 1.68 * | 0.850 | 730 |
| PGLSBM | 0.633 | 4.91 | 4 | 0.043 | 10.3 | 0.669 | −1.36 | 2.79 | 0.662 | 44.99 |
| PGLSOU | 0.597 | 4.46 | 4 | 0.063 | 10.6 ** | 0.534 | −1.43 | 2.89 | 0.946 | 47.07 |
| Juvenile Growth Rate | ||||||||||
| OLS | −0.210 | 0.740 | 4, 6 | 0.644 | −0.161 | 0.075 | 0.033 | 0.349 | 0.161 | 147 |
| PGLSBM | 0.993 | 15.0 | 4 | <0.001 | 1.25 | 0.446 | 1.04 | −0.973 | −0.275 | 9.94 |
| PGLSOU | −0.315 | 10.3 | 4 | <0.001 | 1.25 | 0.449 | 1.05 | −0.975 | −0.278 | 11.94 |
| Population Growth Rate | ||||||||||
| OLS | −0.565 | 0.368 | 4, 7 | 0.821 | 0.125 | 0.394 | 0.037 | 0.271 | 0.373 | 517 |
| PGLSBM | 0.663 | 3.81 | 4 | 0.107 | 0.274 | 0.101 | −0.027 | −0.085 | 0.016 | 19.61 |
| PGLSOU | 0.494 | 2.39 | 4 | 0.189 | 0.273 | 0.111 | −0.027 | 0.064 | . . . | 17.18 |
| Size at Death | ||||||||||
| OLS | . . . | . . . | . . . | . . . | . . . | . . . | . . . | . . . | . . . | . . . |
| PGLSBM | 0.659 | 1.74 | 2 | 0.175 | 2.10 | −0.249 | 0.337 | . . . | . . . | 11.09 |
| PGLSOU | 0.621 | 1.59 | 2 | 0.075 | 2.50* | −0.943 | . . . | . . . | . . . | 11.05 |
| Interclutch Interval | ||||||||||
| OLS | 0.119 | 1.20 | 4, 6 | 0.501 | 8.06 | 0.282 | 0.521 | 0.300 | 0.483 | 234 |
| PGLSBM | 0.368 | 1.38 | 4 | 0.601 | 5.80 | 1.78 | −0.827 | −1.41 | −2.03 | 29.53 |
| PGLSOU | 0.072 | 0.224 | 4 | 0.930 | 6.51 | 0.115 | −0.976 | −0.430 | . . . | 31.93 |
| Lifespan | ||||||||||
| OLS | 0.042 | 1.09 | 4, 8 | 0.468 | −0.484 | 0.716 | 0.134 | 0.090 | 0.629 | 352 |
| PGLSBM | 0.756 | 3.52 | 3 | 0.070 | 29.0 | 2.45 | −6.65 | −9.22 | . . . | 27.14 |
| PGLSOU | 0.391 | 1.24 | 3 | 0.290 | 31.5 | −0.665 | −6.80 | . . . | . . . | 32.46 |
| Response Variable and Model | Regression Summaries | β (Explanatory Variables) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Adj r2 | F/χ2 | df | p | Intercept | PC1temp | PC2temp | PC1precip | PC2precip | AIC † | |
| Age at First Reproduction | ||||||||||
| OLS | 0.042 | 1.09 | 5, 8 | 0.468 | −0.484 * | 0.104 | 0.031 | 0.017 | −0.143 | 52.1 |
| PGLSBM | 0.290 | 0.687 | 5 | 0.849 | −0.425 | 0.030 | 0.048 | 0.032 | −0.047 | 31.64 |
| PGLSOU | 0.999 | 3.18 | 5 | 0.174 | −0.480 * | 0.098 | 0.032 | −0.021 | −0.140 | 31.66 |
| Size at First Reproduction | ||||||||||
| OLS | 0.810 | 5.34 | 4, 9 | 0.047 | −0.015 | −0.112 * | −0.215 * | 0.347 * | 0.296 * | 25.1 |
| PGLSBM | 0.964 | 16.7 | 4 | <0.001 | 0.048 | −0.120 ** | −0.201 ** | 0.342 ** | −0.329 ** | 15.14 |
| PGLSOU | 0.423 | 2.75 | 4 | 0.139 | −0.136 | −0.004 | −0.183 | 0.112 | . . . | 23.51 |
| Clutch Size | ||||||||||
| OLS | 0.581 | 4.82 | 4, 11 | 0.035 | −0.125 | 0.099 | 0.169 | 0.019 | −0.214 | 32.7 |
| PGLSBM | 0.914 | 11.15 | 4 | <0.001 | −0.414 | 0.260 * | −0.214 | −0.178 | −0.551 * | 34.13 |
| PGLSOU | 0.910 | 7.94 | 4 | 0.003 | −0.128 | 0.101 | 0.169 | 0.016 | −0.216 | 33.83 |
| Juvenile Growth Rate | ||||||||||
| OLS | −0.192 | 0.799 | 4, 5 | 0.674 | −0.196 | 0.069 | −0.033 | −0.234 | −0.051 | . . . |
| PGLSBM | 0.838 | 5.47 | 4 | 0.027 | −0.090 | 0.153 | 0.043 | −0.228 | −0.116 | 11.10 |
| PGLSOU | 0.601 | 2.76 | 4 | 0.238 | −0.101 | 0.130 | 0.022 | −0.212 | −0.095 | 13.10 |
| Population Growth Rate | ||||||||||
| OLS | 0.883 | 12.3 | 4, 6 | 0.076 | −0.017 | 0.023 | 0.004 | −0.053 * | −0.049 * | . . . |
| PGLSBM | 0.951 | 10.6 | 4 | <0.001 | −0.015 | 0.022 | 0.003 | −0.050 * | −0.047 | 7.39 |
| PGLSOU | 0.588 | 3.10 | 4 | 0.102 | 0.013 | −0.003 | −0.006 | −0.011 | . . . | 4.60 |
| Size at Death | ||||||||||
| OLS | . . . | . . . | . . . | . . . | . . . | . . . | . . . | . . . | . . . | . . . |
| PGLSBM | 0.283 | 0.665 | 2 | 0.514 | 0.034 | −0.077 | 0.018 | . . . | . . . | 6.06 |
| PGLSOU | 0.257 | 0.595 | 2 | 0.275 | 0.012 | −0.040 | . . . | . . . | . . . | 0.90 |
| Interclutch Interval | ||||||||||
| OLS | −0.488 | 0.508 | 4, 6 | 0.746 | −0.520 | −0.209 | 0.113 | 0.168 | 0.225 | . . . |
| PGLSBM | 0.677 | 2.36 | 4 | 0.316 | −0.538 | −0.234 | 0.129 | 0.200 | 0.220 | 27.69 |
| PGLSOU | 0.430 | 1.31 | 4 | 0.455 | −0.630 | −0.057 | 0.156 | 0.099 | . . . | 25.53 |
| Lifespan | ||||||||||
| OLS | . . . | . . . | . . . | . . . | . . . | . . . | . . . | . . . | . . . | . . . |
| PGLSBM | 0.868 | 4.82 | 3 | 0.022 | −1.03 | −0.839 | 0.348 | 1.32 | . . . | 22.06 |
| PGLSOU | 0.138 | 0.370 | 3 | 0.691 | −1.39 | −0.392 | 0.369 | . . . | . . . | 24.04 |
| Response Variable and Model | Regression Summaries | β (Explanatory Variables) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adj r2 | F/χ2 | df | p | Intercept | PC1temp | PC2temp | PC1prec | PC2prec | Tempexp | Lat | Species † | AICc | |
| Age at First Reproduction | |||||||||||||
| OLSFull | 0.232 | 2.97 | 17, 111 | <0.001 | 23.7 *** | −0.073 | −0.270 | 0.119 | −0.442 | −0.383 *** | −0.196 | ✓ | 629 |
| OLSBest | 0.265 | 21.0 | 2, 111 | <0.001 | 18.9 *** | . . . | . . . | . . . | . . . | −0.377 *** | −0.069 *** | . . . | 602 |
| Size at First Reproduction | |||||||||||||
| OLSFull | 0.832 | 38.7 | 19, 144 | <0.001 | 3.81 *** | −0.018 | −0.117 * | −0.002 | 0.028 | −0.005 | −0.055 *** | ✓ | 97.8 |
| Clutch Size | |||||||||||||
| OLSFull | 0.563 | 9.33 | 18, 116 | <0.001 | 46.9 *** | −0.026 | −1.24 | 0.387 | 0.684 * | −0.180 * | −0.691 ** | ✓ | 674 |
| Juvenile Growth Rate | |||||||||||||
| OLSFull | 0.395 | 7.03 | 13, 120 | <0.001 | −3.37 * | 0.232 * | 0.374 * | 0.123 ** | −0.060 | −0.005 | 0.084 * | ✓ | 95.4 |
| Population Growth Rate | |||||||||||||
| OLSFull | 0.123 | 2.52 | 13, 140 | 0.004 | 4.26 | −0.192 | −0.044 | 0.002 | 0.011 | 0.034 | −0.010 | ✓ | 509 |
| OLSBest | 0.022 | 4.16 | 1, 140 | 0.043 | −0.152 | . . . | . . . | . . . | . . . | 0.041 * | . . . | . . . | 509 |
| Size at Death | |||||||||||||
| OLSFull | 0.936 | 82.7 | 5, 28 | <0.001 | −0.656 | −0.353 | 0.295 | . . . | . . . | −0.040 *** | 0.074 | ✓ | 8.73 |
| OLSBest | 0.938 | 107.1 | 5, 28 | <0.001 | 0.243 | −0.406 | 0.220 | . . . | . . . | −0.040 *** | 0.056 | . . . | 5.39 |
| Interclutch Interval | |||||||||||||
| OLSFull | 0.812 | 17.0 | 10, 37 | <0.001 | −42.5 | 2.94 ** | −4.05 *** | 8.11 * | 7.71 * | −0.560 *** | 1.49 * | ✓ | 218 |
| OLSBest | 0.798 | 25.3 | 6, 37 | <0.001 | 4.69 | 1.40 | −1.64 | −0.184 | 1.46 | −0.490 *** | 0.433 ** | . . . | 210 |
| Lifespan | |||||||||||||
| OLSFull | 0.724 | 12.1 | 9, 38 | <0.001 | 9.32 | 6.48 | −2.20 | −9.33 | −13.0 | −1.85 *** | 1.81 | ✓ | 336 |
| OLSBest | 0.747 | 19.7 | 6, 38 | <0.001 | 5080 *** | −35.1 ** | −364 *** | −142 *** | 40.3 ** | −1.91 *** | −93.4 *** | . . . | 325 |
| Life History Trait | Species Identity | Population/Latitude | Temperature |
|---|---|---|---|
| Age at First Reproduction | ✗ | ✓ | ✓ |
| Size at First Reproduction | ✓ | ✓ | ✗ |
| Clutch Size | ✓ | ✓ | ✓ |
| Juvenile Growth Rate | ✓ | ✓ | ✗ |
| Population Growth Rate | ✗ | ✗ | ✓ |
| Size at Death | ✗ | ✗ | ✓ |
| Interclutch Interval | ✗ | ✓ | ✓ |
| Lifespan | ✗ | ✓ | ✓ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bowman, Jr., L.L.; Post, D.M. The Evolution of Life History Traits and Their Thermal Plasticity in Daphnia. Hydrobiology 2023, 2, 55-74. https://doi.org/10.3390/hydrobiology2010005
Bowman, Jr. LL, Post DM. The Evolution of Life History Traits and Their Thermal Plasticity in Daphnia. Hydrobiology. 2023; 2(1):55-74. https://doi.org/10.3390/hydrobiology2010005
Chicago/Turabian StyleBowman, Jr., Larry L., and David M. Post. 2023. "The Evolution of Life History Traits and Their Thermal Plasticity in Daphnia" Hydrobiology 2, no. 1: 55-74. https://doi.org/10.3390/hydrobiology2010005
APA StyleBowman, Jr., L. L., & Post, D. M. (2023). The Evolution of Life History Traits and Their Thermal Plasticity in Daphnia. Hydrobiology, 2(1), 55-74. https://doi.org/10.3390/hydrobiology2010005






