Abstract
Introduction: Bacterial infections, especially those caused by multidrug-resistant strains, remain a major health concern. This study investigates 4-chlorobenzyl p-coumarate, assessing its antibacterial mechanism, pharmacokinetic profile, and potential to modulate antimicrobial resistance. Methods: In silico studies were conducted, including molecular docking, molecular dynamics simulations, and pharmacokinetic predictions, alongside in vitro assays assessing efflux pump inhibition, antibiotic modulation, and bacterial DNA analysis. Results: The compound showed higher binding affinity and complex stability with the enzyme phosphatidylglycerol phosphate synthase, while also exhibiting reduced residue fluctuations and better flexibility with the NAD+-dependent DNA ligase. Molecular interactions with the efflux proteins MepA and NorA were also observed. Pharmacokinetic predictions indicated a favorable profile, including suitability for oral administration. Experimentally, the compound inhibited the MepA and NorA efflux pumps, modulated the activity of the antibiotics ciprofloxacin and norfloxacin, and reduced DNA concentration in treated cells. Conclusions: The findings suggest that the compound acts through dual mechanisms, with a prediction of activity by disrupting phosphatidylglycerol synthesis and DNA replication while inhibiting and modulating MepA and NorA efflux pumps.
1. Introduction
Bacterial infections caused by Gram-positive bacteria pose a significant public health challenge due to their high transmissibility, treatment complexity, and increasing resistance to antimicrobial agents. Among these microorganisms, species of the genera Staphylococcus and Enterococcus stand out as high-priority clinical pathogens, associated with elevated morbidity and mortality rates in both nosocomial and community-acquired infections [1]. These bacteria are particularly known for their capacity to acquire and disseminate resistance genes, thereby compromising the efficacy of available treatments [2].
Resistance genes commonly found in Staphylococcus and Enterococcus species, such as mecA, vanA, mepA, and norA, are associated with diverse antimicrobial resistance mechanisms. The mecA gene encodes the penicillin-binding protein PBP2a, which has a low affinity for β-lactam antibiotics, rendering these agents ineffective. The vanA gene mediates the alteration of peptidoglycan precursor termini, reducing the binding of glycopeptide antibiotics. Additionally, mepA and norA genes regulate the expression of MepA and NorA efflux pumps, which actively extrude fluoroquinolones and other compounds from the bacterial cell, thereby lowering their intracellular concentrations. These mechanisms, whether acting independently or synergistically, confer multidrug-resistant (MDR) phenotypes to the pathogens [3,4].
p-Coumaric acid, a phenolic compound widely present in plant-derived foods, including fruits (such as apples and strawberries), vegetables (such as potatoes, tomatoes, and carrots) and grains such as barley and white beans, has been investigated for its broad spectrum of biological activities, including antioxidant, anti-inflammatory, antidiabetic, antineoplastic, and neuro and hepatoprotective properties [5]. Studies have highlighted its antibacterial potential, showing inhibitory effects on the viability of various pathogenic microorganisms [6,7]. The compound’s flexible chemical structure allows interactions with multiple molecular targets, making it a promising candidate for the development of novel antimicrobial therapies [7,8].
Semisynthesis and molecular modifications represent effective approaches to improve the bioactive properties of natural compounds. These strategies allow for increased potency, specificity, and stability of substances with therapeutic potential, facilitate access to otherwise unreachable therapeutic targets through natural biosynthesis, in addition to facilitating the overcoming of pharmacokinetic limitations [9]. In the antimicrobial context, such modifications have yielded substantial progress in the development of new therapeutic options to combat bacterial resistance, as in the case of chemical-structural optimization in azithromycin, minocycline, amoxicillin and plazomicin [10].
Several studies describe the antibacterial activity of coumaric acid derivatives and their structural analogues. For example, Molokoane et al. [11] investigated the antibacterial activity of dodecyl-p-coumarate, obtained from Artemisia afra extract, against different bacterial strains, observing good activity against Enterococcus faecalis, with a minimum inhibitory concentration (MIC) of 62.5 mg/mL. Similarly, Li et al. [12] evaluated p-coumaric acid against Alicyclobacillus acidoterrestris, demonstrating high inhibitory capacity against vegetative cells, with MIC and minimum bactericidal concentration (MBC) values of 0.2 mg/mL, whereas for spores, the MIC and MBC values were 0.2 mg/mL and >1.6 mg/mL, respectively. The compound 3′-hydroxy-4′-methoxystyrenyl-(E)-p-coumarate also exhibited antibacterial activity against Escherichia coli ATCC 25922 and Salmonella typhimurium ATCC 14028 at concentrations of 5 and 10 mg/mL [13]. Additionally, (–)-borneol p-coumarate showed significant efficacy against Staphylococcus aureus and MRSA, with MIC values ranging from 2 to 8 µg/mL [14]. In another example, 4-methoxycinnamyl p-coumarate displayed notable antibacterial activity against Cutibacterium acnes, with inhibition zones of 18.67 ± 1.15 mm at 0.5 mg/mL and 14.00 ± 1.00 mm at 0.25 mg/mL [15].
4-Chlorobenzyl p-coumarate (4CpC) is a semisynthetic derivative of p-coumaric acid, obtained through esterification with an aryl halide in the presence of acetone. Our research group previously demonstrated that this compound exhibited antifungal activity against various Candida strains [16]. Furthermore, 4CpC displayed in vitro antibacterial activity against Gram-positive pathogens, such as methicillin-resistant Staphylococcus aureus (MRSA) and Enterococcus faecalis, showing a bactericidal effect after 24 h of exposure to planktonic cells. Microscopic evidence indicated that the compound compromised the integrity of the bacterial cell envelope and promoted disorganization of the genetic material. In addition, 4CpC was able to disrupt the biofilm architecture, penetrating the extracellular matrix and reaching the cells located within the inner layers of the structure [17]. Furthermore, the substance displayed favorable toxicological parameters in in silico predictions, showed limited in vitro cytotoxicity toward human gingival fibroblast cells (HGF), and demonstrated low systemic toxicity in vivo in Galleria mellonella larvae [17].
These findings highlight the potential of 4CpC as a promising candidate for the treatment of bacterial infections. The present study aims to generate additional data to elucidate the mechanism of action, pharmacokinetics profile and 4CpC efflux pump inhibitory activity.
2. Materials and Methods
2.1. Bacterial Strains
For the MDR, efflux pump modulation and inhibition assays, the Staphylococcus aureus strains K2068 and 1199B were employed. The strains 1199B and K2068 are bacteria with characterized efflux pumps (NorA and MepA, respectively). Both strains overexpress these efflux pumps and are clinical isolated strains originally obtained from Wayne State University School of Medicine (Detroit, MI, USA) and are currently maintained on Heart Infusion Agar (HIA, Difco Ltd., São Paulo, Brazil) slants at 4 °C, as well as in glycerol stocks at −80 °C, in the microbiological collection of the Laboratory of Microbiology and Molecular Biology (LMBM), Regional University of Cariri (URCA), Brazil. For DNA assays, the standard strain, isolated from urine, Enterococcus faecalis ATCC 29212, was used. This strain was obtained from Laborclin Ltd. (Pinhais, PR, Brazil) and preserved in glycerol (ACS Científica®, Sumaré, Brazil) at −80 °C at the Federal University of Paraíba, Brazil.
2.2. Synthesis and Characterization of the Compound
The synthesis and characterization of the compound 4CpC were carried out as described by Melo et al. [16]. The spectra and other information about the compound are described in the Supplementary Materials.
2.3. Molecular Docking Analysis
The three-dimensional (3D) structure of 4CpC was constructed using Marvin Sketch v.24.1.3 and subsequently converted to the sdf format. Control drug molecules were retrieved from the PubChem database (Accessed on: 25 March 2025; https://pubchem.ncbi.nlm.nih.gov/) [18]. The molecular structures were imported into Spartan 14 v.1.1.4 for energy minimization employing molecular mechanics and the AM1 semi-empirical method, in order to obtain the most stable conformations. The resulting optimized structures were then used as input for molecular docking simulations performed in Molegro Virtual Docker (MVD) 2013 v.6.0.1.
The target enzymes were downloaded from the Protein Data Bank (Accessed on: 25 March 2025; http://www.rcsb.org) in pdb format [19]. Protein and ligand structures were prepared by correcting the protonation states of amino acid residues and removing water molecules and cofactors from the structures.
The selected mechanisms are consolidated in the literature as important targets in the inhibition of microorganisms and are crucial for their survival, being related to the cell membrane, cell wall and DNA. Reference substances are available for these mechanisms, which reinforces their importance and use in the present study. Proteins involved in key bacterial processes included the enzymes phosphatidylglycerol phosphate synthase (PgsA) (PDB ID: 7DRJ), NAD+-dependent DNA ligase (PDB ID: 3JSN) and penicillin-binding protein 4 (PBP4) (PDB ID: 6BSR) were selected.
For the study of factors related to antimicrobial resistance related to the efflux pumps MepA and NorA, these were obtained from the Alpha Fold platform (Available at: https://alphafold.ebi.ac.uk/entry/Q8NYB0; Accessed on: 30 March 2025), corresponding to: Multidrug export protein MepA (AF-Q8NYB0-F1-v4) and Quinolone resistance protein NorA (AF-P0A0J7-F1-v4).
The preparation of the models obtained in Alpha Fold was performed previously with the visualization and checking of the 3D structure in the software PyMol 2.0, Schrödinger, LLC. (2016), in order to verify the integrity of the structure. Missing hydrogens were added with the PyRX tool [20]. Subsequently, the protein was subjected to energy minimization using GROMACS 2024.1 [21], with the AMBER99SB-ILDN force field [22], to relax possible structural stresses. 2000 minimization steps were performed with the conjugate gradient method, keeping the main structure fixed to preserve the native conformation.
In the targets that had co-crystallized, ligand templates were created to demarcate their active site as well, as previously, the enzyme validation process was performed by redocking through the evaluation of the RMSD (Root Mean Square Deviation) values. The active site was also identified based on the important residues for the active site that are described in the PDB reference article, in addition to using molecular pocket prediction tools such as the user-friendly Protein Plus platform (Available at: https://proteins.plus/help/tutorial; Accessed on: 10 March 2025) [23].
The docking procedure was performed using a grid with a 15 Å radius and a resolution of 0.30 Å to adequately encompass the macromolecular binding sites. All other parameters followed the default settings provided by the software package (Score function: MolDock Score; Ligand evaluation: Internal ES, Internal Hbond, Sp2-Sp2 Torsions, all verified; Number of runs: 30 executions; algorithm: MolDock SE; maximum interactions: 1500; maximum population size: 50; maximum steps: 300; neighbor distance factor: 1.00; maximum number of returned poses: 5).
Interactions were visualized in Discovery Studio Visualizer v20.1.0.19295—BIOVIA software was used (Accessed on: 2 April 2025; https://discover.3ds.com/discovery-studio-visualizer-download).
Computational Methodology for Michael Addition Feasibility
To evaluate the feasibility of a Michael-type covalent reaction between the ligand and nucleophilic residues, truncated side-chain models of Lysine (Ace–Lys–NMe) and Glutamic Acid (Ace–Glu–NMe) were employed. The electrophilic portion of the ligand was represented by the 4CpC moiety, retaining the α,β-unsaturated system responsible for potential Michael reactivity [24].
Single-point energy calculations were performed on Spartan’14 (Wavefunction Inc., Irvine, CA, USA) using DFT/B3LYP/6-31G* employing the CPCM (water) solvation model. The energy of occurrence (ΔE) was obtained as the difference between the initial and final states, with negative values interpreted as indicative of thermodynamic favorability for covalent addition.
2.4. Molecular Dynamics Methodology
Molecular dynamics (MD) simulations were performed using GROMACS 2023.1 software [25,26,27]. The ligand topology was generated using the Automated Topology Builder (ATB) platform v. 3.0 [28] with the GROMOS 54a7 force field a extensively validated for simulations of protein-ligand complexes, particularly when dealing with small organic molecules such as phenolic derivatives [29,30], while the protein topology was constructed in GROMACS employing the GROMOS96 54a7 force field. The SPC water model was used for solvation [31], and Cl− and Na+ ions were added to neutralize the system.
Following energy minimization, the system was equilibrated at 300 K under constant number of particles, volume, and temperature (NVT ensemble) using the V-rescale algorithm for 100 ps, followed by pressure equilibration at 1 atm under the constant number of particles, pressure, and temperature (NPT ensemble) using the Parrinello-Rahman algorithm for an additional 100 ps [32]. The production MD run was carried out for 100 ns (50 million steps). Structural stability and flexibility were assessed through root-mean-square deviation (RMSD) and root-mean-square fluctuation (RMSF) analyses, respectively, along with evaluation of Coulombic and Lennard-Jones interaction energies. All plots were generated using Grace software version 5.1.25 (Grace Development Team, Accessed on: 16 April 2025; http://plasma-gate.weizmann.ac.il/Grace/).
2.5. In Silico Pharmacokinetic Evaluation
To design the pharmacokinetic predictions of the compound, the OSRA 2.1.4 platform was initially used to generate the structural conformation in the required format by converting the compound’s chemical structure into SMILES format. Subsequently, the resulting structure was submitted to the SwissADME (Accessed on: 20 August 2024;http://www.swissadme.ch) [33] and pkCSM (Accessed on: 20 August 2024; https://biosig.lab.uq.edu.au/pkcsm/) [34] electronic interfaces to obtain the predictive models.
2.6. Determination of Minimum Inhibitory Concentration (MIC)
For the MIC assay, cells from the K2068 and 1199B strains were harvested after bacterial growth in HIA medium at 37 °C for 24 h. Standardized inocula were prepared using a 0.5 McFarland standard (approximately 108 CFU/mL) and diluted 1:9 in Brain Heart Infusion (BHI) (HiMedia Laboratories Pvt. Ltd., Mumbai, India) broth [35]. Subsequently, 100 µL of each suspension was transferred to 96-well microdilution plates. Serial microdilutions of the compounds 4CpC, ethidium bromide (EtBr), ciprofloxacin, norfloxacin, and the efflux pump inhibitor carbonyl cyanide m-chlorophenyl hydrazone (CCCP) (Sigma-Aldrich®, St. Louis, MO, USA) were performed starting from a concentration of 1024 µg/mL, using 100 µL per well up to the penultimate columns (1:1). The final columns were reserved for growth control. Plates were incubated at 37 °C for 24 h prior to MIC determination [35]. All experiments were conducted in triplicate.
2.7. Assessment of MepA and NorA Efflux Pump Inhibition via the Ethidium Bromide Assay
Bacterial inocula of K2068 and 1199B were prepared as described in Section 2.6. 4CpC, at subinhibitory concentration (CIM/8), and CCCP were added to the bacterial inocula. The sub-inhibitory concentration MIC/8 was selected by our research team with the aim of avoiding the inhibitory concentration of the tested compound and more promptly identifying its potential to potentiate antibiotic activity. In pilot assays conducted in our laboratory, as well as in our previously published studies, the MIC/8 was the most relevant concentration, showing activity without any significant inhibitory effect against the tested strains. Subsequently, 100 μL of the resulting mixture was transferred to 96-well microdilution plates, where EtBr was microdiluted in concentrations ranging from 1024 μg/mL to 0.5 μg/mL. The last wells were used as the growth control. After 24 h of incubation, 20 μL of resazurin (7-hydroxy-3H-phenoxazin-3-one 10-oxide) (Sigma-Aldrich®, St. Louis, MO, USA) was added to each well to assess bacterial viability through color change. A color shift from blue to pink indicated bacterial growth, while the persistence of the blue color indicated growth inhibition [36]. All experiments were performed in triplicate.
2.8. Evaluation of the Modulatory Activity of 4CpC on Antibiotic Resistance
The suspensions of strains K2068 and 1199B were prepared and standardized as described in item 2.6. 4CpC was added at a concentration equivalent to 1/8 of its MIC against the tested strains, and CCCP was used as a positive control. In 96-well microtiter plates, 100 µL of each treatment solution was dispensed per well, followed by the addition of either ciprofloxacin or norfloxacin at concentrations ranging from 1024 µg/mL to 0.5 µg/mL. Plates were incubated at 37 °C for 24 h. Subsequently, 20 µL of resazurin solution (0.01% w/v in sterile distilled water) was added to each well to assess bacterial growth through color change [36]. All experiments were performed in triplicate.
2.9. Analysis of the Compound’s Effect on DNA
A standardized bacterial culture of E. faecalis at approximately 108 CFU/mL was treated with 4CpC at 16 µg/mL, corresponding to the previously determined MIC for this strain [17], and incubated for 3 h at 37 °C. Bacterial DNA was extracted immediately after cell treatment using the QIAamp® Mini Kit (Qiagen GmbH, Hilden, Germany). Subsequently, the extracted DNA solutions were subjected to 1% agarose gel electrophoresis with ethidium bromide staining and visualized under an ultraviolet transilluminator, as well as to spectroscopic analysis at a wavelength of 260 nm. Cells treated with 0.5% dimethyl sulfoxide (DMSO—Dinâmica® Química Contemporânea Ltd., Indaiatuba, Brazil) were used to assess potential vehicle interference in the assay results. Untreated cells were used as a negative control [37]. The experiment was performed in triplicate.
2.10. Statistical Analysis
For the statistical analysis of the data and production of graphs, GraphPad Prism 9.2.0 statistical software (San Diego, CA, USA) was employed. The evaluations were performed by one-way ANOVA analysis of variance, followed by the Bonferroni post hoc test. Results were considered significant at a confidence level of 95% (p < 0.05).
3. Results
3.1. Molecular Docking
Molecular docking simulations were performed to evaluate the probability of interaction of the 4CpC compound with targets related to antimicrobial activity. Prior to performing the molecular docking simulations, the redocking procedure was performed in order to validate whether the program is positioning the poses correctly and whether there are structural differences between the ligand and its most stable pose. This metric was evaluated by the RMSD (Root Mean Square Deviation) value, with a limit value of up to 2 Å being considered. The target evaluated by redocking corresponded to the enzyme related to the cell wall mechanism of penicillin-binding protein 4 (PBP4) (PDB ID: 6BSR), since this is the only target that presents a co-crystallized ligand, which corresponds to penicillin G, with the RMSD value corresponding to 0.242, validating the methodology used. Table 1 shows the binding energy values of the 4CpC compound according to the MolDock score algorithm for the targets under study.

Table 1.
Binding energy values of the 4CpC compound with the targets under study, according to the MolDock Score algorithm.
The results demonstrate that compound 4CpC presented negative values for all targets under study, revealing interaction, and that its most stable pose demonstrated lower values when compared to the control compounds under study for the targets related to the membrane through the enzyme phosphatidylglycerol phosphate synthase (PDB ID: 7DRJ), to the target related to DNA through the enzyme NAD+-dependent DNA ligase (PDB ID: 3JSN) and to the resistance factors related to the efflux pumps MepA and NorA. The high binding affinity and multitarget potential observed for 4CpC can be attributed to the complementary interactions between its functional groups and the active sites of target proteins, as well as to the recognized ability of phenolic compounds to modulate multiple biological targets, widely described in the literature [38]. Figure 1, Figure 2, Figure 3, Figure 4 and Figure 5 demonstrate the molecular interactions of the 4CpC compound with the targets under study.
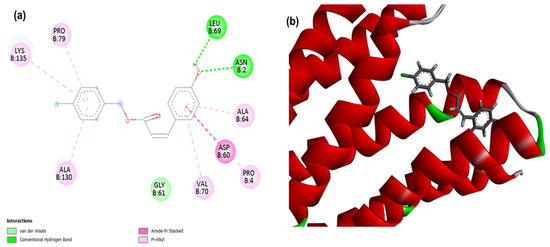
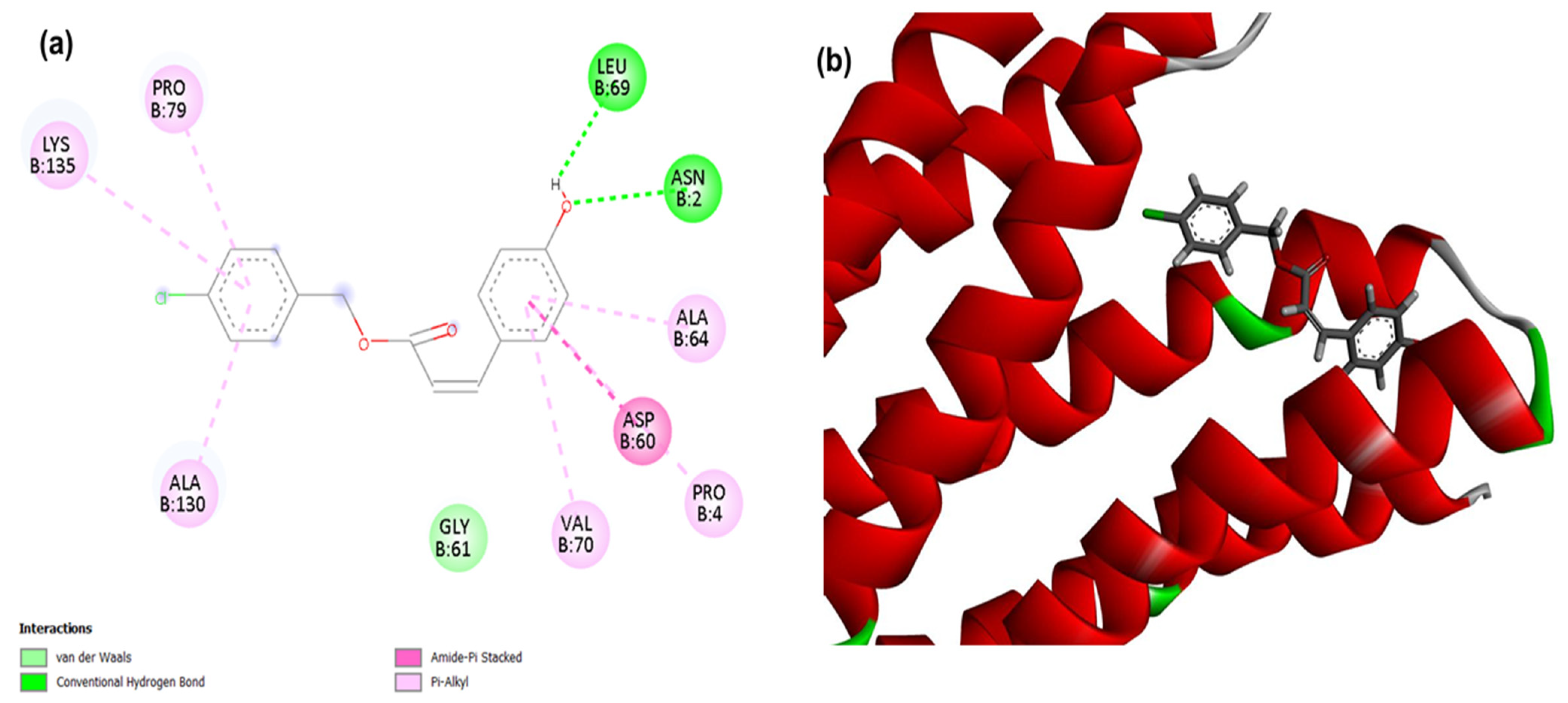
Figure 1.
2D (a) and 3D (b) interactions of compound 4CpC with the target PgsA. Residues: Lys (Lysine), Pro (Proline), Leu (Leucine), Asn (Asparagine), Ala (Alanine), Asp (Aspartic Acid) and Val (Valine).
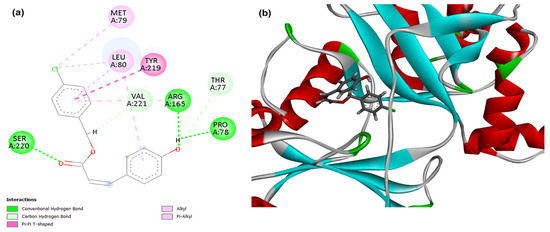
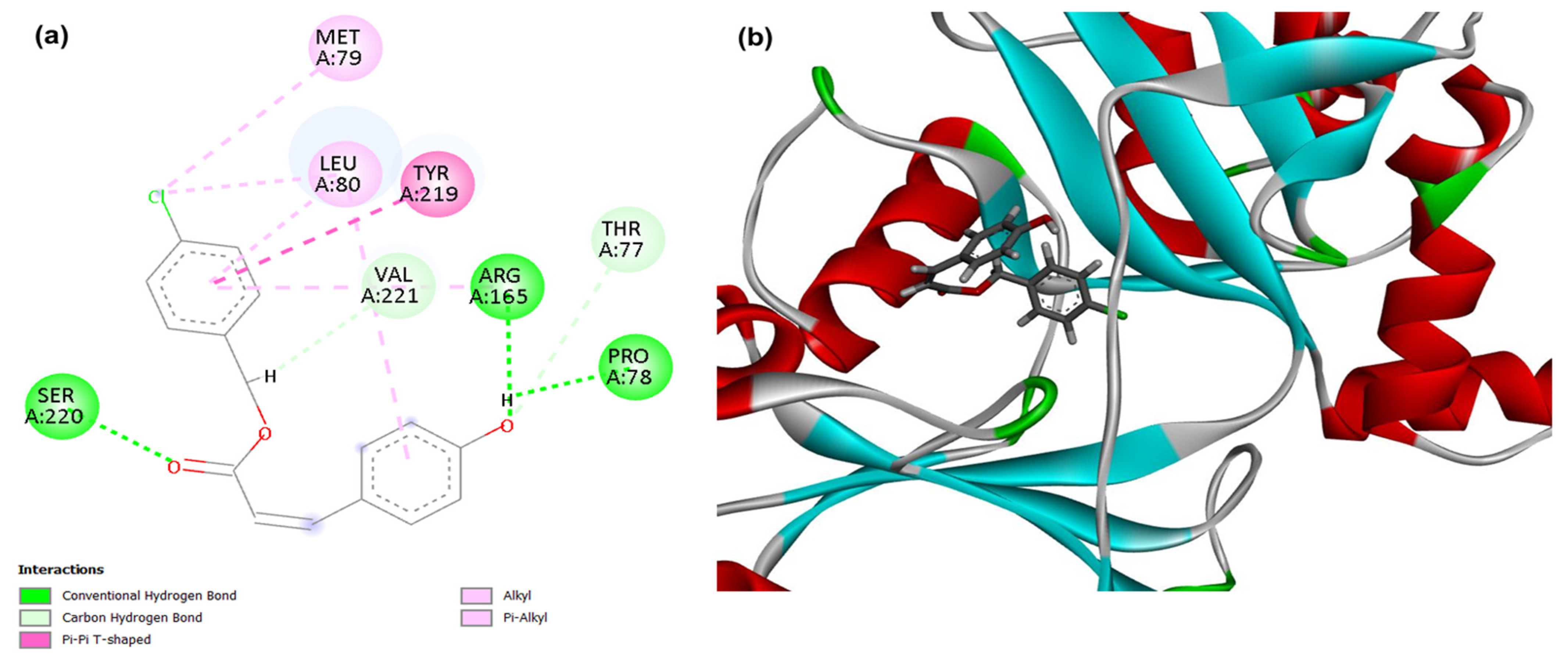
Figure 2.
2D (a) and 3D (b) interactions of compound 4CpC with the target NAD+-dependent DNA ligase. Residues: Ser (Serine), Met (Methionine), Leu (Leucine), Tyr (Tyrosine), Val (Valine), Arg (Arginine), Thr (Threonine), Pro (Proline), Asp (Aspartic Acid), Arg (Arginine), Gln (Glutamine) and Phe (Phenylalanine).
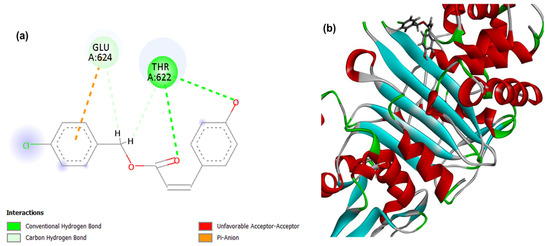
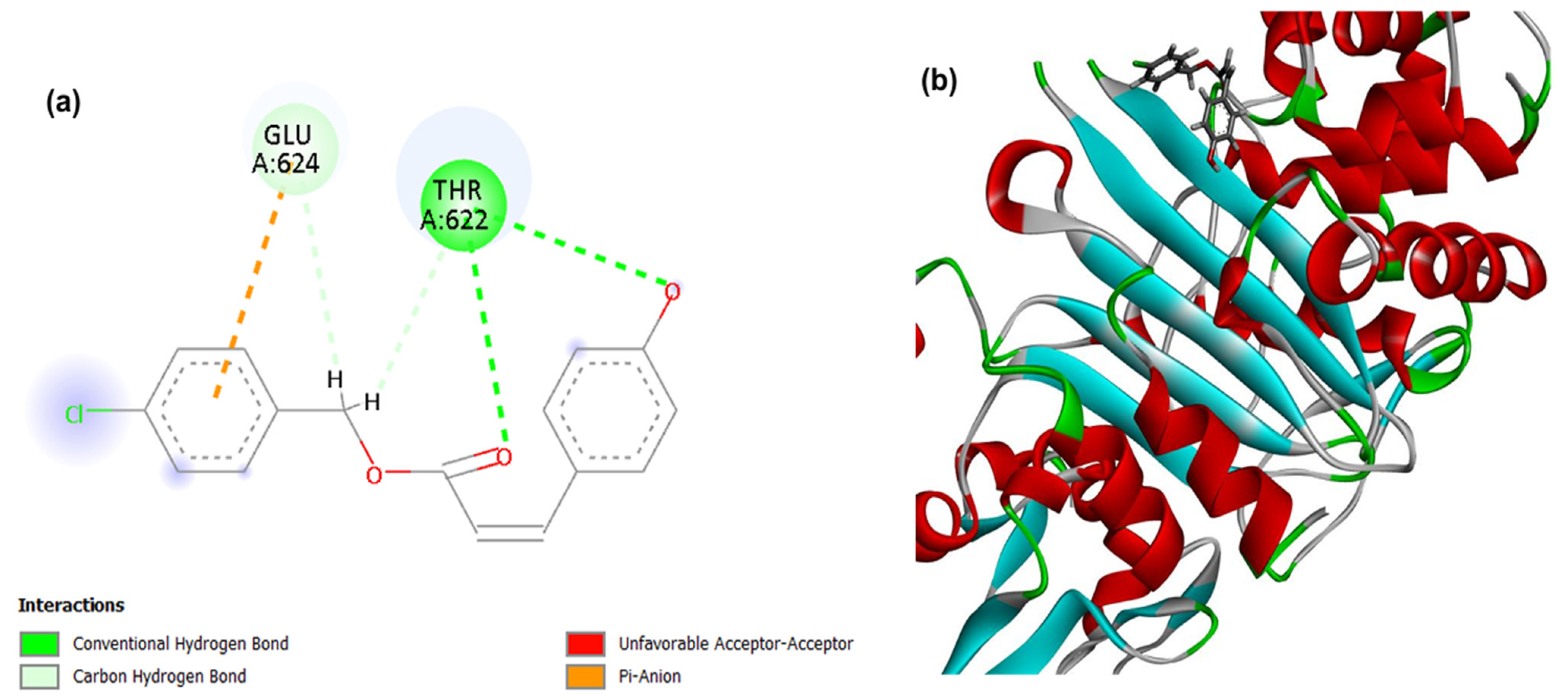
Figure 3.
2D (a) and 3D (b) interactions of compound 4CpC with the target PBP4. Residues: Glu (Glutamic Acid), Gln (Glutamine), Thr (Threonine), Tyr (Tyrosine), Val (Valine), Ser (Serine), Asn (Asparagine), Gln (Glutamine), Thr (Threonine) and Gly (Glycine).
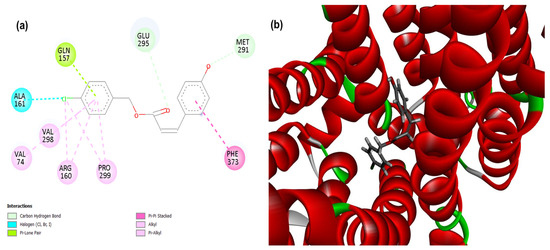
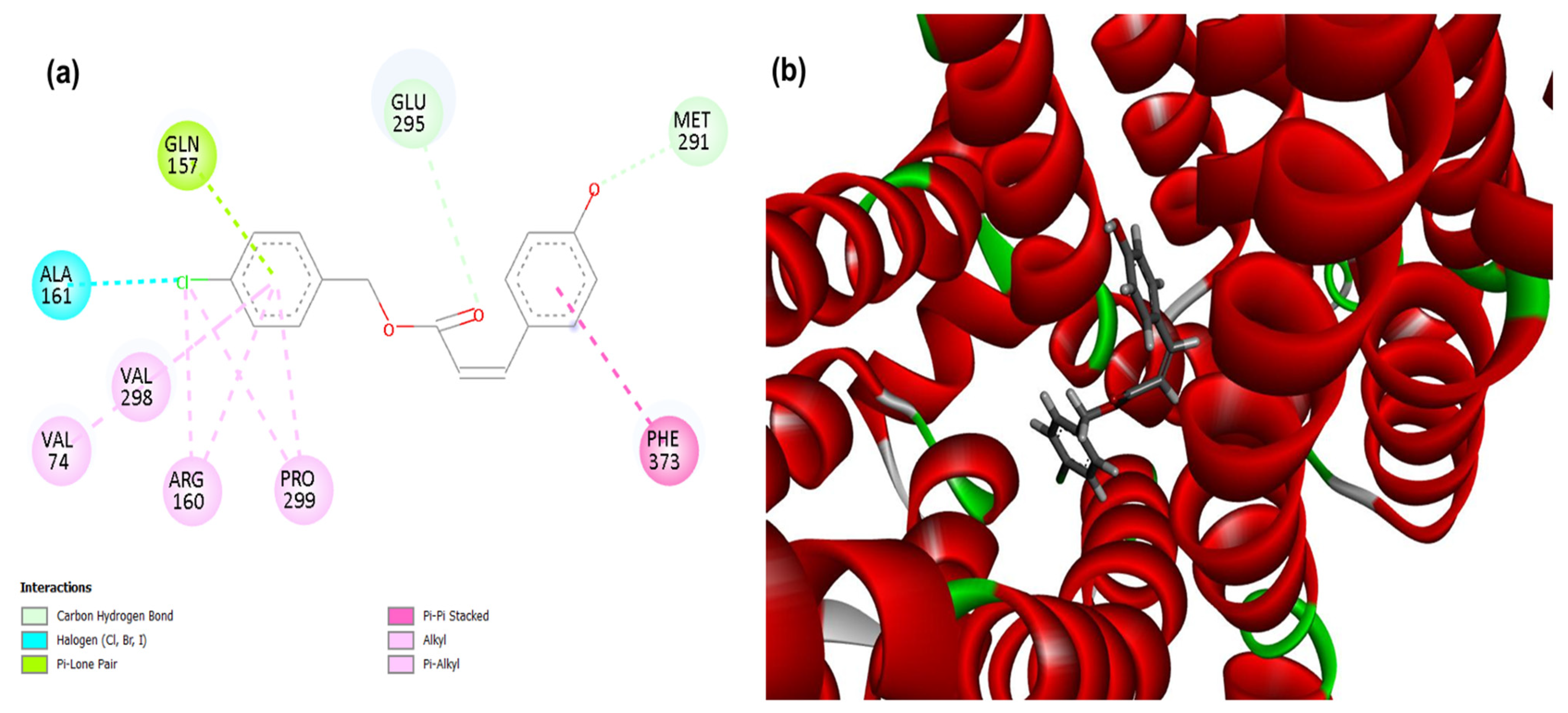
Figure 4.
2D (a) and 3D (b) interactions of compound 4CpC with the target efflux pump MepA (from the MATE family, in Staphylococcus aureus). Residues: Gln (Glutamine), Glu (Glutamic Acid), Met (Methionine), Phe (Phenylalanine), Pro (Proline), Arg (Arginine), Val (Valine) and Ala (Alanine), Ser (Serine) and Thr (Threonine).
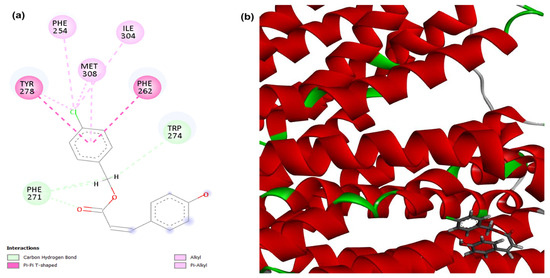
Figure 5.
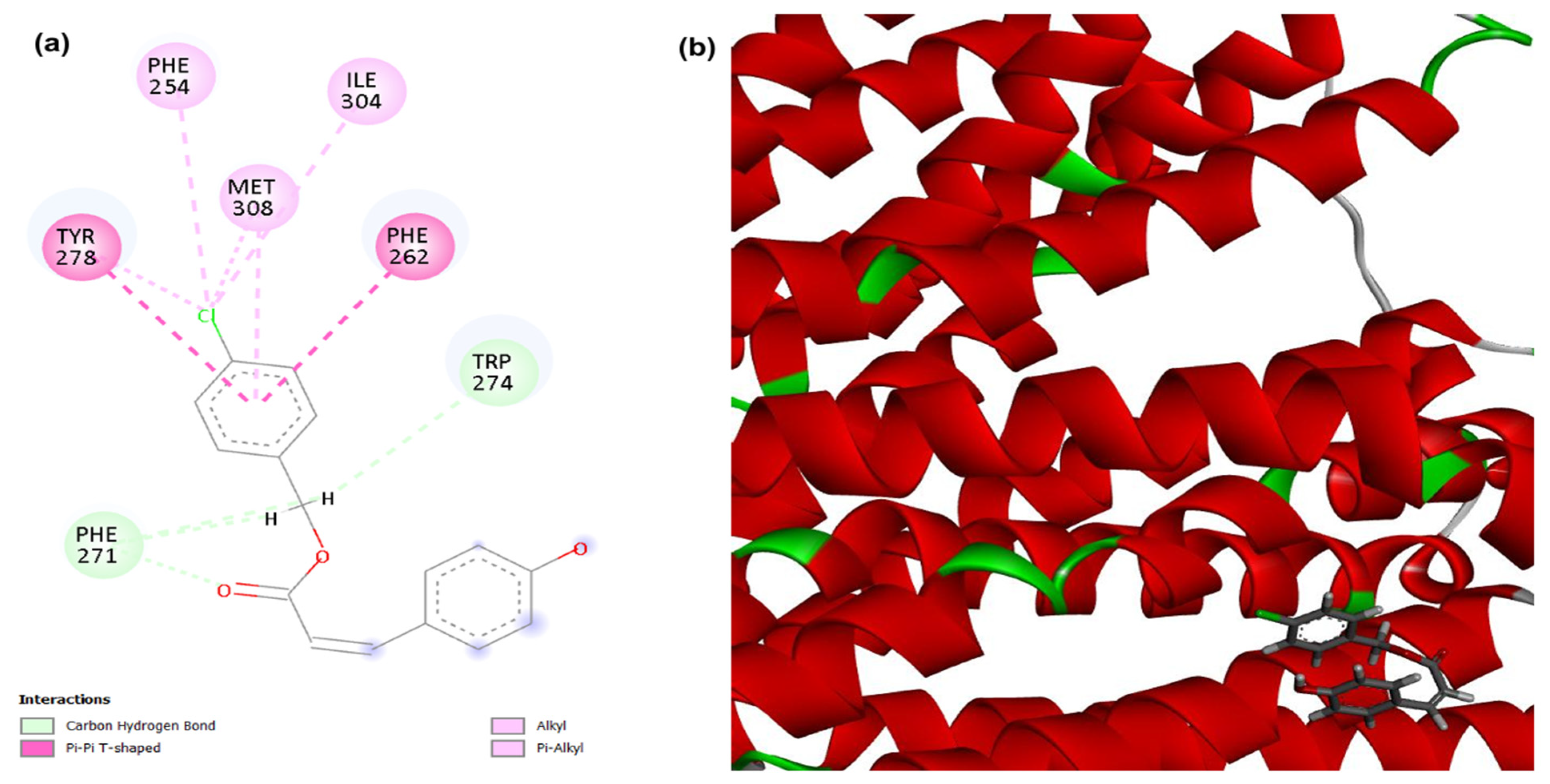
2D (a) and 3D (b) interactions of compound 4CpC with the target efflux pump NorA. Residues: Tyr (Tyrosine), Phe (Phenylalanine), Met (Methionine), Ile (Isoleucine), Trp (Tryptophan), Gly (Glycine) and Ser (Serine).
For PgsA, a membrane-related target, it was observed that the compound 4CpC presented the lowest energy, corresponding to −113,343 (Table 1), demonstrating a stronger and higher affinity interaction in the static model to the target when compared to the control compound cerulenin, which obtained score values corresponding to −102,591. Figure 1 shows that the interactions of compound 4CpC with the membrane-related target occurred in the most voluminous groups of the structure (benzene rings), and it was observed that the compound established critical interactions for the maintenance of enzymatic activity, such as the hydrophobic pi-alkyl interactions (dashed line in light pink) of residues Lysine 135 (Lys135) and Alanine 64 (Ala64), as well as the Pi-stacked Amide interaction (dashed line in light pink) established by the Asp60 residue [39]. These interactions are justified by the hydrophobic packing favored by the pairing of the ring’s π system with halogen groups.
It is important to highlight that the interaction involving the nucleophilic residue Lys135 plays a crucial role in the target PgsA, as it contributes to the partial overlap of the CDP-DAG (cytidine diphosphatediacylglycerol) binding site with that of phosphatidylglycerol-phosphate (PGP), so that the electrostatic interaction between this residue and the α-phosphoryl moiety of CDP-DAG provides the positioning of the substrate in a suitable way for catalysis [39]. Other important nucleophilic residues are observed in the bimetallic binding site located in the active site of the PgsA target, with the aspartic acid residue (Asp60) well positioned for coordination of the CDP-DAG bond mediated by Zn1 and Zn2, being indispensable for the activity of PgsA from S. aureus (SaPgsA), since Zn2 may be involved in the scission of the α-β phosphoric anhydride bond of CDP-DAG, an important step to ensure the irreversibility of catalysis [39,40]. Thus, it is noteworthy that the aromatic groups of compound 4CpC, when complexed with the target PgsA through residues Lys135 and Asp60, can act as nucleophilic sites and participate in Michael addition reactions, since the primary amino group (-NH2) of Lys135 can attack electrophilic centers of the ligand, as an α,β-unsaturation, while the carboxylate (-COO−) of Asp60 also presents nucleophilic potential in certain reactions, including Michael addition [41].
For the target adenylation domain related to the DNA replication process, NAD+-dependent DNA ligase, it was observed that the compound 4CpC presented the lowest energy corresponding to −105,397 (Table 1), demonstrating this form a stronger and higher affinity interaction in the static model to the target when compared to the control compound levofloxacin, which obtained score values corresponding to −101,643. Figure 2 demonstrates that the interactions of compound 4CpC with the studied target included, in addition to the bulky groups, interactions in the central ester group, and it was observed that the compound also established critical interactions for the maintenance of enzymatic activity, such as the hydrophobic interaction of the alkyl type (dashed line in light pink) of residues Leucine 80 (Leu80) with the chlorobenzene group [42]. This is a typical interaction between nonpolar side chains and the aromatic system, stabilizing the complex and promoting hydrophobic packing of the ligand within the active site, thus justifying the high binding affinity of the 4CpC compound to the target.
For PBP4, a cell wall-related target, it was observed that the PDB ligand, penicillin G, presented the lowest energy corresponding to −108.045, demonstrating a stronger and higher affinity interaction in the static model to the target when compared to the compound 4CpC that presented score values corresponding to −70.920 (Table 1). Figure 3 demonstrates that the interactions of the compound 4CpC with the cell wall-related target occurred in the most voluminous groups of the structure (benzene rings), and in the central ester group, and it was observed that the compound established a critical interaction for the maintenance of enzymatic activity, as is the case of the conventional-hydrogen interaction (dashed line in dark green) through the Threonine 622 residue (Thr622), providing strong directional anchoring that stabilizes the pose [43].
A probability of modulation of the MepA efflux pump was observed, since the compound 4CpC presented score values corresponding to −123.85, demonstrating a stronger and higher affinity interaction in the static model to the target when compared to the control compound CCCP (−86.926) (Table 1). In addition, the occurrence of critical interactions (Figure 4) that are present in the active site of this structure was observed, such as the interaction in the halogen group (dashed line in blue) through the residue Alanine 161 (Ala161) and the carbon-hydrogen interaction (dashed line in light green) through the residue Methionine 291 (Met291) [44]. The observed interactions play a key role in the geometric complementarity and proper orientation of the ligand within the efflux channel. In particular, the halogen Ala161 interaction significantly contributes to the higher binding affinity demonstrated by the 4CpC compound, as the halogen atom acts as an electropositive region (σ-hole) capable of interacting with electronegative atoms, stabilizing the complex [45]. This interaction helps firmly anchor the compound to the active site and may increase its target specificity.
There was also a probability of modulation of the NorA efflux pump, as the 4CpC compound presented score values corresponding to −108.877, demonstrating a stronger and higher affinity interaction in the static model to the target when compared to the control compound CCCP (−93.487) (Table 1). Regarding the molecular interactions of compound 4CpC with the target related to the encoded region of the NorA efflux pump (Figure 5), carbon-hydrogen interactions (dashed line in light green) were observed with the ester group through residues Tryptophan 274 (Trp274), this interaction also being observed in the control compound CCCP, and two interactions through residue Phe 271. Hydrophobic interactions were visualized in the chlorobenzene group, with two T-shaped Pi-pi interactions (dashed line in dark pink) being observed through residues Tyrosine 278 (Tyr278) and Phenylalanine 262 (Phe 262), and four alkyl and pi-alkyl interactions (dashed line in light pink) through residues Isoleucine 304 (Ile304), Methionine 308 (Met308), Phenylalanine 254 (Phe254) and Tyrosine 278 (Tyr278) [46]. It is important to mention that the mentioned interactions, especially the Trp 274 carbon-hydrogen interaction, play an essential role in the geometric complementarity and proper orientation of the compound 4CpC within the efflux channel and consequently explain its greater affinity for the target in question.
Computational Analysis of Michael Addition Feasibility
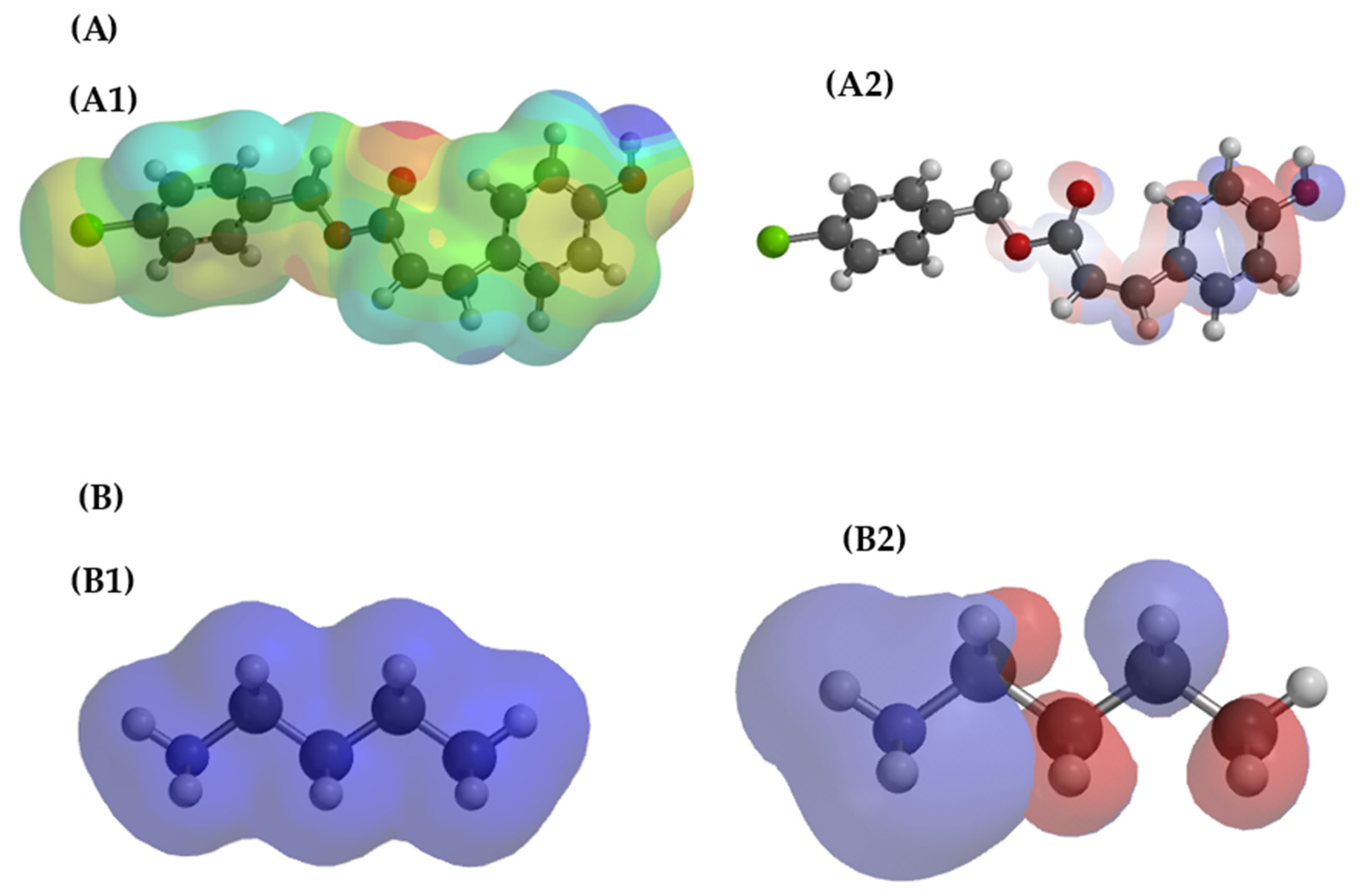
Covalent addition was analyzed using the Michael Addition mechanism, in which the ε-amino group of lysine acts as a nucleophile attacking the β-carbon of 4CpC on target PgSA. Calculations were performed by DFT (B3LYP/6-31G*) in water, indicating that the formation of the adduct with lysine is energetically favorable (ΔE = −18.0 kcal·mol−1). In contrast, aspartate showed positive energy (ΔE = +5.81 kcal·mol−1), demonstrating that the reaction is unfavorable for the carboxylate group. Thus, the results support that lysine is the residue most likely to participate in the Michael reaction with 4CpC. Figure 6 presents the theoretical analyses relating to the electrostatic potential maps and the HOMO–LUMO frontier orbitals for 4CpC (A) and for the lysine side chain (B), demonstrating the nature of the interaction between these molecules.
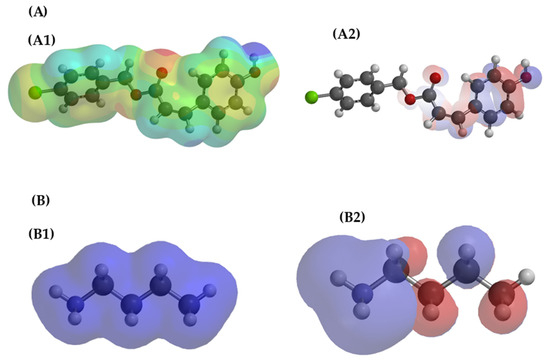
Figure 6.
Theoretical representations referring to the electrostatic potential map (EPM) (A1,B1) and the distribution of HOMO–LUMO frontier orbitals (A2,B2) for 4CpC (A) and the lysine side chain (B). In the electrostatic potential map (EPM), the red regions indicate higher electron density, while the blue regions represent electron deficiency. In the HOMO–LUMO orbitals, the colors (red and blue) represent the opposite phases of the wave functions, with HOMO indicating the regions of higher probability of electron donation and LUMO those of electron acceptance.
It is observed that 4CpC exhibits regions of negative electrostatic potential concentrated in the carbonyl and hydroxyl oxygens, indicating electronegative character and the ability to accept electrons, while lysine presents positive regions near the amine group, characteristic of a good electron donor. The overlap between the HOMO of lysine, located in the amine group, and the LUMO of 4CpC, associated with the oxygenated regions, favors the transfer of electron density and the formation of electrostatic interactions and hydrogen bonds between the two species [47]. The negative value of ΔE (−18.0 kcal·mol−1) obtained in the calculation confirms that this interaction is energetically favorable, indicating stabilization of the system and the formation of a more stable complex, resulting from the electronic complementarity and affinity between the regions of opposite charges of the two molecules.
3.2. Molecular Dynamics Simulations
Molecular Dynamics (MD) simulations have been conducted with the aim of investigating the behavior of molecules over time and providing detailed information about their motions, interactions and stability at the atomic level. This approach is widely used to assess the stability of protein-ligand complexes, as well as to estimate the binding free energy [48].
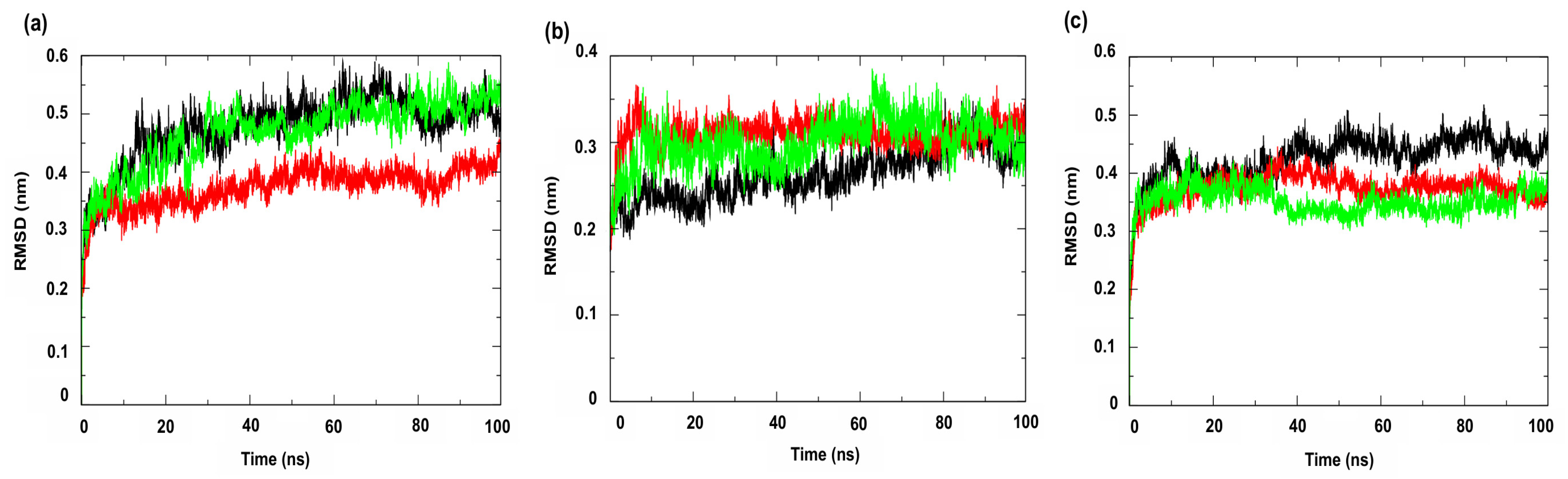
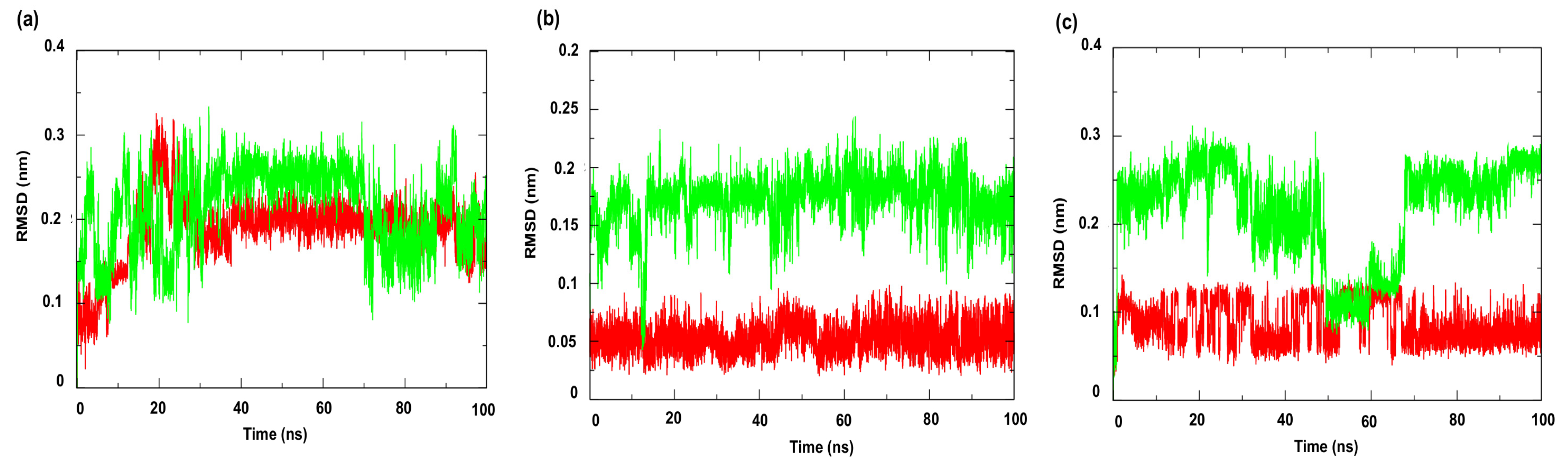
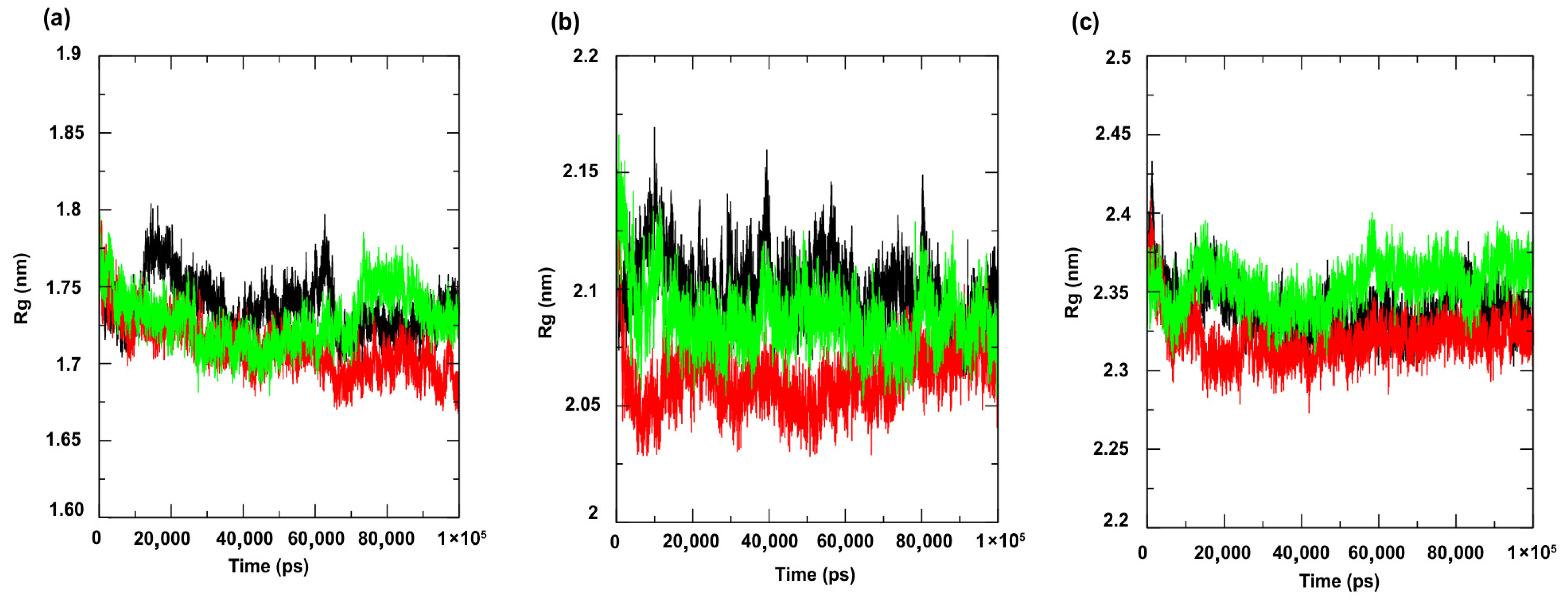
Figure 7 shows the RMSD (Root Mean Square Deviation) profiles of the complexes formed with the compounds under study. This metric evaluates the average of the differences in the positions of the alpha carbon atoms (Cα) of the protein throughout the simulation, compared to a reference structure (usually the initial structure or an experimental structure, such as those available in the PDB) [49]. The results indicated greater stability for the complexes involving the enzyme PgsA, a membrane-associated target, when complexed with the ligand 4CpC (red line), which presented lower RMSD values compared to the control compound (green line) (Figure 7a), indicating that the stable interaction of 4CpC with the protein phosphatidylglycerol phosphate synthase (PgsA) denotes that the compound may interfere with the catalytic activity of the enzyme, which is essential for the biosynthesis of phospholipids in bacterial membranes, which may result in a prolonged inhibitory effect and compromise membrane integrity [39]. It was also observed that 4CpC has a greater capacity to remain bound to the active site of the protein even in the presence of solvents and in the face of sudden variations in temperature and pressure, which was observed in the metrics related to the RMSD value of the complex related to the ligand (Figure 8a–c), since the RMSD values of the ligand for 4CpC remained consistently lower than those observed for the positive controls in all targets studied. These results indicate that 4CpC, in addition to binding to the target with high affinity, also has the potential to possibly exert a sustained effect on its function under physiological conditions.
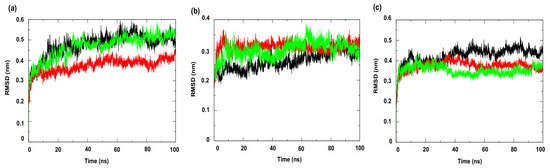
Figure 7.
Root Mean Square Deviation—Protein (RMSD-Protein) of the α-Carbon. Protein: (a) Cell membrane enzyme: PgsA; (b) Enzyme involved in DNA replication: NAD+-dependent DNA ligase; (c) Cell wall enzyme: PBP4. Complex: protein (black line); 4CpC compound + protein (red line) and positive control + protein (green line). Control compounds: (a) cerulenin; (b) levofloxacin and (c) penicillin.
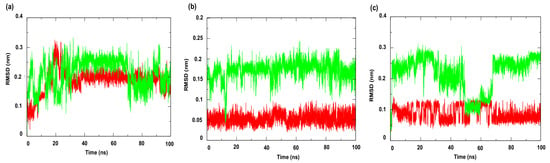
Figure 8.
Root Mean Square Deviation—Ligand (RMSD-Ligand) of the α-Carbon. Protein: (a) Cell membrane enzyme: PgsA; (b) Enzyme involved in DNA replication: NAD+-dependent DNA ligase; (c) Cell wall enzyme: PBP4. Complex: 4CpC compound + protein (red line) and positive control + protein (green line). Control compounds: (a) cerulenin; (b) levofloxacin and (c) penicillin.
The evaluation of the structural packing of the protein through the analysis of the radius of gyration (Figure 9a–c) revealed small fluctuations in the values of this metric, with overlapping of some trajectories for all targets under study, indicating the occurrence of discrete conformational changes during the simulation. This is because the radius of gyration measures the degree of compaction of the protein, that is, how distributed the atoms are around the center of mass [50]. When the variation in the radius of gyration metric values is discrete, it means the protein has not undergone major structural expansions or collapses, maintaining its conformational integrity. The overlap of trajectories between replicas or between different complexes also suggests that interactions with the ligand did not cause significant perturbations in the overall structure, reflecting a balanced and energetically stable system, indicating the maintenance of complex activity.
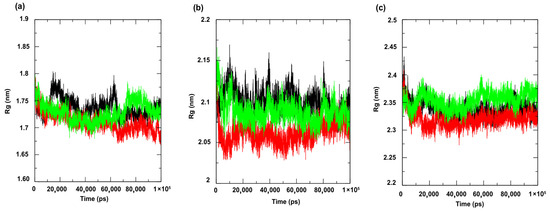
Figure 9.
Radius of gyration. Protein: (a) Cell membrane enzyme: PgsA; (b) Enzyme involved in DNA replication: NAD+-dependent DNA ligase; (c) Cell wall enzyme: PBP4. Complex: protein (black line); 4CpC compound + protein (red line) and positive control + protein (green line). Control compounds: (a) cerulenin; (b) levofloxacin and (c) penicillin.
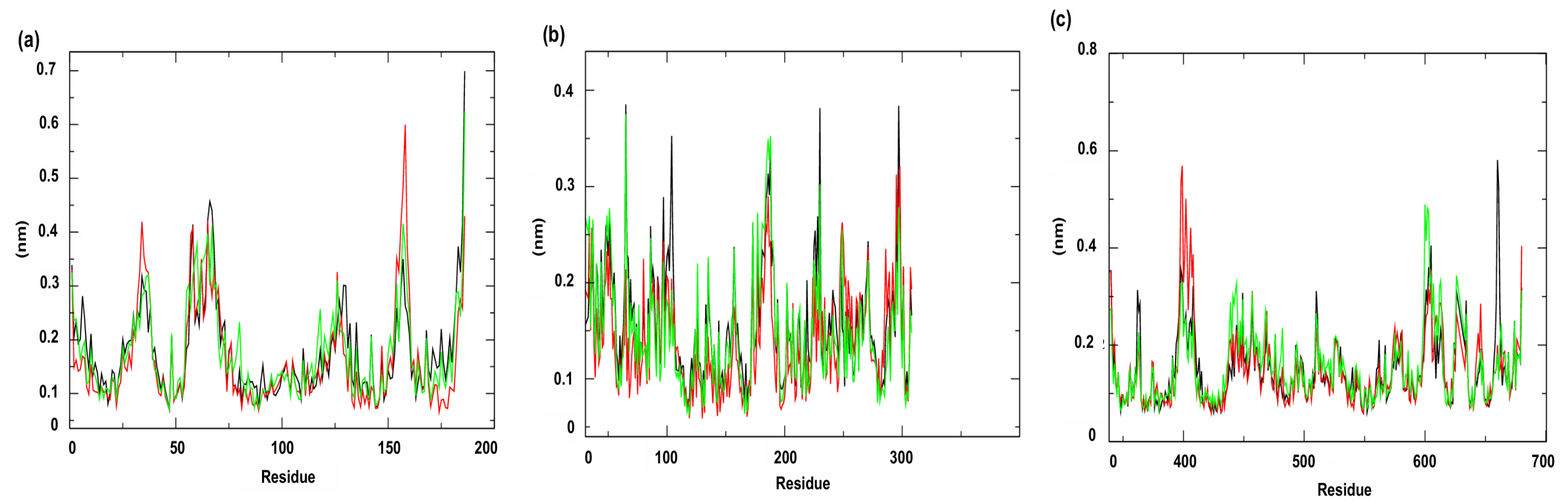
The RMSF (Root Mean Square Fluctuation) analysis (Figure 10), which quantifies the fluidity and variation in each atom (usually the Cα atoms of the protein structure) around its mean position throughout the simulation, allows identifying regions of the protein that present greater mobility during the simulation, reflecting local structural flexibility [51]. RMSF values above 0.3 nm indicate more dynamic regions, which may be associated with loops, terminals or domains subject to conformational modifications, with these values being observed only in the complexes referring to targets related to membrane and cell wall mechanisms. For the membrane target, PgsA (PDB ID: 7DRJ), the largest fluctuations were identified in the residues Methionine 1 (Met1), Phenylalanine 58 (Phe58), Tyrosine 62 (Tyr62), Arginine 65 (Arg65), Lysine 66 (Lys66), Leucine 157 (Leu157) and Arginine 186 (Arg186). Among these, residues Arg65 and Phe58, located in the active site, may be associated with potential conformational adjustments relevant for ligand recognition. For the cell wall-related target, PBP4 (PDB ID: 6BSR), significant fluctuations were observed at residues Threonine 259 (Thr259), Lysine 260 (Lys260), Threonine 284 (Thr284), Glutamic Acid 286 (Glu286), Aspartic Acid 287 (Asp287), Isoleucine 288 (Ile288), Glutamic Acid 289 (Glu289), Lysine 290 (Lys290), Glutamine 316 (Gln316), and Aspartic Acid 317 (Asp317). Importantly, these residues are not part of the enzyme active site, indicating peripheral changes that do not directly affect catalysis but may be related to the global dynamics of the structure.
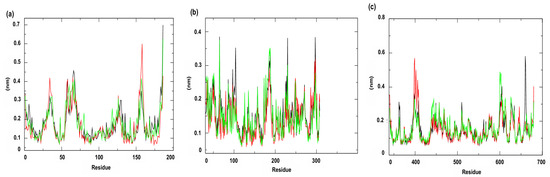
Figure 10.
Root Mean Square Fluctuation (RMSF). Protein: (a) Cell membrane enzyme: PgsA; (b) Enzyme involved in DNA replication: NAD+-dependent DNA ligase; (c) Cell wall enzyme: PBP4. Complex: protein (black line); 4CpC compound + protein (red line) and positive control + protein (green line). Control compounds: (a) cerulenin; (b) levofloxacin and (c) penicillin.
Table 2 presents the contributions of the Coulomb and Lennard-Jones energies for the analyzed complexes. In all targets studied, Coulombic interactions were mainly responsible for the total interaction energy. 4CpC presented more negative Coulomb energy values compared to the control compounds, with −241.229 kJ/mol (4CpC) versus −102.851 kJ/mol (positive control) for the membrane target, −258.000 kJ/mol (4CpC) versus −26.676 kJ/mol (positive control) for the DNA replication-related target and −301.038 kJ/mol (4CpC) versus −124.426 kJ/mol (positive control) for the cell wall target. These results indicate a more favorable electrostatic interaction between 4CpC and the respective targets [52].

Table 2.
Contributions of Coulomb and Lennard-Jones energies for the compound under study with the enzymes.
The more negative coulombic energy values observed for 4CpC compared to the control compounds indicate stronger attractive forces between the charged or polar groups of the ligand (as was observed, the contribution of the ester group, the halogen-substituted benzene ring and the para-substituted phenol group) and those of the target residues. This effect may arise from the presence of functional groups capable of forming multiple hydrogen bonds or strong dipole–dipole interactions, characteristics always present in phenolic compounds, increasing the complementarity between the electronic distributions of the ligand and the active site, as observed in molecular Docking simulations in the PBP4 target with the occurrence and contribution of the conventional hydrogen type interaction through Threonine 622 (Thr622) [38].
This electrostatic stabilization is particularly relevant because Coulombic interactions govern the initial recognition and orientation of the ligand within the binding site, guiding the subsequent van der Waals and hydrophobic contacts that reinforce the stability of the complex [53], which may justify the hydrophobic contribution to the high affinity of the 4CpC compound observed in molecular docking simulations in the membrane-related PgsA targets, where residues Lysine 135 (Lys135), Aspartic acid 60 (Asp60), and Alanine 64 (Ala64) were observed, and in the NAD+-dependent DNA ligase, where hydrophobic interaction was observed through residue Leucine 80 (Leu80), being essential interactions for directing the ligand to the active site.
Therefore, the greater contribution of Coulombic energy to 4CpC suggests a more favorable binding geometry and stronger electronic complementarity with the amino acid residues of the targets, which may explain its higher affinity and possibly the likelihood of biological activity.
3.3. In Silico Pharmacokinetic Predictions
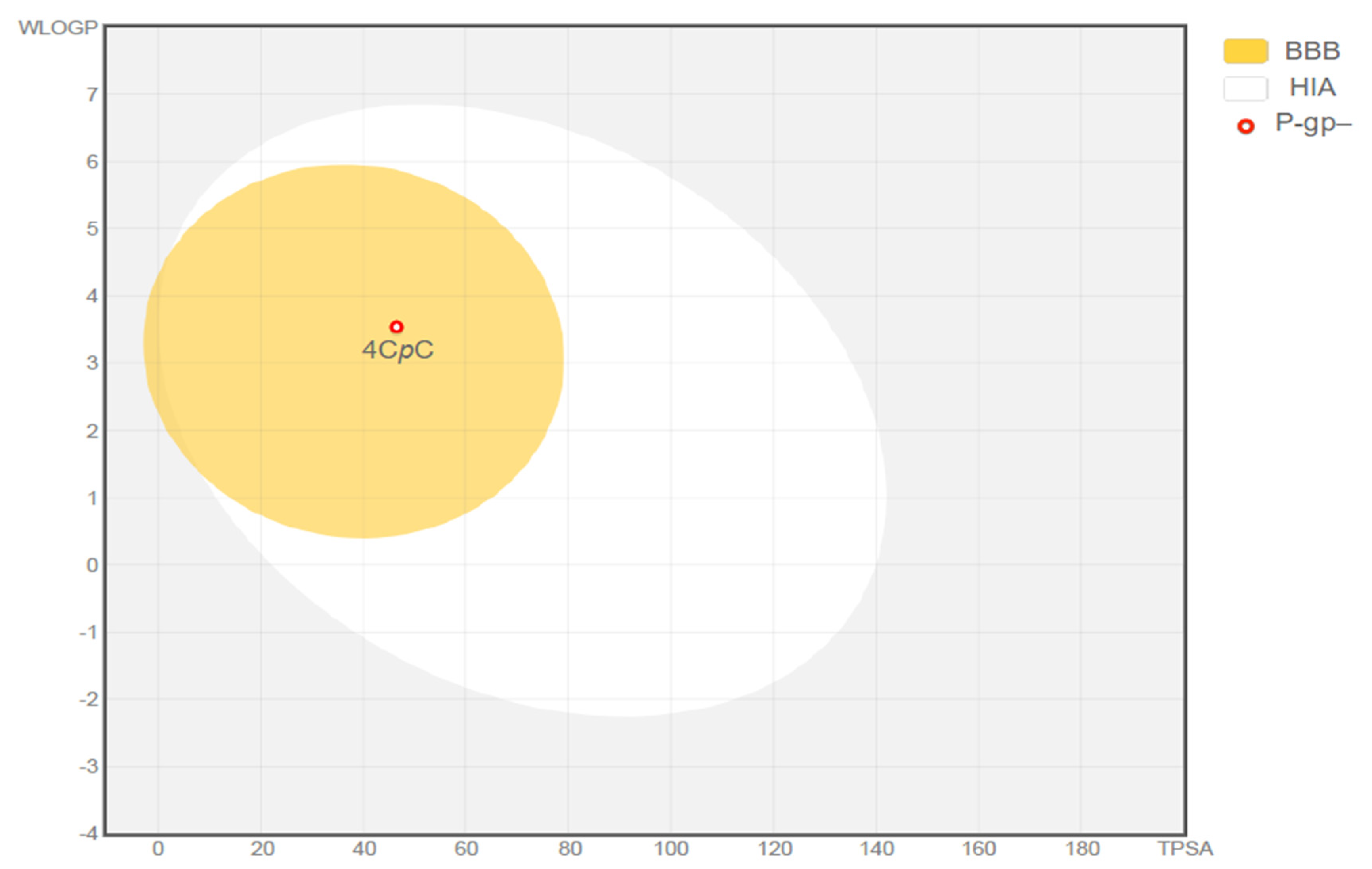
Comprehensive in silico analyses provided physicochemical properties and ADME (absorption, distribution, metabolism, and excretion) profiles of the compound 4CpC. The molecule was evaluated according to Lipinski’s Rule of Five and demonstrated compatibility with oral administration. Using the Boiled-Egg model, 4CpC was predicted to cross the blood–brain barrier (logPS = −1.431 > −2) and exhibited a high probability of gastrointestinal absorption (92.318%) (Figure 11). Additionally, it was not shown to be a possible inhibitor or substrate for P-glycoprotein (P-gP-) and presented a low steady-state volume of distribution (VDss). The compound was also predicted to inhibit specific cytochrome P450 (CYP450) isoforms, while not serving as a substrate for others. Notably, 4CpC was not identified as a substrate for the Organic Cation Transporter 2 (OCT2).
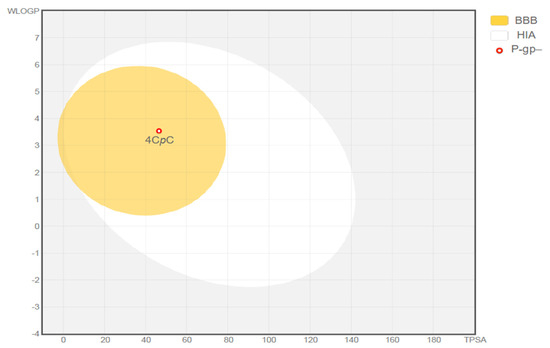
Figure 11.
Representation of the gastrointestinal absorption and brain penetration of 4-chlorobenzyl p-coumarate (4CpC) using the Boiled-Egg model. BBB (Blood–Brain Barrier); HIA (Human Intestinal Absorption); P-gp (P-glycoprotein substrate, positive or negative).
3.4. Minimum Inhibitory Concentration (MIC) Assay
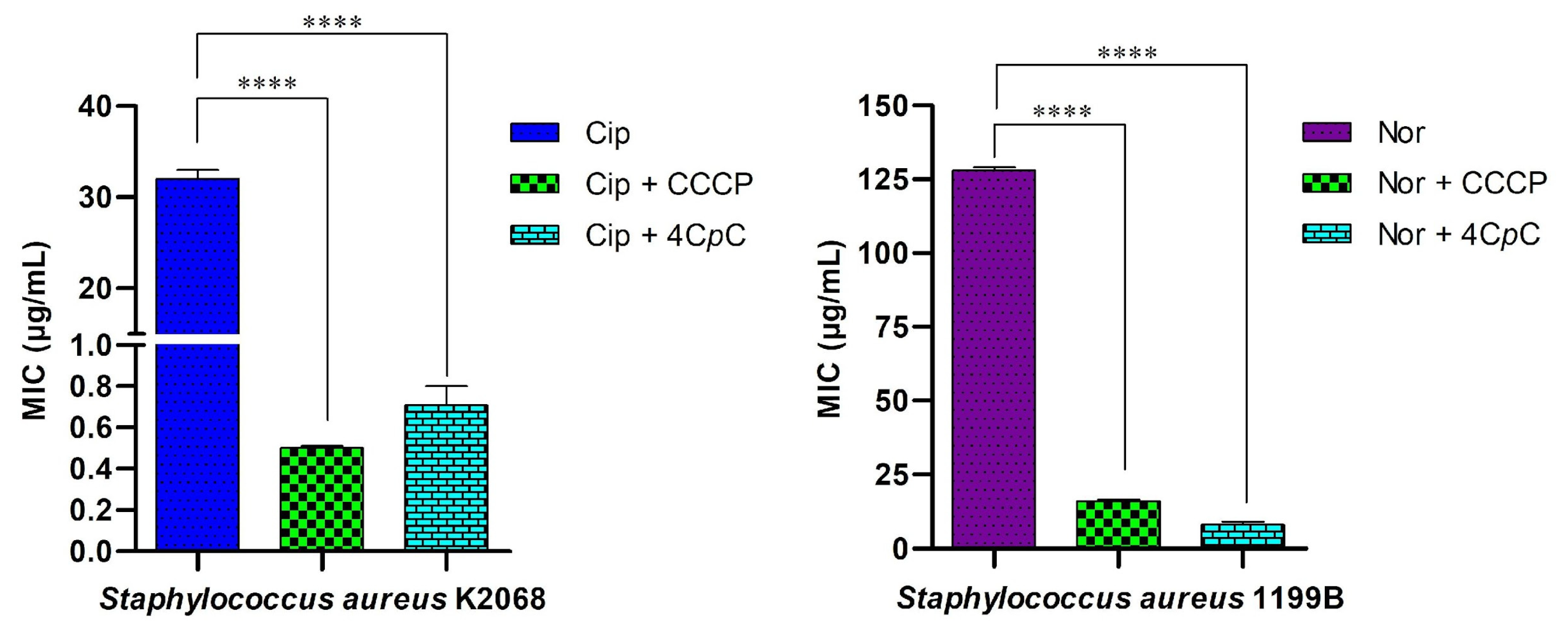
Minimum Inhibitory Concentration (MIC) assays using ethidium bromide (EtBr), as well as the antibiotics ciprofloxacin and norfloxacin, were performed on the K2068 and 1199B strains to confirm the presence of efflux pump activity and its contribution to the observed resistance phenotype. Additional assays were conducted with the compound 4CpC and the efflux pump inhibitor CCCP. 4CpC, when tested alone, exhibited a MIC value of 1024 μg/mL (3554.93 μM) against S. aureus K2068 and S. aureus 1199B. Ciprofloxacin exhibited a MIC of 32 μg/mL (96.57 μM) against strain K2068 (Resistance breakpoint for Staphylococcus spp. ≥ 4 μg/mL [54]), while norfloxacin showed a MIC of 128 μg/mL (400.83 μM) against strain 1199B (Resistance breakpoint for Staphylococcus spp. ≥ 16 μg/mL [54]). CCCP exhibited MIC values of 8 μg/mL (39.09 μM) for both strains, confirming its role as an efflux pump inhibitor.
3.5. Inhibition of MepA and NorA Efflux Pumps: Ethidium Bromide Assay Outcomes
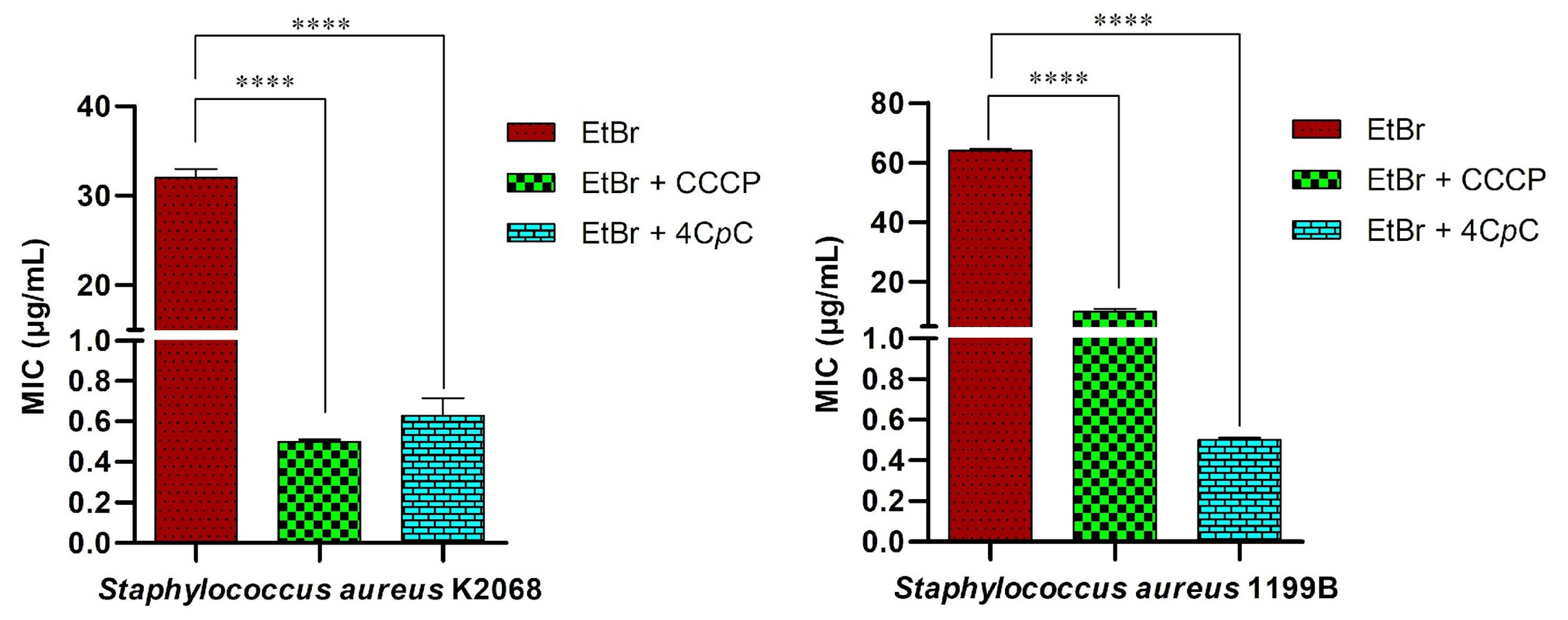
This method is based on evaluating the reduction in the MIC of ethidium bromide (EtBr) when co-administered with the test compound. As shown in Figure 12, the assay demonstrated inhibition of the MepA and NorA efflux pumps, expressed by S. aureus strains K2068 and 1199B, respectively. The known efflux pump inhibitor CCCP (positive control) and the test compound 4CpC were used in combination with EtBr to assess their ability to enhance intracellular accumulation of the dye with a reduction in MIC. 4CpC markedly decreased the MIC of EtBr in both S. aureus strains from 32 μg/mL to 0.62 μg/mL in strain K2068 (p < 0.0001) and from 64 μg/mL to 0.50 μg/mL in strain 1199B (p < 0.0001). Notably, the reduction observed in strain 1199B exceeded that achieved by CCCP. The observed decrease in EtBr MIC in strains harboring efflux pumps suggests effective inhibition of pump activity [55].
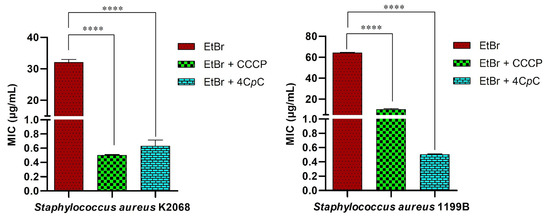
Figure 12.
Ethidium bromide accumulation assay for efflux inhibition analysis. The MIC was assessed in S. aureus K2068, harboring the MepA efflux pump, and S. aureus 1199B, carrying the NorA efflux pump, treated with CCCP and 4CpC in combination with ethidium bromide. **** p < 0.0001 indicates statistically significant differences between groups. Statistical significance was determined using one-way ANOVA followed by Bonferroni’s post hoc test.
3.6. Modulatory Effects of 4CpC on Antibiotic Resistance
This study investigates the ability of the test compound to modulate bacterial resistance to antibiotics. Given that S. aureus strains K2068 and 1199B harbor genes encoding efflux proteins associated with resistance to ciprofloxacin and norfloxacin, a reduction in the MIC of the antibiotic in the presence of the test compound is indicative of efflux pump inhibition [55]. The compound 4CpC significantly reduced the MIC of ciprofloxacin against S. aureus K2068 from 32 μg/mL to 0.70 μg/mL (p < 0.0001), suggesting inhibition of MepA-mediated antibiotic efflux. An even more striking result was observed with the S. aureus 1199B strain, which overexpresses the NorA efflux pump: 4CpC reduced the MIC of norfloxacin fourfold, from 128 μg/mL to 8 μg/mL (p < 0.0001). This reduction exceeded that achieved by the efflux pump inhibitor CCCP, which decreased the MIC threefold from 128 μg/mL to 16 μg/mL (p < 0.0001), as shown in Figure 13. These findings indicate that 4CpC modulates antibiotic resistance to ciprofloxacin and norfloxacin in S. aureus strains K2068 and 1199B, respectively, by acting as an efflux pump inhibitor.
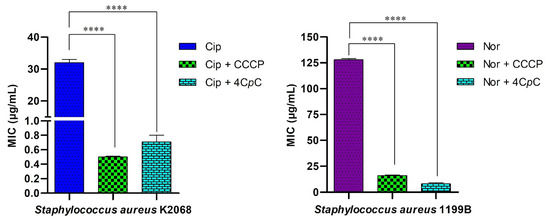
Figure 13.
Modulation of antibiotic activity against efflux pump-expressing strains. 4CpC was evaluated in combination with ciprofloxacin and norfloxacin against Staphylococcus aureus strains K2068 and 1199B, which overexpress the efflux proteins MepA and NorA, respectively. **** p < 0.0001 indicates statistically significant differences between groups. Statistical significance was assessed by one-way ANOVA followed by a Bonferroni post hoc test.
3.7. Effects of the Compound on DNA Integrity
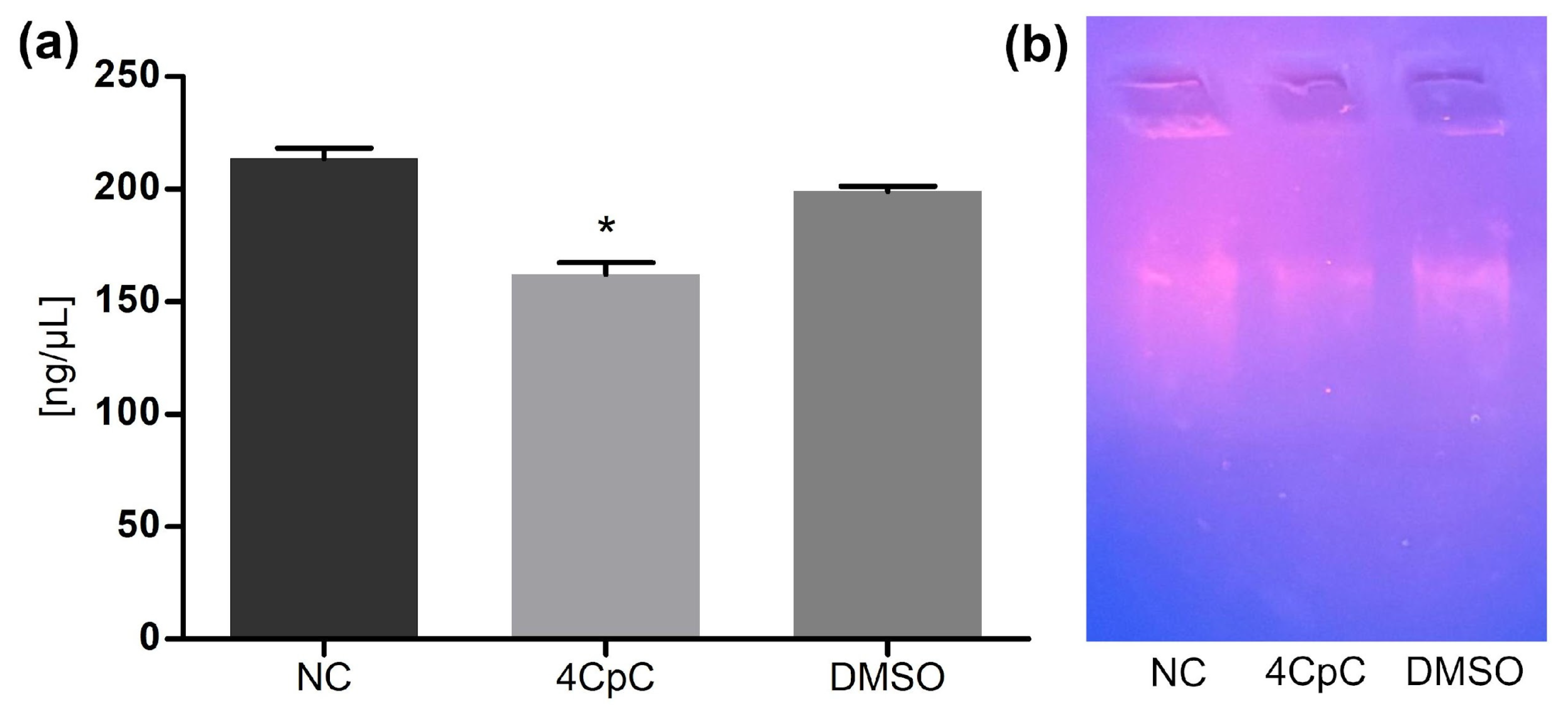
Quantitative analysis of DNA content (Figure 14a) revealed a statistically significant reduction (p < 0.0001) in the samples treated with 4CpC compared to the untreated control. This decrease may reflect a lower number of viable cells after exposure or a partial degradation of genetic material. The DNA concentration in the DMSO control group was similar to that of untreated cells, indicating no solvent interference.
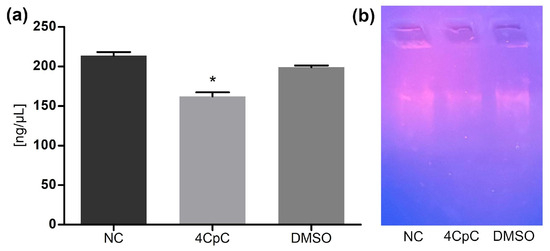
Figure 14.
(a) Impact of 4CpC on DNA concentration; (b) Agarose gel electrophoresis of genomic DNA from E. faecalis. * p < 0.0001 indicates statistically significant differences between groups. Statistical significance was assessed by one-way ANOVA followed by a Bonferroni post hoc test. NC (negative control); 4CpC (4-chlorobenzyl p-coumarate); DMSO (dimethyl sufoxide).
Agarose gel electrophoresis (Figure 14b) showed a visible reduction in the intensity of the genomic DNA band in the 4CpC-treated sample relative to the control, but without the appearance of specific fragmentation patterns. This result suggests that the compound may indirectly affect DNA integrity, possibly as a downstream consequence of membrane disruption or cell lysis, as previously described for 4CpC [17].
Altogether, these findings demonstrate that treatment with 4CpC leads to a measurable reduction in total DNA yield; however, they do not provide direct evidence of binding to DNA or inhibition of replication machinery. Further experiments employing DNA-binding fluorophores are needed to confirm whether 4CpC interacts directly with nucleic acids.
4. Discussion
This study represents the first report involving the compound 4CpC, a derivative of p-coumaric acid, integrating analyses for prediction of its mode of action and assays on the modulation of antimicrobial resistance to assess its ability to inhibit efflux mechanisms, as reported herein.
Molecular docking analyses predicted the docking poses and provided the binding energies between 4CpC and reference compounds with selected molecular targets. Penicillin-binding protein 4 (PBP4) is a selected target that exhibits enzymatic activity in peptidoglycan biosynthesis by linking new N-acetylglucosamine–N-acetylmuramic acid–pentapeptide fragments to the bacterial cell wall in dividing cells [43]. DNA ligase promotes the joining of DNA strand fragments by catalyzing the formation of phosphodiester bonds between nucleotides [42]. Phosphatidylglycerol phosphate synthase (PgsA) catalyzes a key step in the biosynthetic pathway of phosphatidylglycerol, one of the main phospholipids that constitute the cytoplasmic membrane of bacteria such as S. aureus and E. faecalis [39]. MepA and NorA are transmembrane proteins that function as efflux pumps, expelling a variety of compounds, including antibiotics, from the bacterial cell, thereby contributing to microbial resistance [44,46].
Molecular docking analysis revealed that 4CpC binds to all tested enzymes, showing the highest affinity for the membrane-associated enzyme PgsA, followed by DNA ligase. These findings parallel those of Lou et al. [7], who reported that the 4CpC precursor, p-coumaric acid, disrupts bacterial membranes and interacts with DNA. Likewise, Singha et al. [56] demonstrated that p-coumaric acid inhibits phosphatidylglycerol synthesis by targeting CDP-diacylglycerol–glycerol-3-phosphate 3-phosphatidyltransferase in bacterial membranes.
Molecular dynamics simulations demonstrated stable interactions between 4CpC and its molecular targets, particularly with PgsA, which showed the lowest deviation values. These results suggest fewer, but more stable, ligand–protein contacts, leading to faster stabilization of the binding complex. Fluctuations in the residues were lower for DNA ligase, indicating a less flexible structure within a balanced dynamic profile that does not undergo significant conformational changes, thereby contributing to longer residence time and greater interaction stability.
4CpC is a semisynthetic compound that is soluble in organic solvents and exhibits moderate water solubility. These physicochemical properties suggest that the molecule possesses characteristics compatible with both membrane interaction and intracellular activity. In a study conducted by our research group [17], 4CpC was shown to damage the cell membranes of S. aureus and E. faecalis, in addition to causing disorganization of the genetic material. These findings support the in silico results presented herein.
The potential inhibition of DNA ligase by 4CpC in bacteria could disrupt DNA replication and repair processes. This enzyme is essential for the synthesis of the discontinuous DNA strand and for the ligation of DNA fragments (Okazaki fragments). Disruption of these processes leads to genomic instability, rendering bacteria more vulnerable to adverse environmental conditions and ultimately resulting in cell death [57]. This represents a promising therapeutic target that remains underexplored.
By binding to PgsA, 4CpC could impair the biosynthesis of phospholipids that are structurally essential for membrane integrity, triggering osmotic disturbances that interfere with normal cellular function. Furthermore, its interaction with PBP, albeit to a lesser extent, could disrupt the cell wall synthesis process, ultimately leading to lysis and cell death.
The binding affinity of the test compound for the MepA and NorA efflux pumps suggests that the compound may interact with these pump structures and inhibit their normal function. Consequently, this may enhance the effectiveness of co-administered antibiotics. These findings are consistent with the in vitro results reported in this study.
4CpC exhibited more pronounced electrostatic (Coulomb) binding energies, which favored the formation and temporal stability of the complex, as observed in the RMSD analysis. Coulomb interactions are specific and selectively attract defined groups toward the target site, enhancing ligand fit within the active site while reducing off-target binding, which could otherwise trigger adverse side effects.
The results obtained from docking and molecular dynamics simulations suggest a dual mechanism of action for 4CpC in bacterial cells, targeting both membrane biosynthesis and DNA replication. Additionally, the compound exhibits sufficient binding energy to potentially inhibit resistance mechanisms mediated by MepA and NorA efflux proteins.
Lipinski’s rule of five was applied to evaluate the compound’s suitability for oral activity. For medicinal chemistry purposes, the substance must meet at least three of these four criteria [58]. The compound 4CpC has a molecular mass of 288.73 Da, 1 hydrogen bond donor, 3 hydrogen bond acceptors, and a consensus LogPo/w (the arithmetic mean of values predicted by five methods for lipophilicity) of 3.49, thus fulfilling all of Lipinski’s parameters. Across the three solubility models (ESOL, Ali, and SILICOS-IT), the compound showed a LogS between −6 and −4, indicating moderate water solubility.
The Boiled-Egg model provides an intuitive graphical prediction of molecular absorption based on two key physicochemical parameters: lipophilicity (logP) and topological polar surface area (TPSA), which together describe a compound’s balance between lipid affinity and polarity [33]. According to this model, the compound falls within the “white” region, suggesting a high likelihood of gastrointestinal absorption.
Caco-2 cells, derived from human colorectal epithelial adenocarcinoma, are widely used as an in vitro model to assess the intestinal permeability of orally administered compounds [59]. The compound demonstrated permeability characteristics indicative of efficient absorption in the small intestine. This finding is consistent with its low TPSA, which falls within the range typically associated with favorable oral bioavailability. Regarding dermal permeability, the compound exhibited characteristics consistent with low potential for transdermal absorption, as its permeability coefficient falls within the range typically associated with limited skin penetration.
P-glycoprotein (Pgp) is a key efflux transporter that limits the bioavailability of xenobiotics by actively exporting them out of cells, thereby playing a protective role but also potentially reducing therapeutic drug concentrations [60]. In this context, it is desirable for drug candidates to exhibit minimal interaction with this protein to avoid unfavorable pharmacokinetic outcomes. The compound 4CpC is predicted neither to be a substrate nor an inhibitor of Pgp.
In accordance with the Boiled-Egg model, 4CpC is capable of crossing the blood–brain barrier (BBB), as it falls within the physicochemical space represented by the “yolk” region. This suggests that the compound may be bioavailable within the central nervous system (CNS), supported by its logPS value of −1.431, which is above the threshold of −2. This property is particularly relevant for the potential treatment of infections affecting the CNS.
The steady-state volume of distribution (VDss) reflects the theoretical volume in which the total administered dose of a drug would need to be uniformly distributed to achieve the same concentration as observed in plasma [61]. In this context, the compound exhibited a profile consistent with limited tissue distribution and a tendency to remain predominantly within the plasma compartment.
Another parameter evaluated was metabolic prediction. Among the main metabolic systems is the mitochondrial cytochrome P450 (CYP450) system, a superfamily of enzymes involved in the metabolism of a wide range of drugs. Computational assessments indicated that the compound is a potential inhibitor of CYP1A2, CYP2C19, and CYP2C9 isoforms. This inhibition suggests a reduced biotransformation rate. On one hand, this effect may enhance the pharmacological performance of the compound by prolonging its bioavailability; on the other hand, it may also lead to the occurrence of undesirable effects due to substance accumulation. Moreover, the compound 4CpC was predicted neither to inhibit nor to serve as a substrate for CYP2D6 and CYP3A4. These findings are relevant in the context of pharmacokinetic interactions, particularly in identifying specific isoforms affected by compounds with enzymatic inducer or inhibitor activity [62].
To assess the excretion profile of 4CpC, its potential as a substrate for the Organic Cation Transporter 2 (OCT2), a renal uptake transporter involved in the elimination of both endogenous and exogenous compounds, was investigated. The results indicated that 4CpC is not a substrate for OCT2. This finding is relevant to preventing potential drug–drug interactions when co-administered with other compounds that are OCT2 substrates. Additionally, the total clearance of 4CpC was estimated (log ml/min/kg = −0.196), a proportionality constant reflecting the combined hepatic and renal elimination pathways, thereby enabling dose predictions to achieve steady-state concentrations [63]. Further information on ADME predictions is described in the Supplementary Materials.
The result of the MIC of 4CpC against the tested S. aureus strains corroborates the high level of bacterial resistance, consistent with the presence of active efflux mechanisms. However, in a previous study [17], it was reported that 4CpC demonstrated antibacterial activity against S. aureus strains NCTC 12981 and 13373, with MIC values of 8 μg/mL for both. The strain 13373 carries a target-site alteration as a genetically encoded resistance mechanism. In line with the present study, in an investigation involving p-coumaric acid, the precursor of 4CpC, Silveira et al. [64] observed MIC values exceeding 1024 μg/mL against S. aureus strains 1199 and 1199B, both of which express the NorA efflux pump, and strain IS-58, which produces the MepA pump. Based on the in silico analyses conducted in this study, together with the data reported by Falcão et al. [17], it is proposed that 4CpC may also exert its activity at the intracellular level. This intracellular mechanism of action may render 4CpC liable to efflux systems.
Ethidium bromide (EtBr) is a toxic and mutagenic agent that intercalates between DNA base pairs and is resisted exclusively through efflux pump-mediated mechanisms [65]. Although it can exert toxic effects on cells, including bacterial cells, it is neither considered nor employed as an antimicrobial agent. EtBr is commonly used in research to assess a bacterium’s ability to expel the compound, thereby indicating the presence of efflux pumps. It acts by stimulating these pumps, with efflux activity being relatively proportional to the intracellular concentration of EtBr [66].
The compound 4CpC, at subinhibitory concentrations, exhibited promising activity by reducing the MIC of EtBr in both tested strains, with a more pronounced effect against S. aureus 1199B. This indicates a potential inhibitory effect of 4CpC on EtBr efflux mechanisms, leading to increased intracellular retention of the compound and potentiation of its activity. It also suggests that structural variations among bacterial efflux systems may significantly influence the interaction and activity of efflux modulators.
The modulation of bacterial resistance to antibiotics is evidenced by the ability of certain compounds to restore or enhance the effectiveness of antimicrobials against resistant strains. Considering the effects of 4CpC observed in the EtBr efflux assay, the modulation of ciprofloxacin and norfloxacin activity was evaluated in resistant S. aureus strains. 4CpC acted as a modulator of resistance to ciprofloxacin and norfloxacin in S. aureus, potentially functioning as an inhibitor of the MepA and NorA efflux pumps. The combination of antibacterial agents with efflux pump inhibitors may elicit a synergistic effect between the compounds. This interaction can lead to a reduction in the MICs of the antibiotics, suggesting a reversal of the bacterial resistance phenotype and an enhancement of antibiotic efficacy.
In an in silico study, phenolic acids were subjected to molecular docking to assess their potential binding affinity to the NorA efflux pump. Among the tested compounds, p-coumaric acid exhibited one of the most favorable binding energies (−6.3 kcal/mol), indicating a strong interaction with target protein, even surpassing that of ciprofloxacin (−4.9 kcal/mol), the reference drug used in the study. Subsequently, molecular dynamics simulations were performed to analyze the conformational stability and flexibility of the complexes formed between NorA and the phenolic acids. The results indicated that p-coumaric acid anchored NorA more stably than the ciprofloxacin-NorA complex [67]. These results strengthen the evidence for the potential inhibitory activity of the 4CpC derivative against the NorA efflux pump.
The observed reduction in total DNA content following exposure to 4CpC in E. faecalis suggests an alteration in cellular processes involving nucleic acids. Spectrophotometric quantification at 260 nm revealed a marked decrease in absorbance values in the treated group compared with the untreated control, suggesting a reduction in nucleic acid content. This analytical method, which exploits the intrinsic UV absorbance of nitrogenous bases, is sensitive to conformational changes such as denaturation, strand breaks, or complex formation between DNA and intercalating molecules, and has been widely validated for accurate nucleic acid determination [68]. Complementary analysis by agarose gel electrophoresis (1% w/v), followed by ethidium bromide staining, demonstrated a pronounced reduction in the intensity of genomic DNA bands in 4CpC-treated samples. Since ethidium bromide intercalates between DNA base pairs, a decrease in fluorescence intensity under UV illumination reflects molecular damage, loss of double-stranded integrity, or conformational alterations—consistent with standard methodologies for DNA integrity assessment [69]. Nevertheless, these findings should be interpreted with caution, since the effect may be secondary to cell membrane disruption or decreased cell viability rather than a direct interaction with DNA or its replication enzymes.
Together with the results from molecular docking and dynamics simulations, these findings suggest that 4CpC may establish π–π stacking or hydrogen-bond interactions with the phosphate backbone or nucleobase regions, destabilizing the double-helix structure. Consequently, bacterial DNA emerges as a plausible molecular target for 4CpC. Although molecular docking predicted a favorable binding energy between 4CpC and NAD+-dependent DNA ligase, such computational evidence alone cannot confirm biological interference. Therefore, the hypothesis that 4CpC affects DNA replication remains speculative. Future studies will include selectivity assays—such as competitive binding with ethidium bromide, electrophoretic mobility shift assays, or comparison with non-DNA targets—to clarify whether the compound directly interacts with DNA or acts through indirect mechanisms.
Collectively, the data presented here suggest that 4CpC possesses a dual mode of action against bacteria, a predictive effect that may represent an advantage, since compounds targeting a single site may have their effects more readily circumvented by bacterial resistance mechanisms. The findings demonstrate that this molecule inhibits MepA and NorA pump function, supporting the hypothesis that it acts as an efflux pump inhibitor modulating antibiotic resistance.
5. Conclusions
4-Chlorobenzyl p-coumarate (4CpC) exhibited binding affinity and stability with the enzymes phosphatidylglycerol phosphate synthase and NAD+-dependent DNA ligase, as well as with the efflux pumps MepA and NorA. It demonstrated a potential inhibitory effect on efflux mechanisms, modulating resistance in S. aureus, and showed some activity on bacterial DNA.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/futurepharmacol5040071/s1, Scheme S1: Synthesis of 4-chlorobenzyl p-coumarate; Method: Synthesis of 4-chlorobenzyl p-coumarate; Figure S1: 1H NMR spectrum (500 MHz, CDCl3) of 4-chlorobenzyl p-coumarate; Figure S2: 1H NMR spectrum (500 MHz, CDCl3) of 4-chlorobenzyl p-coumarate; Figure S3: 13C NMR-APT spectrum (100 MHz, CDCl3) of 4-chlorobenzyl p-coumarate; Table S1: In silico data estimated by the pkCSM and SwissADME platform.
Author Contributions
Conceptualization, É.P.F., J.J.E., D.P.d.S. and R.D.d.C.; methodology, É.P.F.; validation, É.P.F. and R.D.d.C.; formal analysis, É.P.F.; investigation, É.P.F., N.F.d.S., K.K.G.M., J.E.R., W.L.d.S.S., J.F.B., L.S., M.T.S., J.C.R.G. and H.D.M.C.; data curation, É.P.F.; writing—original draft preparation, É.P.F.; writing—review and editing, J.J.E., N.F.d.S., K.K.G.M., J.C.R.G., W.L.d.S.S., J.F.B., L.S., M.T.S., J.C.R.G., H.D.M.C., D.P.d.S. and R.D.d.C.; visualization, É.P.F. and R.D.d.C.; supervision, R.D.d.C.; project administration, R.D.d.C. All authors have read and agreed to the published version of the manuscript.
Funding
This study was partially funded by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES), Finance Code 001. Additional support was provided by the Fundação de Apoio à Pesquisa do Estado da Paraíba (FAPESQ).
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author(s).
Acknowledgments
We would like to thank the financial and operational support of the Federal University of Paraíba (UFPB).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- World Health Organization. WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research Development and Strategies to Prevent and Control Antimicrobial Resistance; WHO: Geneva, Switzerland, 2024; 72p. [Google Scholar]
- Baran, A.; Kwiatkowska, A.; Potocki, L. Antibiotics and Bacterial Resistance-A Short Story of an Endless Arms Race. Int. J. Mol. Sci. 2023, 24, 5777. [Google Scholar] [CrossRef]
- Brdová, D.; Ruml, T.; Viktorová, J. Mechanism of Staphylococcal Resistance to Clinically Relevant Antibiotics. Drug Resist. Updat. 2024, 77, 101147. [Google Scholar] [CrossRef]
- Sinha, S.; Aggarwal, S.; Singh, D.V. Efflux Pumps: Gatekeepers of Antibiotic Resistance in Staphylococcus aureus Biofilms. Microb. Cell 2024, 11, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, P.S.; Victorelli, F.D.; Fonseca-Santos, B.; Chorilli, M. A Review of Analytical Methods for P-Coumaric Acid in Plant-Based Products, Beverages, and Biological Matrices. Crit. Rev. Anal. Chem. 2019, 49, 21–31. [Google Scholar] [CrossRef]
- Kot, B.; Wicha, J.; Piechota, M.; Wolska, K.; Grużewska, A. Antibiofilm Activity of Trans-Cinnamaldehyde, p-Coumaric, and Ferulic Acids on Uropathogenic Escherichia Coli. Turkish J. Med. Sci. 2015, 45, 919–924. [Google Scholar] [CrossRef]
- Lou, Z.; Wang, H.; Rao, S.; Sun, J.; Ma, C.; Li, J. P-Coumaric Acid Kills Bacteria through Dual Damage Mechanisms. Food Control 2012, 25, 550–554. [Google Scholar] [CrossRef]
- Li, Y.; Li, J.; Qian, B.; Cheng, L.; Xu, S.; Wang, R. De Novo Biosynthesis of P-Coumaric Acid in E. Coli with a Trans-Cinnamic Acid 4-Hydroxylase from the Amaryllidaceae Plant Lycoris Aurea. Molecules 2018, 23, 3185. [Google Scholar] [CrossRef] [PubMed]
- Ribaudo, G. Natural Products Chemistry: Advances in Synthetic, Analytical and Bioactivity Studies. Molecules 2023, 28, 5577. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D. Opportunities for Natural Products in 21st Century Antibiotic Discovery. Nat. Prod. Rep. 2017, 34, 694–701. [Google Scholar] [CrossRef]
- Molokoane, T.L.; Kemboi, D.; Siwe-Noundou, X.; Famuyide, I.M.; McGaw, L.J.; Tembu, V.J. Extractives from Artemisia Afra with Anti-Bacterial and Anti-Fungal Properties. Plants 2023, 12, 3369. [Google Scholar] [CrossRef]
- Li, J.; Zhao, N.; Xu, R.; Li, G.; Dong, H.; Wang, B.; Li, Z.; Fan, M.; Wei, X. Deciphering the Antibacterial Activity and Mechanism of P-Coumaric Acid against Alicyclobacillus Acidoterrestris and Its Application in Apple Juice. Int. J. Food Microbiol. 2022, 378, 109822. [Google Scholar] [CrossRef]
- Chefo Kengne, N.; Wangso, H.; Silvère Gade, I.; Laya, A.; Paul Bayang, J.; Bargui Koubala, B.; Laurent, S.; Henoumont, C.; Talla, E. Antibacterial and Antioxidant Activities of a New Phenolic Coumpound from the Roots Barks of Cassia Arereh Delile (Fabaceae). Results Chem. 2024, 11, 101802. [Google Scholar] [CrossRef]
- Li, H.; Song, X.; Li, H.; Zhu, L.; Cao, S.; Liu, J. Sesquiterpenes and Monoterpenes from the Leaves and Stems of Illicium Simonsii and Their Antibacterial Activity. Molecules 2022, 27, 1115. [Google Scholar] [CrossRef] [PubMed]
- Iawsipo, P.; Sangnim, T.; Thalang, P.P.N.; Chernchom, R.; Bantao, R.; Luangpraditkun, K. Bioactivities and Promising Active Compounds of Etlingera Pavieana (Pierre Ex Gagnep) R.M.Sm. Rhizome Extract for Dermatological Applications. Nat. Resour. Hum. Heal. 2025, 5, 634–647. [Google Scholar] [CrossRef] [PubMed]
- Vieira Melo, A.K.; da Nóbrega Alves, D.; Queiroga Gomes da Costa, P.C.; Pereira Lopes, S.; Pergentino de Sousa, D.; Queiroga Sarmento Guerra, F.; Vieira Sobral, M.; Gomes Moura, A.P.; Scotti, L.; Dias de Castro, R. Antifungal Activity, Mode of Action, and Cytotoxicity of 4-Chlorobenzyl p-Coumarate: A Promising New Molecule. Chem. Biodivers. 2024, 21, e202400330. [Google Scholar] [CrossRef]
- Falcão, É.P.; Alves, D.D.N.; Lopes, S.P.; Lazarini, J.G.; Rosalen, P.L.; de Sousa, D.P.; de Castro, R.D. Synthesis, Antimicrobial Activity, and Toxicological Evaluation of a p-Coumaric Acid Derivative as a Potential New Antibacterial Agent. J. Appl. Microbiol. 2025, 136, lxaf065. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2025 Update. Nucleic Acids Res. 2025, 53, D1516–D1525. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Dallakyan, S.; Olson, A.J. Small-Molecule Library Screening by Docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar] [CrossRef]
- Van der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS Documentation, GROMACS 2024.1. Available online: https://manual.gromacs.org/documentation/2024.1/manual-2024.1.pdf (accessed on 18 April 2025).
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved Side-chain Torsion Potentials for the Amber Ff99SB Protein Force Field. Proteins 2010, 78, 1950–1958. [Google Scholar] [CrossRef]
- Gandhi, R.G.; Sharanya, C.S.; Jayanandan, A.; Haridas, M.; Edwin Hillary, V.; Rajiv Gandhi, S.; Sridharan, G.; Sivasubramanian, R.; Silva Vasconcelos, A.B.; Montalvão, M.M.; et al. Multitargeted Molecular Docking and Dynamics Simulation Studies of Flavonoids and Volatile Components from the Peel of Citrus Sinensis L. (Osbeck) against Specific Tumor Protein Markers. J. Biomol. Struct. Dyn. 2024, 42, 3051–3080. [Google Scholar] [CrossRef]
- Lonsdale, R.; Harvey, J.N.; Mulholland, A.J. A Practical Guide to Modelling Enzyme-Catalysed Reactions. Chem. Soc. Rev. 2012, 41, 3025–3038. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindah, E. Gromacs: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; van der Spoel, D.; van Drunen, R. GROMACS: A Message-Passing Parallel Molecular Dynamics Implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- GROMACS. GROMACS 2023.1 Manual-Release Notes. Available online: https://manual.gromacs.org/documentation/current/release-notes/2023/2023.1.html (accessed on 18 April 2025).
- Stroet, M.; Caron, B.; Visscher, K.M.; Geerke, D.P.; Malde, A.K.; Mark, A.E. Automated Topology Builder Version 3.0: Prediction of Solvation Free Enthalpies in Water and Hexane. J. Chem. Theory Comput. 2018, 14, 5834–5845. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Freitag, F.; Gattin, Z.; Haberkern, H.; Jaun, B.; Siwko, M.; Vyas, R.; Vangunsteren, W.F.; Dolenc, J. Validation of the GROMOS 54A7 Force Field Regarding Mixed α/β-Peptide Molecules. Helv. Chim. Acta 2012, 95, 2562–2577. [Google Scholar] [CrossRef]
- Schmid, N.; Eichenberger, A.P.; Choutko, A.; Riniker, S.; Winger, M.; Mark, A.E.; Van Gunsteren, W.F. Definition and Testing of the GROMOS Force-Field Versions 54A7 and 54B7. Eur. Biophys. J. 2011, 40, 843–856. [Google Scholar] [CrossRef] [PubMed]
- Bondi, A. Van Der Waals Volumes and Radii. J. Phys. Chem. 1964, 68, 441–451. [Google Scholar] [CrossRef]
- Lemkul, J. From Proteins to Perturbed Hamiltonians: A Suite of Tutorials for the GROMACS-2018 Molecular Simulation Package [Article v1.0]. Living J. Comput. Mol. Sci. 2019, 1, 1–53. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Pires, D.E.; Blundell, T.L.; Ascher, D.B. PkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef] [PubMed]
- Freitas, P.R.; de Araújo, A.C.J.; Araújo, I.M.; de Almeida, R.S.; Borges, J.A.O.; Paulo, C.L.R.; Oliveira-Tintino, C.D.M.; Miranda, G.M.; Araújo-Neto, J.B.; Nascimento, I.J.S.; et al. Thiazine-Derived Compounds in Inhibiting Efflux Pump in Staphylococcus aureus K2068, MepA Gene Expression, and Membrane Permeability Alteration. Biomed. Pharmacother. 2024, 179, 117291. [Google Scholar] [CrossRef] [PubMed]
- De Almeida, R.S.; Freitas, P.R.; de Araujo, A.C.J.; Tintino, S.R.; Ribeiro-Filho, J.; Miranda, G.M.; Sigueira, G.M.; Gonçalves, S.A.; Carvalho, D.T.; de Souza, T.B.; et al. Liposomal Formulation with Thiazolic Compounds against Bacterial Efflux Pumps. Biomed. Pharmacother. 2024, 180, 117600. [Google Scholar] [CrossRef] [PubMed]
- Misba, L.; Zaidi, S.; Khan, A.U. A Comparison of Antibacterial and Antibiofilm Efficacy of Phenothiazinium Dyes between Gram Positive and Gram Negative Bacterial Biofilm. Photodiagnosis Photodyn. Ther. 2017, 18, 24–33. [Google Scholar] [CrossRef]
- Zhao, L.; Zhou, Y.; Yue, W.; Shen, Q.; Ke, J.; Ma, Y.; Zhang, L.; Bian, H. Natural Phenolics as Multitarget Antimicrobials for Food Preservation: Mechanisms of Action. Food Chem. X 2025, 31, 103056. [Google Scholar] [CrossRef]
- Yang, B.; Yao, H.; Li, D.; Liu, Z. The Phosphatidylglycerol Phosphate Synthase PgsA Utilizes a Trifurcated Amphipathic Cavity for Catalysis at the Membrane-Cytosol Interface. Curr. Res. Struct. Biol. 2021, 3, 312–323. [Google Scholar] [CrossRef]
- Luévano-Martínez, L.A.; Dunca, A.L. Origin and Diversification of the Cardiolipin Biosynthetic Pathway in the Eukarya Domain. Biochem. Soc. Trans. 2020, 48, 1035–1046. [Google Scholar] [CrossRef]
- Mamatha Jyothi, R.S.; Sripathi, M.P.; Thirupathi, P. Recent Advances in Base-Assisted Michael Addition Reactions. Curr. Org. Chem. 2022, 26, 1264–1293. [Google Scholar] [CrossRef]
- Han, S.; Chang, J.S.; Griffor, M. Structure of the Adenylation Domain of NAD+-Dependent DNA Ligase from Staphylococcus aureus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2009, 65, 1078–1082. [Google Scholar] [CrossRef]
- Moon, T.M.; D’Andréa, É.D.; Lee, C.W.; Soares, A.; Jakoncic, J.; Desbonnet, C.; Garcia-Solache, M.; Rice, L.B.; Page, R.; Peti, W. The Structures of Penicillin-Binding Protein 4 (PBP4) and PBP5 from Enterococci Provide Structural Insights into -Lactam Resistance. J. Biol. Chem. 2018, 293, 18574–18585. [Google Scholar] [CrossRef]
- Schindler, B.D.; Patel, D.; Seo, S.M.; Kaatz, G.W. Mutagenesis and Modeling to Predict Structural and Functional Characteristics of the Staphylococcus aureus MepA Multidrug Efflux Pump. J. Bacteriol. 2013, 195, 523–533. [Google Scholar] [CrossRef]
- Kolár, M.H.; Hobza, P. Computer Modeling of Halogen Bonds and Otherσ-Hole Interactions. Chem. Rev. 2016, 116, 5155–5187. [Google Scholar] [CrossRef] [PubMed]
- Brawley, D.N.; Sauer, D.B.; Li, J.; Zheng, X.; Koide, A.; Jedhe, G.S.; Suwatthee, T.; Song, J.; Liu, Z.; Arora, P.S.; et al. Structural Basis for Inhibition of the Drug Efflux Pump NorA from Staphylococcus aureus. Nat. Chem. Biol. 2022, 18, 706–712. [Google Scholar] [CrossRef]
- Bendjeddou, A.; Abbaz, T.; Gouasmia, A.K.; Villemin, D. Molecular Structure, HOMO-LUMO, MEP and Fukui Function Analysis of Some TTF-Donor Substituted Molecules Using DFT (B3LYP) Calculations. Int. Res. J. Pure Appl. Chem. 2016, 12, 1–9. [Google Scholar] [CrossRef]
- Aghajani, J.; Farnia, P.; Farnia, P.; Ghanavi, J.; Velayati, A.A. Molecular Dynamic Simulations and Molecular Docking as a Potential Way for Designed New Inhibitor Drug without Resistance. Tanaffos 2022, 21, 1–14. [Google Scholar]
- Fukutani, T.; Miyazawa, K.; Iwata, S.; Satoh, H. G-RMSD: Root Mean Square Deviation Based Method for Three-Dimensional Molecular Similarity Determination. Bull. Chem. Soc. Jpn. 2021, 94, 655–665. [Google Scholar] [CrossRef]
- Lobanov, M.Y.; Bogatyreva, N.S.; Galzitskaya, O.V. Radius of Gyration as an Indicator of Protein Structure Compactness. Mol. Biol. 2008, 42, 623–628. [Google Scholar] [CrossRef]
- Shukla, R.; Tripathi, T. Molecular Dynamics Simulation of Protein and Protein–Ligand Complexes. In Computer-Aided Drug Design; Singh, D.B., Ed.; Springer: Singapore, 2020. [Google Scholar]
- Mehta, N.A.; Levin, D.A. Electrospray Molecular Dynamics Simulations Using an Octree-Based Coulomb Interaction Method. Phys. Rev. E 2019, 99, 033302. [Google Scholar] [CrossRef]
- Robalo, J.R.; Streacker, L.M.; Mendes de Oliveira, D.; Imhof, P.; Ben-Amotz, D.; Verde, A.V. Hydrophobic but Water-Friendly: Favorable Water-Perfluoromethyl Interactions Promote Hydration Shell Defects. J. Am. Chem. Soc. 2019, 141, 15856. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. M100 Performance Standards for Antimicrobial Susceptibility Testing, 31st ed.; CLSI: Wayne, PA, USA, 2021. [Google Scholar]
- Tintino, S.R.; Oliveira-Tintino, C.D.M.; Campina, F.F.; Costa, M.S.; Cruz, R.P.; Pereira, R.L.S.; Andrade, J.C.; Sousa, E.O.; Siqueira-Junior, J.P.; Coutinho, H.D.M.; et al. Cholesterol and Ergosterol Affect the Activity of Staphylococcus aureus Antibiotic Efflux Pumps. Microb. Pathog. 2017, 104, 133–136. [Google Scholar] [CrossRef] [PubMed]
- Singha, K.; Banerjee, A.; Jana, A.; Bandyopadhyay, P.; Maiti, S.; Pati, B.R.; Mohapatra, P.K. Das Molecular Exposition of Broad-Spectrum Antibacterial Efficacy by p-Coumaric Acid from an Edible Mushroom Termitomyces Heimii: In Vitro and in Silico Approach. Syst. Microbiol. Biomanufacturing 2023, 3, 750–764. [Google Scholar] [CrossRef]
- Van Eijk, E.; Wittekoek, B.; Kuijper, E.J.; Smits, W.K. DNA Replication Proteins as Potential Targets for Antimicrobials in Drug-Resistant Bacterial Pathogens. J. Antimicrob. Chemother. 2017, 72, 1275–1284. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 2012, 64, 4–17. [Google Scholar] [CrossRef]
- Pires, D.E.; Blundell, T.L.; Ascher, D.B. PkCSM: Predicting Small-Molecule Pharmacokinetic Properties Using Graph-Based Signatures (Theory-How to Enterpret PkCSM Result). Available online: https://biosig.lab.uq.edu.au/pkcsm/theory (accessed on 20 August 2024).
- Mohamed, A.A.; El-Kadi, A. P-Glycoprotein Effects on Drugs Pharmacokinetics and Drug-Drug- Interactions and Their Clinical Implications. Libyan J. Pharm. Clin. Pharmacol. 2012, 1, 1–13. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, H.; Fan, Y.; Chen, X.; Yang, Y.; Zhu, L.; Zhao, J.; Chen, Y.; Zhang, Y. In Silico Prediction of Human Intravenous Pharmacokinetic Parameters with Improved Accuracy. J. Chem. Inf. Model. 2019, 59, 3968–3980. [Google Scholar] [CrossRef] [PubMed]
- Zaretzki, J.; Matlock, M.; Swamidass, S.J. XenoSite: Accurately Predicting Cyp-Mediated Sites of Metabolism with Neural Networks. J. Chem. Inf. Model. 2013, 53, 3373–3383. [Google Scholar] [CrossRef] [PubMed]
- Grime, K.H.; Barton, P.; McGinnity, D.F. Application of in Silico, in Vitro and Preclinical Pharmacokinetic Data for the Effective and Efficient Prediction of Human Pharmacokinetics. Mol. Pharm. 2013, 10, 1191–1206. [Google Scholar] [CrossRef]
- Silveira, Z.d.S.; Macêdo, N.S.; Dantas, D.d.M.; Barbosa, C.R.d.S.; Oliveira-Tintino, C.D.d.M.; Tintino, S.R.; Alencar, G.G.; Silva, E.; Nunes, M.; Machado, M. Evaluation of the Antibacterial and Inhibitory Activity of the NorA and TetK Efflux Pumps of Staphylococcus aureus by p -Coumaric Acid. Microb. Pathog. 2025, 200, 107318. [Google Scholar] [CrossRef] [PubMed]
- De Sousa, J.N.; de Andrade Ferreira Barreto, J.; de Alcântara Oliveira, F.A.; Haydée Lima Ferreira, J.; Dias Rufino Arcanjo, D.; Emidio Sampaio Nogueira, C.; Machado Marinho, M.; dos Santos, H.S.; Maria Lins Rolim, H.; de Siqueira-Júnior, J.P.; et al. ADMET Study and Inhibition of Staphylococcus aureus Efflux Pumps by a Synthetic P-Aminochalcone. Results Chem. 2024, 7, 101449. [Google Scholar] [CrossRef]
- Pal, S.; Misra, A.; Banerjee, S.; Dam, B. Adaptation of Ethidium Bromide Fluorescence Assay to Monitor Activity of Efflux Pumps in Bacterial Pure Cultures or Mixed Population from Environmental Samples. J. King Saud Univ.-Sci. 2020, 32, 939–945. [Google Scholar] [CrossRef]
- Singh, K.; Coopoosamy, R.M.; Gumede, N.J.; Sabiu, S. Computational Insights and In Vitro Validation of Antibacterial Potential of Shikimate Pathway-Derived Phenolic Acids as NorA Efflux Pump Inhibitors. Molecules 2022, 27, 2601. [Google Scholar] [CrossRef] [PubMed]
- García-Alegría, A.M.; Anduro-Corona, I.; Pérez-Martínez, C.J.; Corella-Madueño, M.A.G.; Rascón-Durán, M.L.; Astiazaran-Garcia, H. Quantification of DNA through the Nanodrop Spectrophotometer: Methodological Validation Using Standard Reference Material and Sprague Dawley Rat and Human DNA. Int. J. Anal. Chem. 2020, 2020, 8896738. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Sambrook, J. Analysis of DNA by Agarose Gel Electrophoresis. Cold Spring Harb. Protoc. 2019, 2019, 6–15. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).