Activity and Silencing of Transposable Elements in C. elegans
Abstract
1. Introduction
2. Transposable Elements in the C. elegans Genome
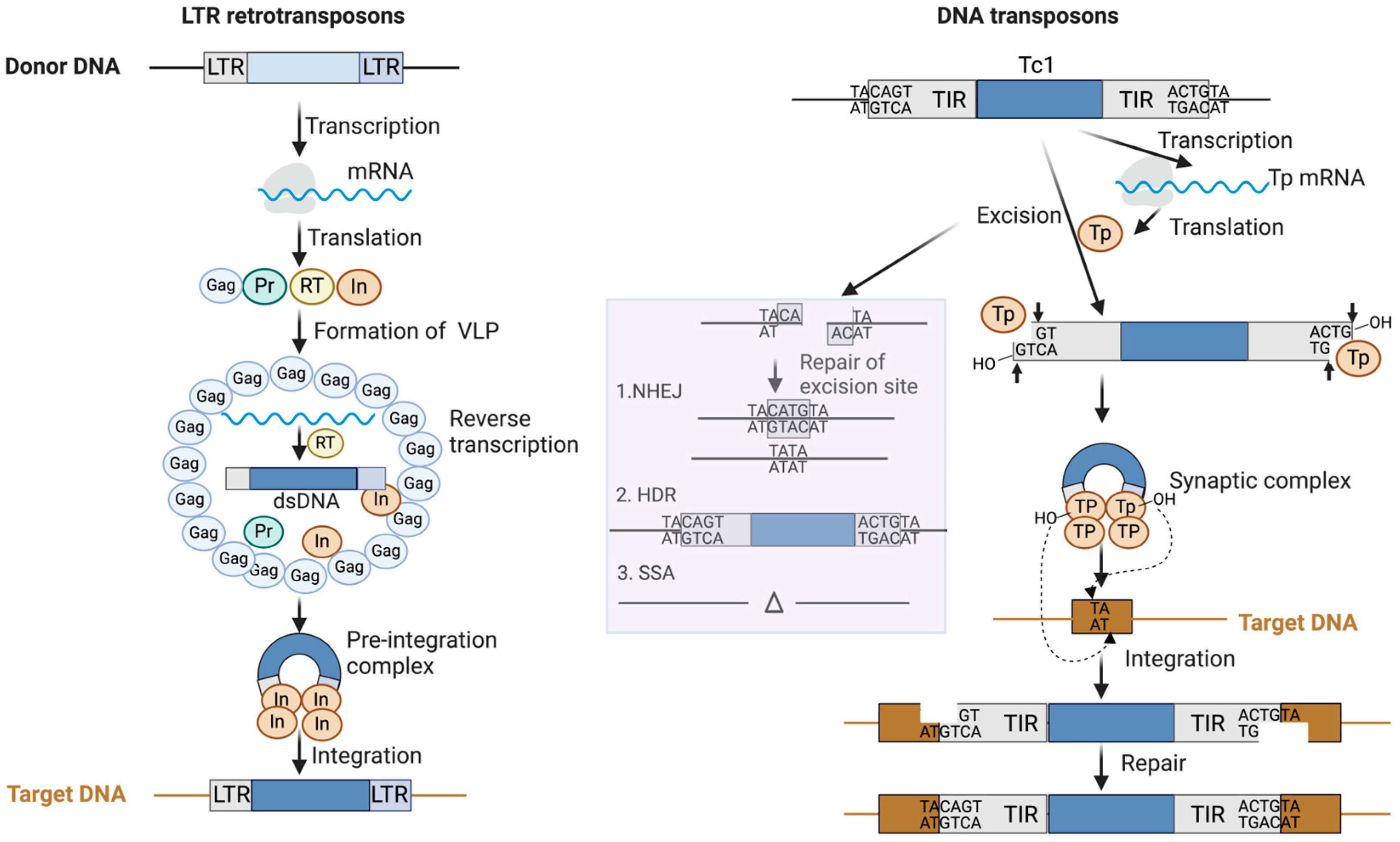
3. Mechanisms of Transposition in C. elegans
4. Silencing of Transposable Elements
5. Other Mechanisms of Transposon Regulation: Adaptation and Domestication
6. Conclusions and Future Directions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- McClintock, B. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA 1950, 36, 344–355. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Laricchia, K.M.; Zdraljevic, S.; Cook, D.E.; Andersen, E.C. Natural Variation in the Distribution and Abundance of Transposable Elements Across the Caenorhabditis elegans Species. Mol. Biol. Evol. 2017, 34, 2187–2202. [Google Scholar] [CrossRef] [PubMed]
- Capy, P.; Gasperi, G.; Biémont, C.; Bazin, C. Stress and transposable elements: Co-evolution or useful parasites? Heredity 2000, 85, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Saksouk, N.; Simboeck, E.; Déjardin, J. Constitutive heterochromatin formation and transcription in mammals. Epigenetics Chromatin 2015, 8, 3. [Google Scholar] [CrossRef]
- Ho, J.W.K.; Jung, Y.L.; Liu, T.; Alver, B.H.; Lee, S.; Ikegami, K.; Sohn, K.-A.; Minoda, A.; Tolstorukov, M.Y.; Appert, A.; et al. Comparative analysis of metazoan chromatin organization. Nature 2015, 512, 449–452. [Google Scholar] [CrossRef] [PubMed]
- McMurchy, A.N.; Stempor, P.; Gaarenstroom, T.; Wysolmerski, B.; Dong, Y.; Aussianikava, D.; Appert, A.; Huang, N.; Kolasinska-Zwierz, P.; Sapetschnig, A.; et al. A team of heterochromatin factors collaborates with small RNA pathways to combat repetitive elements and germline stress. eLife 2017, 6, e21666. [Google Scholar] [CrossRef]
- Ahringer, J.; Gasser, S.M. Repressive Chromatin in Caenorhabditis elegans: Establishment, Composition, and Function. Genetics 2018, 208, 491–511. [Google Scholar] [CrossRef] [PubMed]
- Fedoroff, N.V. Transposable Elements, Epigenetics, and Genome Evolution. Science 2012, 338, 758–767. [Google Scholar] [CrossRef]
- Sánchez-Gracia, A.; Maside, X.; Charlesworth, B. High rate of horizontal transfer of transposable elements in Drosophila. Trends Genet. 2005, 21, 200–2003. [Google Scholar] [CrossRef]
- Zhang, H.-H.; Peccoud, J.; Xu, M.-R.; Zhang, X.-G.; Gilbert, C. Horizontal transfer and evolution of transposable elements in vertebrates. Nat. Commun. 2019, 11, 1362. [Google Scholar] [CrossRef] [PubMed]
- Brenner, S. The Genetics of Caenorhabditis elegans. Genetics 1974, 77, 71–94. [Google Scholar] [CrossRef] [PubMed]
- The, C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 1998, 282, 2012–2018. [Google Scholar]
- Eide, D.J.; Anderson, P. Novel Insertion Mutation in Caenorhabditis elegans. Mol. Cell. Biol. 1985, 5, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Eide, D.; Anderson, P. Transposition of Tc1 in the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 1985, 82, 1756–1760. [Google Scholar] [CrossRef] [PubMed]
- Emmons, S.W.; Yesner, L.; Ruan, K.-S.; Katzenberg, D. Evidence for a transposon in Caenorhabditis elegans. Cell 1983, 32, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Hunter, S.E.; Finnegan, E.F.; Zisoulis, D.G.; Lovci, M.T.; Melnik-Martinez, K.V.; Yeo, G.W.; Pasquinelli, A.E. Functional Genomic Analysis of the let-7 Regulatory Network in Caenorhabditis elegans. PLoS Genet. 2013, 9, e1003353. [Google Scholar] [CrossRef] [PubMed]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef]
- Arata, Y.; Jurica, P.; Parrish, N.; Sako, Y. Comprehensive identification of potentially functional genes for transposon mobility in the C. elegans genome. bioRxiv, 2023; 2023.08.08.552548. [Google Scholar] [CrossRef]
- Oosumi, T.; Garlick, B.; Belknap, W.R. Identification of putative nonautonomous transposable elements associated with several transposon families in Caenorhabditis elegans. J. Mol. Evol. 1996, 43, 11–18. [Google Scholar] [CrossRef]
- Moerman, D.G.; Waterston, R.H. Spontaneous unstable unc-22 IV mutations in C. elegans var. Bergerac. Genetics 1984, 108, 859–877. [Google Scholar] [CrossRef]
- Collins, J.; Saari, B.; Anderson, P. Activation of a transposable element in the germ line but not the soma of Caenorhabditis elegans. Nature 1987, 328, 726–728. [Google Scholar] [CrossRef] [PubMed]
- Ketting, R.F.; Haverkamp, T.H.; van Luenen, H.G.; Plasterk, R.H. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 1999, 99, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Tabara, H.; Sarkissian, M.; Kelly, W.G.; Fleenor, J.; Grishok, A.; Timmons, L.; Fire, A.; Mello, C.C. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 1999, 99, 123–132. [Google Scholar] [CrossRef]
- Egilmez, N.K.; Reis, R.J.S. Age-dependent somatic excision of transposable element Tc1 in Caenorhabditis elegans. Mutat. Res. 1994, 316, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Daigle, A.T.; Deiss, T.C.; Melde, R.H.; Bergthorsson, U.; Katju, V. Bergerac strains of Caenorhabditis elegans revisited: Expansion of Tc1 elements imposes a significant genomic and fitness cost. G3 2022, 12, jkac214. [Google Scholar] [CrossRef]
- Collins, J.; Forbes, E.; Anderson, P. The Tc3 family of transposable genetic elements in Caenorhabditis elegans. Genetics 1989, 121, 47–55. [Google Scholar] [CrossRef]
- Mori, I.; Moerman, D.G.; Waterston, R.H. Analysis of a mutator activity necessary for germline transposition and excision of Tc1 transposable elements in Caenorhabditis elegans. Genetics 1988, 120, 397–407. [Google Scholar] [CrossRef]
- Rezsohazy, R.; van Luenen, H.G.A.M.; Durbin, R.M.; Plasterk, R.H.A. Tc7, a Tc1-hitch hiking transposon in Caenorhabditis elegans. Nucleic Acids Res. 1997, 25, 4048–4054. [Google Scholar] [CrossRef]
- Yuan, J.Y.; Finney, M.; Tsung, N.; Horvitz, H.R. Tc4, a Caenorhabditis elegans transposable element with an unusual fold-back structure. Proc. Natl. Acad. Sci. USA 1991, 88, 3334–3338. [Google Scholar] [CrossRef]
- Li, W.; Shaw, J.E. A variant Tc4 transposable element in the nematode C. elegans could encode a novel protein. Nucleic Acids Res. 1993, 21, 59–67. [Google Scholar] [CrossRef]
- Collins, J.J.; Anderson, P. The Tc5 family of transposable elements in Caenorhabditis elegans. Genetics 1994, 137, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Levitt, A.; Emmons, S.W. The Tc2 transposon in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 1989, 86, 3232–3236. [Google Scholar] [CrossRef]
- Dupeyron, M.; Baril, T.; Bass, C.; Hayward, A. Phylogenetic analysis of the Tc1/mariner superfamily reveals the unexplored diversity of pogo-like elements. Mob. DNA 2019, 11, 21. [Google Scholar] [CrossRef] [PubMed]
- Plasterk, R.H.A.; Izsvák, Z.; Ivics, Z. Resident aliens: The Tc1/mariner superfamily of transposable elements. Trends Genet. 1999, 15, 326–332. [Google Scholar] [CrossRef] [PubMed]
- Bannert, N.; Kurth, R. Retroelements and the human genome: New perspectives on an old relation. Proc. Natl. Acad. Sci. USA 2004, 101 (Suppl. S2), 14572–14579. [Google Scholar] [CrossRef] [PubMed]
- Britten, R.J. Active gypsy/Ty3 retrotransposons or retroviruses in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 1995, 92, 599–601. [Google Scholar] [CrossRef] [PubMed]
- Frame, I.G.; Cutfield, J.F.; Poulter, R.T. New BEL-like LTR-retrotransposons in Fugu rubripes, Caenorhabditis elegans, and Drosophila melanogaster. Gene 2001, 263, 219–230. [Google Scholar] [CrossRef]
- Ganko, E.W.; Bhattacharjee, V.; Schliekelman, P.; McDonald, J.F. Evidence for the contribution of LTR retrotransposons to C. elegans gene evolution. Mol. Biol. Evol. 2003, 20, 1925–1931. [Google Scholar] [CrossRef]
- Ganko, E.W.; Fielman, K.T.; McDonald, J.F. Evolutionary history of Cer elements and their impact on the C. elegans genome. Genome Res. 2001, 11, 2066–2074. [Google Scholar] [CrossRef]
- Youngman, S.; van Luenen, H.G.; Plasterk, R.H. Rte-1, a retrotransposon-like element in Caenorhabditis elegans. FEBS Lett. 1996, 380, 1–7. [Google Scholar] [CrossRef]
- Malik, H.S.; Eickbush, T.H. NeSL-1, an Ancient Lineage of Site-Specific Non-LTR Retrotransposons From Caenorhabditis elegans. Genetics 2000, 154, 193–203. [Google Scholar] [CrossRef]
- Fischer, S.E.J.; Ruvkun, G. Caenorhabditis elegans ADAR editing and the ERI-6/7/MOV10 RNAi pathway silence endogenous viral elements and LTR retrotransposons. Proc. Natl. Acad. Sci. USA 2020, 117, 5987–5996. [Google Scholar] [CrossRef]
- Sun, B.; Kim, H.; Mello, C.C.; Priess, J.R. The CERV protein of Cer1, a C. elegans LTR retrotransposon, is required for nuclear export of viral genomic RNA and can form giant nuclear rods. PLoS Genet. 2023, 19, e1010804. [Google Scholar] [CrossRef]
- Palopoli, M.F.; Rockman, M.V.; TinMaung, A.; Ramsay, C.; Curwen, S.; Aduna, A.; Laurita, J.; Kruglyak, L. Molecular basis of the copulatory plug polymorphism in Caenorhabditis elegans. Nature 2008, 454, 1019–1022. [Google Scholar] [CrossRef]
- Dennis, S.; Sheth, U.; Feldman, J.L.; English, K.A.; Priess, J.R. C. elegans Germ Cells Show Temperature and Age-Dependent Expression of Cer1, a Gypsy/Ty3-Related Retrotransposon. PLoS Pathog. 2012, 8, e1002591. [Google Scholar] [CrossRef]
- van Luenen, H.; Colloms, S.; Plasterk, R. The mechanism of transposition of Tc3 in C. elegans. Cell 1994, 79, 293–301. [Google Scholar] [CrossRef]
- Vos, J.C.; De Baere, I.; Plasterk, R.H. Transposase is the only nematode protein required for in vitro transposition of Tc1. Genes Dev. 1996, 10, 755–761. [Google Scholar] [CrossRef]
- Schukkink, R.F.; Plasterk, R.H. TcA, the putative transposase of the C. elegans Tc1 transposon, has an N-terminal DNA binding domain. Nucleic Acids Res. 1990, 18, 895–900. [Google Scholar] [CrossRef]
- Vos, J.C.; Plasterk, R.H. Tc1 transposase of Caenorhabditis elegans is an endonuclease with a bipartite DNA binding domain. EMBO J. 1994, 13, 6125–6132. [Google Scholar] [CrossRef]
- Vos, J.C.; van Luenen, H.G.; Plasterk, R.H. Characterization of the Caenorhabditis elegans Tc1 transposase in vivo and in vitro. Genes Dev. 1993, 7, 1244–1253. [Google Scholar] [CrossRef]
- Fischer, S.E.J.; Wienholds, E.; Plasterk, R.H.A. Continuous Exchange of Sequence Information Between Dispersed Tc1 Transposons in the Caenorhabditis elegans Genome. Genetics 2003, 164, 127–134. [Google Scholar] [CrossRef]
- Plasterk, R.H. The origin of footprints of the Tc1 transposon of Caenorhabditis elegans. EMBO J. 1991, 10, 1919–1925. [Google Scholar] [CrossRef] [PubMed]
- Zwaal, R.R.; Broeks, A.; van Meurs, J.; Groenen, J.T.; Plasterk, R.H. Target-selected gene inactivation in Caenorhabditis elegans by using a frozen transposon insertion mutant bank. Proc. Natl. Acad. Sci. USA 1993, 90, 7431–7435. [Google Scholar] [CrossRef]
- Ketting, R.F.; Fischer, S.E.; Plasterk, R.H. Target choice determinants of the Tc1 transposon of Caenorhabditis elegans. Nucleic Acids Res. 1997, 25, 4041–4047. [Google Scholar] [CrossRef] [PubMed][Green Version]
- van Luenen, H.G.; Plasterk, R.H. Target site choice of the related transposable elements Tc1 and Tc3 of Caenorhabditis elegans. Nucleic Acids Res. 1994, 22, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Duret, L.; Marais, G.; Biémont, C. Transposons but Not Retrotransposons Are Located Preferentially in Regions of High Recombination Rate in Caenorhabditis elegans. Genetics 2000, 156, 1661–1669. [Google Scholar] [CrossRef]
- Das, P.P.; Bagijn, M.P.; Goldstein, L.D.; Woolford, J.R.; Lehrbach, N.J.; Sapetschnig, A.; Buhecha, H.R.; Gilchrist, M.J.; Howe, K.L.; Stark, R.; et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol. Cell 2008, 31, 79–90. [Google Scholar] [CrossRef]
- Ketting, F.R.; Cochella, L. Concepts and functions of small RNA pathways in C. elegans. Curr. Top. Dev. Biol. 2020, 144, 45–89. [Google Scholar] [CrossRef]
- Shen, E.-Z.; Chen, H.; Ozturk, A.R.; Tu, S.; Shirayama, M.; Tang, W.; Ding, Y.-H.; Dai, S.-Y.; Weng, Z.; Mello, C.C. Identification of piRNA Binding Sites Reveals the Argonaute Regulatory Landscape of the C. elegans Germline. Cell 2018, 172, 937–951.e18. [Google Scholar] [CrossRef]
- McEnany, J.; Meir, Y.; Wingreen, N.S. piRNAs of Caenorhabditis elegans broadly silence nonself sequences through functionally random targeting. Nucleic Acids Res. 2022, 50, 1416–1429. [Google Scholar] [CrossRef]
- Phillips, C.M.; Montgomery, T.A.; Breen, P.C.; Ruvkun, G. MUT-16 promotes formation of perinuclear Mutator foci required for RNA silencing in the C. elegans germline. Gene Dev. 2012, 26, 1433–1444. [Google Scholar] [CrossRef] [PubMed]
- Vastenhouw, N.L.; Fischer, S.E.; Robert, V.J.; Thijssen, K.L.; Fraser, A.G.; Kamath, R.S.; Ahringer, J.; Plasterk, R.H. A Genome-Wide Screen Identifies 27 Genes Involved in Transposon Silencing in C. elegans. Curr. Biol. 2003, 13, 1311–1316. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.; Yan, J.; Pagano, D.J.; Dodson, A.E.; Fei, Y.; Gorham, J.; Seidman, J.G.; Wickens, M.; Kennedy, S. poly(UG)-tailed RNAs in genome protection and epigenetic inheritance. Nature 2020, 582, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Pak, J.; Fire, A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 2007, 315, 241–244. [Google Scholar] [CrossRef] [PubMed]
- Seroussi, U.; Lugowski, A.; Wadi, L.; Lao, R.X.; Willis, A.R.; Zhao, W.; Sundby, A.E.; Charlesworth, A.G.; Reinke, A.W.; Claycomb, J.M. A comprehensive survey of C. elegans argonaute proteins reveals organism-wide gene regulatory networks and functions. eLife 2023, 12, e83853. [Google Scholar] [CrossRef] [PubMed]
- Buckley, B.A.; Burkhart, K.B.; Gu, S.G.; Spracklin, G.; Kershner, A.; Fritz, H.; Kimble, J.; Fire, A.; Kennedy, S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 2012, 489, 447–451. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.E.; Pan, Q.; Breen, P.C.; Qi, Y.; Shi, Z.; Zhang, C.; Ruvkun, G. Multiple small RNA pathways regulate the silencing of repeated and foreign genes in C. elegans. Gene Dev. 2013, 27, 2678–2695. [Google Scholar] [CrossRef]
- Burton, N.O.; Burkhart, K.B.; Kennedy, S. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2011, 108, 19683–19688. [Google Scholar] [CrossRef]
- Mao, H.; Zhu, C.; Zong, D.; Weng, C.; Yang, X.; Huang, H.; Liu, D.; Feng, X.; Guang, S. The Nrde Pathway Mediates Small-RNA-Directed Histone H3 Lysine 27 Trimethylation in Caenorhabditis elegans. Curr. Biol. 2015, 25, 2398–2403. [Google Scholar] [CrossRef]
- Schwartz-Orbach, L.; Zhang, C.; Sidoli, S.; Amin, R.; Kaur, D.; Zhebrun, A.; Ni, J.; Gu, S.G. Caenorhabditis elegans nuclear RNAi factor SET-32 deposits the transgenerational histone modification, H3K23me3. eLife 2020, 9, e54309. [Google Scholar] [CrossRef]
- Ding, Y.-H.; Ochoa, H.J.; Ishidate, T.; Shirayama, M.; Mello, C.C. The nuclear Argonaute HRDE-1 directs target gene re-localization and shuttles to nuage to promote small RNA-mediated inherited silencing. Cell Rep. 2023, 42, 112408. [Google Scholar] [CrossRef] [PubMed]
- Fields, B.D.; Kennedy, S. Chromatin Compaction by Small RNAs and the Nuclear RNAi Machinery in C. elegans. Sci. Rep. 2019, 9, 9030. [Google Scholar] [CrossRef] [PubMed]
- Garrigues, J.M.; Sidoli, S.; Garcia, B.A.; Strome, S. Defining heterochromatin in C. elegans through genome-wide analysis of the heterochromatin protein 1 homolog HPL-2. Genome Res. 2015, 25, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Hisanaga, T.; Romani, F.; Wu, S.; Kowar, T.; Wu, Y.; Lintermann, R.; Fridrich, A.; Cho, C.H.; Chaumier, T.; Jamge, B.; et al. The Polycomb repressive complex 2 deposits H3K27me3 and represses transposable elements in a broad range of eukaryotes. Curr. Biol. 2023, 33, 4367–4380.e9. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Solberg, T.; Maurer-Alcalá, X.X.; Swart, E.C.; Gao, F.; Nowacki, M. A small RNA-guided PRC2 complex eliminates DNA as an extreme form of transposon silencing. Cell Rep. 2022, 40, 111263. [Google Scholar] [CrossRef] [PubMed]
- Snel, B.; Heuvel, S.v.D.; Seidl, M.F. Caenorhabditis elegans MES-3 is a highly divergent ortholog of the canonical PRC2 component SUZ12. iScience 2022, 25, 104633. [Google Scholar] [CrossRef]
- Mazzetto, M.; Gonzalez, L.E.; Sanchez, N.; Reinke, V. Characterization of the distribution and dynamics of chromatin states in the C. elegans germ line reveals substantial H3K4me3 remodeling during oogenesis. Genome Res. 2023, 34, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.V.; Vernaz, G.; Putman, A.L.; Miska, E.A. Taming transposable elements in vertebrates: From epigenetic silencing to domestication. Trends Genet. 2022, 38, 529–553. [Google Scholar] [CrossRef] [PubMed]
- Ponger, L.; Li, W.-H. Evolutionary Diversification of DNA Methyltransferases in Eukaryotic Genomes. Mol. Biol. Evol. 2005, 22, 1119–1128. [Google Scholar] [CrossRef]
- Ashe, A.; Sapetschnig, A.; Weick, E.-M.; Mitchell, J.; Bagijn, M.P.; Cording, A.C.; Doebley, A.-L.; Goldstein, L.D.; Lehrbach, N.J.; Le Pen, J.; et al. piRNAs Can Trigger a Multigenerational Epigenetic Memory in the Germline of C. elegans. Cell 2012, 150, 88–99. [Google Scholar] [CrossRef]
- Quarato, P.; Singh, M.; Bourdon, L.; Cecere, G. Inheritance and maintenance of small RNA-mediated epigenetic effects. Bioessays 2022, 44, e2100284. [Google Scholar] [CrossRef]
- Cecere, G. Small RNAs in epigenetic inheritance: From mechanisms to trait transmission. FEBS Lett. 2021, 595, 2953–2977. [Google Scholar] [CrossRef]
- Özdemir, I.; Steiner, F.A. Transmission of chromatin states across generations in C. elegans. Semin. Cell Dev. Biol. 2022, 127, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.E.J.; Montgomery, T.A.; Zhang, C.; Fahlgren, N.; Breen, P.C.; Hwang, A.; Sullivan, C.M.; Carrington, J.C.; Ruvkun, G. The ERI-6/7 Helicase Acts at the First Stage of an siRNA Amplification Pathway That Targets Recent Gene Duplications. PLoS Genet. 2011, 7, e1002369. [Google Scholar] [CrossRef]
- Newman, M.A.; Ji, F.; Fischer, S.E.; Anselmo, A.; Sadreyev, R.I.; Ruvkun, G. The surveillance of pre-mRNA splicing is an early step in C. elegans RNAi of endogenous genes. Gene Dev. 2018, 32, 670–681. [Google Scholar] [CrossRef]
- Sijen, T.; Plasterk, R.H.A. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature 2003, 426, 310–314. [Google Scholar] [CrossRef] [PubMed]
- Makeyeva, Y.V.; Shirayama, M.; Mello, C.C. Cues from mRNA splicing prevent default Argonaute silencing in C. elegans. Dev. Cell 2021, 56, 2636–2648.e4. [Google Scholar] [CrossRef]
- Akay, A.; Di Domenico, T.; Suen, K.M.; Nabih, A.; Parada, G.E.; Larance, M.; Medhi, R.; Berkyurek, A.C.; Zhang, X.; Wedeles, C.J.; et al. The Helicase Aquarius/EMB-4 Is Required to Overcome Intronic Barriers to Allow Nuclear RNAi Pathways to Heritably Silence Transcription. Dev. Cell 2017, 42, 241–255.e6. [Google Scholar] [CrossRef]
- Carr, B.; Anderson, P. Imprecise Excision of the Caenorhabditis elegans Transposon Tel Creates Functional 5′ Splice Sites. Mol. Cell. Biol. 1994, 14, 3426–3433. [Google Scholar] [CrossRef] [PubMed]
- Rushforth, A.M.; Anderson, P. Splicing removes the Caenorhabditis elegans transposon Tc1 from most mutant pre-mRNAs. Mol. Cell. Biol. 1996, 16, 422–429. [Google Scholar] [CrossRef]
- Kurhanewicz, N.A.; Dinwiddie, D.; Bush, Z.D.; Libuda, D.E. Elevated Temperatures Cause Transposon-Associated DNA Damage in C. elegans Spermatocytes. Curr. Biol. 2020, 30, 5007–5017.e4. [Google Scholar] [CrossRef] [PubMed]
- Rogers, A.K.; Phillips, C.M. RNAi pathways repress reprogramming of C. elegans germ cells during heat stress. Nucleic Acids Res. 2020, 48, 4256–4273. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.S.; Kaletsky, R.; Lesnik, C.; Cota, V.; Blackman, E.; Parsons, L.R.; Gitai, Z.; Murphy, C.T. The role of the Cer1 transposon in horizontal transfer of transgenerational memory. Cell 2021, 184, 4697–4712.e18. [Google Scholar] [CrossRef] [PubMed]
- Carelli, F.N.; Cerrato, C.; Dong, Y.; Appert, A.; Dernburg, A.; Ahringer, J. Widespread transposon co-option in the Caenorhabditis germline regulatory network. Sci. Adv. 2022, 8, eabo4082. [Google Scholar] [CrossRef]
- Garrigues, J.M.; Tsu, B.V.; Daugherty, M.D.; Pasquinelli, A.E. Diversification of the Caenorhabditis heat shock response by Helitron transposable elements. eLife 2019, 8, e51139. [Google Scholar] [CrossRef]

| Element | Class | Order | Superfamily | Family | Copy Number | Length (bp) | Catalytic Motif | IR/TIR Length (bp) | Target Site (Duplicaton) |
|---|---|---|---|---|---|---|---|---|---|
| Cer1 | Class I | LTR | Gypsy | Gypsy | 1 | 8865 | DDE | 492 | |
| Tc1 | Class II | TIR | Tc1/mariner | Tc1 | 32 | 1611 | DD34E | 54 | TA (TA-TA) |
| Tc2 | Class II | TIR | Tc1/mariner-Tc2/pogo group | Pogo | 4 | 2074 | DD35D | 24 | TA (TA-TA) |
| Tc3 | Class II | TIR | Tc1/mariner | Tc1 | 22 | 2335 | DD34E | 462 | TA (TA-TA) |
| Tc4/Tc4v | Class II | TIR | Tc1/mariner- Tc4 group | Tc4 | 10 | 1605/3483 | DD37D | 774 | CTNAG (TNA-TNA) |
| Tc5 | Class II | TIR | Tc1/mariner- Tc4 group | Tc4 | 4 | 3171 | DD37D | 491 | CTNAG (TNA-TNA) |
| Tc7 (Tc1 MITE) | Class II | TIR | Tc1/mariner | Tc1 | 11 | 921 | n/a | 345 | TA (TA-TA) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fischer, S.E.J. Activity and Silencing of Transposable Elements in C. elegans. DNA 2024, 4, 129-140. https://doi.org/10.3390/dna4020007
Fischer SEJ. Activity and Silencing of Transposable Elements in C. elegans. DNA. 2024; 4(2):129-140. https://doi.org/10.3390/dna4020007
Chicago/Turabian StyleFischer, Sylvia E. J. 2024. "Activity and Silencing of Transposable Elements in C. elegans" DNA 4, no. 2: 129-140. https://doi.org/10.3390/dna4020007
APA StyleFischer, S. E. J. (2024). Activity and Silencing of Transposable Elements in C. elegans. DNA, 4(2), 129-140. https://doi.org/10.3390/dna4020007







