Transposon and Transgene Tribulations in Mosquitoes: A Perspective of piRNA Proportions
Abstract
1. Why Think of Fruit Flies When Mosquitoes Are Humanity’s Bigger Problem?
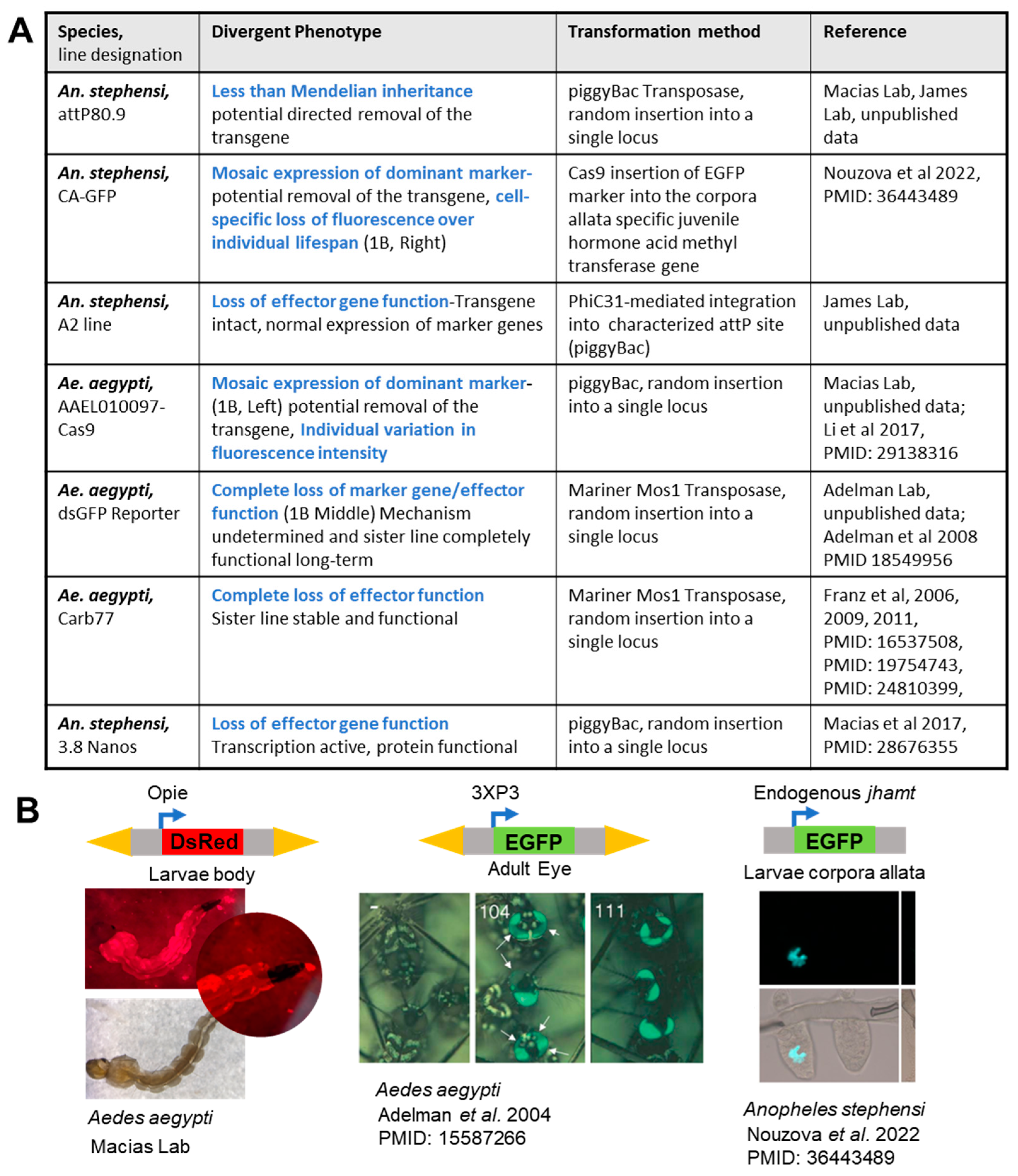
2. The Challenges in Stable Transgene Expression in Mosquitoes
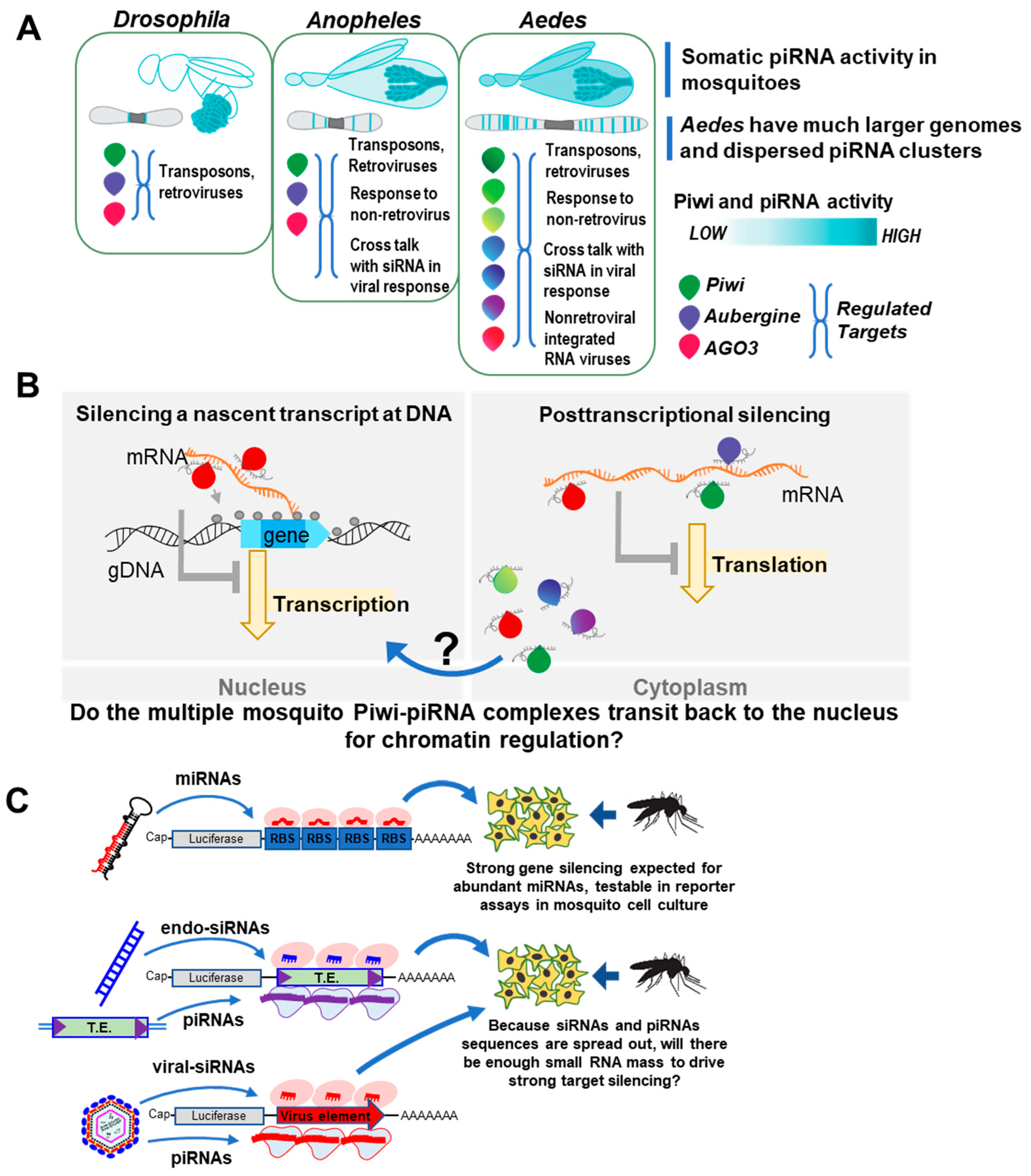
3. Recapping What Makes Mosquito piRNA Pathways Special
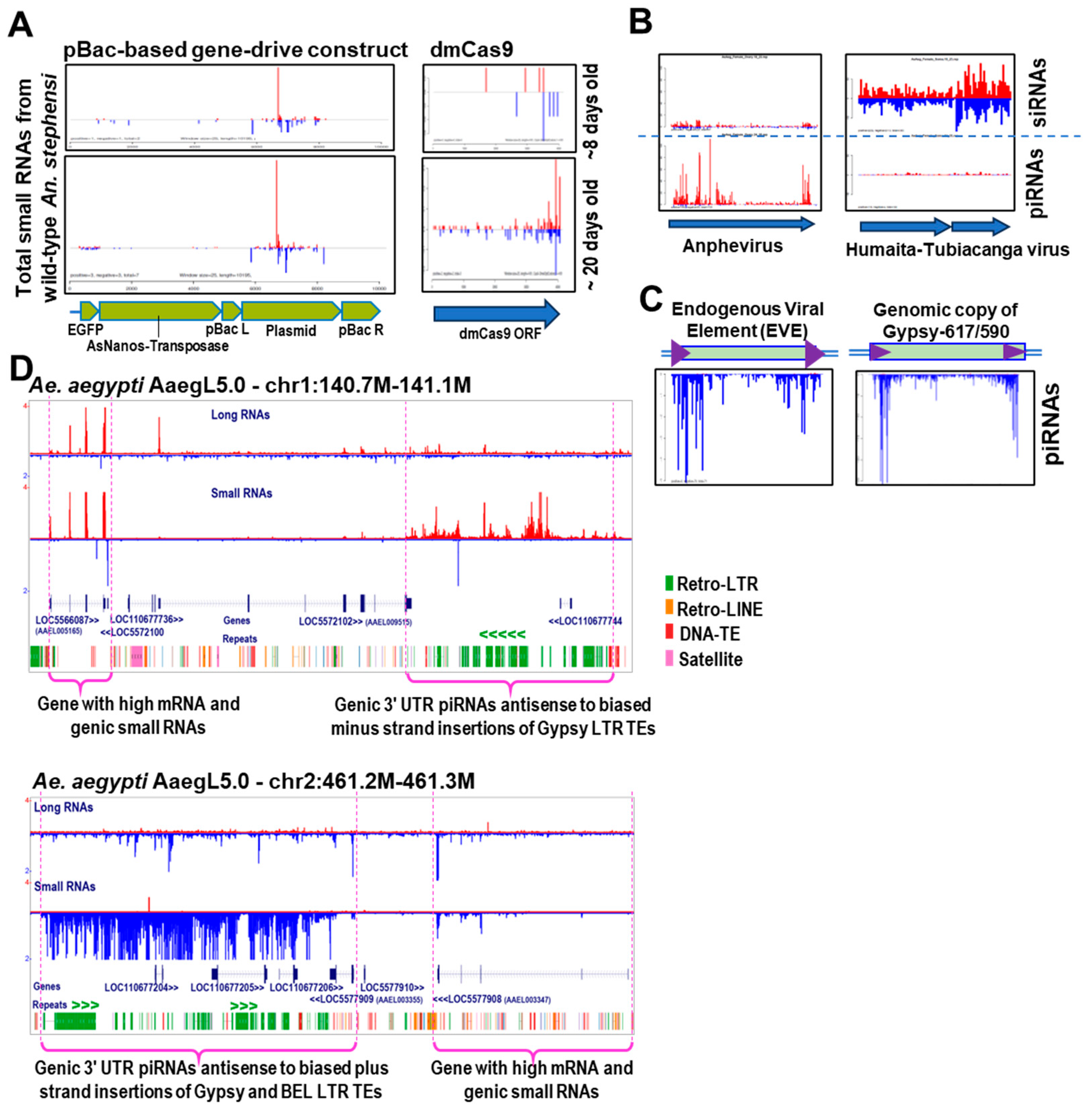
4. Nuclear versus Cytoplasmic Piwi-piRNA Complexes: What Do Mosquitoes Favor?
5. The Importance of Biochemical Silencing Capacity Assays for Mosquito Small RNAs
6. Are Mosquito piRNA Pathways “Overreacting” to Transgenes?
7. Are Mosquito Transgenes Triggering Co-Suppression via siRNAs and piRNAs?
8. Can We Mask Transgenes from Mosquito RNAi Responses?
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hall, J.C.; Rosbash, M.; Young, M.W. Nobel Prize in Physiology and Medicine 2017. Available online: https://www.nobelprize.org/prizes/medicine/2017/summary/.
- Lewis, E.B.; Nüsslein-Volhard, C.; Wieschaus, E.F. Nobel Prize in Physiology and Medicine 1995. Available online: https://www.nobelprize.org/prizes/medicine/1995/summary/.
- Morgan, T.H. Nobel Prize in Physiology and Medicine 1933. Available online: https://www.nobelprize.org/prizes/medicine/1933/summary/.
- Muller, H.J. Nobel Prize in Physiology and Medicine 1946. Available online: https://www.nobelprize.org/prizes/medicine/1946/summary/.
- Hoffman, J. The Nobel Prize in Physiology or Medicine 2011. Available online: https://www.nobelprize.org/prizes/medicine/2011/summary/.
- Carlson, J.; Olson, K.; Higgs, S.; Beaty, B. Molecular genetic manipulation of mosquito vectors. Annu. Rev. Entomol. 1995, 40, 359–388. [Google Scholar] [CrossRef] [PubMed]
- Kovach, M.J.; Carlson, J.O.; Beaty, B.J. A Drosophila metallothionein promoter is inducible in mosquito cells. Insect Mol. Biol. 1992, 1, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Beaty, B.J.; Prager, D.J.; James, A.A.; Jacobs-Lorena, M.; Miller, L.H.; Law, J.H.; Collins, F.H.; Kafatos, F.C. From Tucson to genomics and transgenics: The vector biology network and the emergence of modern vector biology. PLoS Negl. Trop. Dis. 2009, 3, e343. [Google Scholar] [CrossRef] [PubMed]
- George, P.; Jensen, S.; Pogorelcnik, R.; Lee, J.; Xing, Y.; Brasset, E.; Vaury, C.; Sharakhov, I.V. Increased production of piRNAs from euchromatic clusters and genes in Anopheles gambiae compared with Drosophila melanogaster. Epigenetics Chromatin 2015, 8, 50. [Google Scholar] [CrossRef] [PubMed]
- Bolshakov, V.N.; Topalis, P.; Blass, C.; Kokoza, E.; della Torre, A.; Kafatos, F.C.; Louis, C. A comparative genomic analysis of two distant diptera, the fruit fly, Drosophila melanogaster, and the malaria mosquito, Anopheles gambiae. Genome Res. 2002, 12, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Behura, S.K.; Haugen, M.; Flannery, E.; Sarro, J.; Tessier, C.R.; Severson, D.W.; Duman-Scheel, M. Comparative genomic analysis of Drosophila melanogaster and vector mosquito developmental genes. PLoS ONE 2011, 6, e21504. [Google Scholar] [CrossRef] [PubMed]
- Lehane, M.J. The Biology of Blood-Sucking in Insects, 2nd ed.; Cambridge University Press: Cambridge, UK, 2005; 336p. [Google Scholar]
- Gamez, S.; Srivastav, S.; Akbari, O.S.; Lau, N.C. Diverse Defenses: A Perspective Comparing Dipteran Piwi-piRNA Pathways. Cells 2020, 9, 2180. [Google Scholar] [CrossRef] [PubMed]
- Macias, V.M.; Palatini, U.; Bonizzoni, M.; Rasgon, J.L. Leaning into the Bite: The piRNA Pathway as an Exemplar for the Genetic Engineering Need in Mosquitoes. Front. Cell. Infect. Microbiol. 2020, 10, 614342. [Google Scholar] [CrossRef] [PubMed]
- Sundby, A.E.; Molnar, R.I.; Claycomb, J.M. Connecting the Dots: Linking Caenorhabditis elegans Small RNA Pathways and Germ Granules. Trends Cell Biol. 2021, 31, 387–401. [Google Scholar] [CrossRef]
- Wu, P.H.; Zamore, P.D. Defining the functions of PIWI-interacting RNAs. Nat. Rev. Mol. Cell Biol. 2021, 22, 239–240. [Google Scholar] [CrossRef]
- Shang, R.; Lee, S.; Senavirathne, G.; Lai, E.C. microRNAs in action: Biogenesis, function and regulation. Nat. Rev. Genet. 2023, 24, 816–833. [Google Scholar] [CrossRef] [PubMed]
- Tabara, H.; Sarkissian, M.; Kelly, W.G.; Fleenor, J.; Grishok, A.; Timmons, L.; Fire, A.; Mello, C.C. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 1999, 99, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Napoli, C.; Lemieux, C.; Jorgensen, R. Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans. Plant Cell 1990, 2, 279–289. [Google Scholar] [CrossRef] [PubMed]
- van der Krol, A.R.; Brunelle, A.; Tsuchimoto, S.; Chua, N.H. Functional analysis of petunia floral homeotic MADS box gene pMADS1. Genes Dev. 1993, 7, 1214–1228. [Google Scholar] [CrossRef] [PubMed]
- Angenent, G.C.; Franken, J.; Busscher, M.; Colombo, L.; van Tunen, A.J. Petal and stamen formation in petunia is regulated by the homeotic gene fbp1. Plant J. 1993, 4, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Pal-Bhadra, M.; Bhadra, U.; Birchler, J.A. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol. Cell 2002, 9, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Kennerdell, J.R.; Carthew, R.W. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 1998, 95, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- Jensen, S.; Gassama, M.P.; Heidmann, T. Taming of transposable elements by homology-dependent gene silencing. Nat. Genet. 1999, 21, 209–212. [Google Scholar] [CrossRef]
- Rubin, G.M.; Spradling, A.C. Vectors for P element-mediated gene transfer in Drosophila. Nucleic Acids Res. 1983, 11, 6341–6351. [Google Scholar] [CrossRef][Green Version]
- Hazelrigg, T.; Levis, R.; Rubin, G.M. Transformation of white locus DNA in Drosophila: Dosage compensation, zeste interaction, and position effects. Cell 1984, 36, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Nagarkar-Jaiswal, S.; DeLuca, S.Z.; Lee, P.T.; Lin, W.W.; Pan, H.; Zuo, Z.; Lv, J.; Spradling, A.C.; Bellen, H.J. A genetic toolkit for tagging intronic MiMIC containing genes. eLife 2015, 4, e08469. [Google Scholar] [CrossRef] [PubMed]
- Kapetanaki, M.G.; Loukeris, T.G.; Livadaras, I.; Savakis, C. High frequencies of Minos transposon mobilization are obtained in insects by using in vitro synthesized mRNA as a source of transposase. Nucleic Acids Res. 2002, 30, 3333–3340. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Venken, K.J.; He, Y.; Hoskins, R.A.; Bellen, H.J. P[acman]: A BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 2006, 314, 1747–1751. [Google Scholar] [CrossRef] [PubMed]
- Bischof, J.; Maeda, R.K.; Hediger, M.; Karch, F.; Basler, K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 2007, 104, 3312–3317. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Sun, J.; Housden, B.E.; Hu, Y.; Roesel, C.; Lin, S.; Liu, L.-P.; Yang, Z.; Mao, D.; Sun, L.; et al. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc. Natl. Acad. Sci. USA 2013, 110, 19012–19017. [Google Scholar] [CrossRef] [PubMed]
- Gratz, S.J.; Ukken, F.P.; Rubinstein, C.D.; Thiede, G.; Donohue, L.K.; Cummings, A.M.; O’Connor-Giles, K.M. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 2014, 196, 961–971. [Google Scholar] [CrossRef] [PubMed]
- Port, F.; Chen, H.M.; Lee, T.; Bullock, S.L. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA 2014, 111, E2967–E2976. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Kwak, S.J.; Kim, J.; Kim, A.K.; Noh, H.M.; Kim, J.S.; Yu, K. RNA-guided genome editing in Drosophila with the purified Cas9 protein. G3 2014, 4, 1291–1295. [Google Scholar] [CrossRef]
- Perrimon, N.; Noll, E.; McCall, K.; Brand, A. Generating lineage-specific markers to study Drosophila development. Dev. Genet. 1991, 12, 238–252. [Google Scholar] [CrossRef]
- Brand, A.H.; Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993, 118, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Kondo, S.; Booker, M.; Perrimon, N. Cross-species RNAi rescue platform in Drosophila melanogaster. Genetics 2009, 183, 1165–1173. [Google Scholar] [CrossRef] [PubMed]
- Dietzl, G.; Chen, D.; Schnorrer, F.; Su, K.C.; Barinova, Y.; Fellner, M.; Gasser, B.; Kinsey, K.; Oppel, S.; Scheiblauer, S.; et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 2007, 448, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Perkins, L.A.; Holderbaum, L.; Tao, R.; Hu, Y.; Sopko, R.; McCall, K.; Yang-Zhou, D.; Flockhart, I.; Binari, R.; Shim, H.-S.; et al. The Transgenic RNAi Project at Harvard Medical School: Resources and Validation. Genetics 2015, 201, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Fish, M.P.; Groth, A.C.; Calos, M.P.; Nusse, R. Creating transgenic Drosophila by microinjecting the site-specific phiC31 integrase mRNA and a transgene-containing donor plasmid. Nat. Protoc. 2007, 2, 2325–2331. [Google Scholar] [CrossRef]
- Ringrose, L. Transgenesis in Drosophila melanogaster. Methods Mol. Biol. 2009, 561, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Bui, M.; Yang, T.; Bowman, C.S.; White, B.J.; Akbari, O.S. Germline Cas9 expression yields highly efficient genome engineering in a major worldwide disease vector, Aedes aegypti. Proc. Natl. Acad. Sci. USA 2017, 114, E10540–E10549. [Google Scholar] [CrossRef] [PubMed]
- Adelman, Z.N.; Anderson, M.A.; Morazzani, E.M.; Myles, K.M. A transgenic sensor strain for monitoring the RNAi pathway in the yellow fever mosquito, Aedes aegypti. Insect Biochem. Mol. Biol. 2008, 38, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Franz, A.W.; Sanchez-Vargas, I.; Raban, R.R.; Black, W.C.; James, A.A.; Olson, K.E. Fitness impact and stability of a transgene conferring resistance to dengue-2 virus following introgression into a genetically diverse Aedes aegypti strain. PLoS Negl. Trop. Dis. 2014, 8, e2833. [Google Scholar] [CrossRef] [PubMed]
- Franz, A.W.; Sanchez-Vargas, I.; Piper, J.; Smith, M.R.; Khoo, C.C.; James, A.A.; Olson, K.E. Stability and loss of a virus resistance phenotype over time in transgenic mosquitoes harbouring an antiviral effector gene. Insect Mol. Biol. 2009, 18, 661–672. [Google Scholar] [CrossRef]
- Franz, A.W.; Sanchez-Vargas, I.; Adelman, Z.N.; Blair, C.D.; Beaty, B.J.; James, A.A.; Olson, K.E. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti. Proc. Natl. Acad. Sci. USA 2006, 103, 4198–4203. [Google Scholar] [CrossRef] [PubMed]
- Macias, V.M.; Jimenez, A.J.; Burini-Kojin, B.; Pledger, D.; Jasinskiene, N.; Phong, C.H.; Chu, K.; Fazekas, A.; Martin, K.; Marinotti, O.; et al. nanos-Driven expression of piggyBac transposase induces mobilization of a synthetic autonomous transposon in the malaria vector mosquito, Anopheles stephensi. Insect Biochem. Mol. Biol. 2017, 87, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.E.; Cook, K.R.; Hawley, R.S. The joy of balancers. PLoS Genet. 2019, 15, e1008421. [Google Scholar] [CrossRef] [PubMed]
- Holsopple, J.M.; Cook, K.R.; Popodi, E.M. Identification of novel split-GAL4 drivers for the characterization of enteroendocrine cells in the Drosophila melanogaster midgut. G3 2022, 12, jkac102. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, S.L.; Shimell, M.; Tauscher, P.; Daly, S.M.; Shimmi, O.; O’Connor, M.B.; Newfeld, S.J. New resources for the Drosophila 4th chromosome: FRT101F enabled mitotic clones and Bloom syndrome helicase enabled meiotic recombination. G3 2022, 12, jkac019. [Google Scholar] [CrossRef] [PubMed]
- Kahsai, L.; Millburn, G.H.; Cook, K.R. Phenotypes Associated with Second Chromosome P Element Insertions in Drosophila melanogaster. G3 2016, 6, 2665–2670. [Google Scholar] [CrossRef] [PubMed]
- Southall, T.D.; Elliott, D.A.; Brand, A.H. The GAL4 System: A Versatile Toolkit for Gene Expression in Drosophila. CSH Protoc. 2008, 2008, pdb top49. [Google Scholar] [CrossRef]
- Poulton, B.C.; Colman, F.; Anthousi, A.; Grigoraki, L.; Adolfi, A.; Lynd, A.; Lycett, G.J. Using the GAL4-UAS System for Functional Genetics in Anopheles gambiae. J. Vis. Exp. 2021, 170, e62131. [Google Scholar] [CrossRef]
- Lynd, A.; Lycett, G.J. Development of the bi-partite Gal4-UAS system in the African malaria mosquito, Anopheles gambiae. PLoS ONE 2012, 7, e31552. [Google Scholar] [CrossRef]
- Zhao, B.; Kokoza, V.A.; Saha, T.T.; Wang, S.; Roy, S.; Raikhel, A.S. Regulation of the gut-specific carboxypeptidase: A study using the binary Gal4/UAS system in the mosquito Aedes aegypti. Insect Biochem. Mol. Biol. 2014, 54, 1–10. [Google Scholar] [CrossRef]
- Kokoza, V.A.; Raikhel, A.S. Targeted gene expression in the transgenic Aedes aegypti using the binary Gal4-UAS system. Insect Biochem. Mol. Biol. 2011, 41, 637–644. [Google Scholar] [CrossRef] [PubMed]
- Folsz, O.; Lin, C.C.; Task, D.; Riabinina, O.; Potter, C.J. The Q-system: A Versatile Repressible Binary Expression System. Methods Mol. Biol. 2022, 2540, 35–78. [Google Scholar] [CrossRef] [PubMed]
- Riabinina, O.; Potter, C.J. The Q-System: A Versatile Expression System for Drosophila. Methods Mol. Biol. 2016, 1478, 53–78. [Google Scholar] [CrossRef] [PubMed]
- Riabinina, O.; Task, D.; Marr, E.; Lin, C.C.; Alford, R.; O’Brochta, D.A.; Potter, C.J. Organization of olfactory centres in the malaria mosquito Anopheles gambiae. Nat. Commun. 2016, 7, 13010. [Google Scholar] [CrossRef] [PubMed]
- Pascini, T.V.; Jeong, Y.J.; Huang, W.; Pala, Z.R.; Sa, J.M.; Wells, M.B.; Kizito, C.; Sweeney, B.; Silva, T.L.A.E.; Andrew, D.J.; et al. Transgenic Anopheles mosquitoes expressing human PAI-1 impair malaria transmission. Nat. Commun. 2022, 13, 2949. [Google Scholar] [CrossRef] [PubMed]
- Macias, V.M.; Ohm, J.R.; Rasgon, J.L. Gene Drive for Mosquito Control: Where Did It Come from and Where Are We Headed? Int. J. Environ. Res. Public Health 2017, 14, 1006. [Google Scholar] [CrossRef]
- Saito, K.; Nishida, K.M.; Mori, T.; Kawamura, Y.; Miyoshi, K.; Nagami, T.; Siomi, H.; Siomi, M.C. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006, 20, 2214–2222. [Google Scholar] [CrossRef] [PubMed]
- Vagin, V.V.; Sigova, A.; Li, C.; Seitz, H.; Gvozdev, V.; Zamore, P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 2006, 313, 320–324. [Google Scholar] [CrossRef]
- Aravin, A.A.; Hannon, G.J.; Brennecke, J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 2007, 318, 761–764. [Google Scholar] [CrossRef]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef]
- Brower-Toland, B.; Findley, S.D.; Jiang, L.; Liu, L.; Yin, H.; Dus, M.; Zhou, P.; Elgin, S.C.R.; Lin, H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007, 21, 2300–2311. [Google Scholar] [CrossRef]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef] [PubMed]
- Onishi, R.; Yamanaka, S.; Siomi, M.C. piRNA- and siRNA-mediated transcriptional repression in Drosophila, mice, and yeast: New insights and biodiversity. EMBO Rep. 2021, 22, e53062. [Google Scholar] [CrossRef] [PubMed]
- Juliano, C.; Wang, J.; Lin, H. Uniting germline and stem cells: The function of Piwi proteins and the piRNA pathway in diverse organisms. Annu. Rev. Genet. 2011, 45, 447–469. [Google Scholar] [CrossRef] [PubMed]
- Chaverra-Rodriguez, D.; Macias, V.M.; Hughes, G.L.; Pujhari, S.; Suzuki, Y.; Peterson, D.R.; Kim, D.; McKeand, S.; Rasgon, J.L. Targeted delivery of CRISPR-Cas9 ribonucleoprotein into arthropod ovaries for heritable germline gene editing. Nat. Commun. 2018, 9, 3008. [Google Scholar] [CrossRef] [PubMed]
- Macias, V.M.; McKeand, S.; Chaverra-Rodriguez, D.; Hughes, G.L.; Fazekas, A.; Pujhari, S.; Jasinskiene, N.; James, A.A.; Rasgon, J.L. Cas9-Mediated Gene-Editing in the Malaria Mosquito Anopheles stephensi by ReMOT Control. G3 2020, 10, 1353–1360. [Google Scholar] [CrossRef] [PubMed]
- Gantz, V.M.; Jasinskiene, N.; Tatarenkova, O.; Fazekas, A.; Macias, V.M.; Bier, E.; James, A.A. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl. Acad. Sci. USA 2015, 112, E6736–E6743. [Google Scholar] [CrossRef]
- Adelman, Z.N.; Jasinskiene, N.; Vally, K.J.; Peek, C.; Travanty, E.A.; Olson, K.E.; Brown, S.E.; Stephens, J.L.; Knudson, D.L.; Coates, C.J.; et al. Formation and loss of large, unstable tandem arrays of the piggyBac transposable element in the yellow fever mosquito, Aedes aegypti. Transgenic Res. 2004, 13, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Ross, P.A.; Endersby-Harshman, N.M.; Hoffmann, A.A. A comprehensive assessment of inbreeding and laboratory adaptation in Aedes aegypti mosquitoes. Evol. Appl. 2019, 12, 572–586. [Google Scholar] [CrossRef]
- Koenraadt, C.J.; Kormaksson, M.; Harrington, L.C. Effects of inbreeding and genetic modification on Aedes aegypti larval competition and adult energy reserves. Parasit. Vectors 2010, 3, 92. [Google Scholar] [CrossRef]
- Mejia, A.J.; Jimenez, L.; Dutra, H.L.C.; Perera, R.; McGraw, E.A. Attempts to use breeding approaches in Aedes aegypti to create lines with distinct and stable relative Wolbachia densities. Heredity 2022, 129, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.R.; Evans, B.R. How Much Does Inbreeding Reduce Heterozygosity? Empirical Results from Aedes aegypti. Am. J. Trop. Med. Hyg. 2017, 96, 157–158. [Google Scholar] [CrossRef] [PubMed]
- Nouzova, M.; Edwards, M.J.; DeGennaro, M.; Leyva, D.; Tose, L.V.; Fernandez-Lima, F.; Noriega, F.G. Genetics tools for corpora allata specific gene expression in Aedes aegypti mosquitoes. Sci. Rep. 2022, 12, 20426. [Google Scholar] [CrossRef] [PubMed]
- Galizi, R.; Doyle, L.A.; Menichelli, M.; Bernardini, F.; Deredec, A.; Burt, A.; Stoddard, B.L.; Windbichler, N.; Crisanti, A. A synthetic sex ratio distortion system for the control of the human malaria mosquito. Nat. Commun. 2014, 5, 3977. [Google Scholar] [CrossRef] [PubMed]
- Vitale, M.; Leo, C.; Courty, T.; Kranjc, N.; Connolly, J.B.; Morselli, G.; Bamikole, C.; Haghighat-Khah, R.E.; Bernardini, F.; Fuchs, S. Comprehensive characterization of a transgene insertion in a highly repetitive, centromeric region of Anopheles mosquitoes. Pathog. Glob. Health 2023, 117, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.O.; Kumon, T.; Yamashita, Y.M. rDNA magnification is a unique feature of germline stem cells. Proc. Natl. Acad. Sci. USA 2023, 120, e2314440120. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.O.; Slicko, A.; Yamashita, Y.M. The retrotransposon R2 maintains Drosophila ribosomal DNA repeats. Proc. Natl. Acad. Sci. USA 2023, 120, e2221613120. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.O.; Watase, G.J.; Warsinger-Pepe, N.; Yamashita, Y.M. Mechanisms of rDNA Copy Number Maintenance. Trends Genet. 2019, 35, 734–742. [Google Scholar] [CrossRef]
- Watase, G.J.; Nelson, J.O.; Yamashita, Y.M. Nonrandom sister chromatid segregation mediates rDNA copy number maintenance in Drosophila. Sci. Adv. 2022, 8, eabo4443. [Google Scholar] [CrossRef]
- Stolyarenko, A.D. Nuclear Argonaute Piwi Gene Mutation Affects rRNA by Inducing rRNA Fragment Accumulation, Antisense Expression, and Defective Processing in Drosophila Ovaries. Int. J. Mol. Sci. 2020, 21, 1119. [Google Scholar] [CrossRef]
- Malone, C.D.; Brennecke, J.; Dus, M.; Stark, A.; McCombie, W.R.; Sachidanandam, R.; Hannon, G.J. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 2009, 137, 522–535. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Munafo, M.; Ciabrelli, F.; Eastwood, E.L.; Fabry, M.H.; Kneuss, E.; Hannon, G.J. piRNA-Guided Genome Defense: From Biogenesis to Silencing. Annu. Rev. Genet. 2018, 52, 131–157. [Google Scholar] [CrossRef] [PubMed]
- Schnettler, E.; Donald, C.L.; Human, S.; Watson, M.; Siu, R.W.C.; McFarlane, M.; Fazakerley, J.K.; Kohl, A.; Fragkoudis, R. Knockdown of piRNA pathway proteins results in enhanced Semliki Forest virus production in mosquito cells. J. Gen. Virol. 2013, 94 Pt 7, 1680–1689. [Google Scholar] [CrossRef]
- Miesen, P.; Girardi, E.; van Rij, R.P. Distinct sets of PIWI proteins produce arbovirus and transposon-derived piRNAs in Aedes aegypti mosquito cells. Nucleic Acids Res. 2015, 43, 6545–6556. [Google Scholar] [CrossRef] [PubMed]
- Girardi, E.; Miesen, P.; Pennings, B.; Frangeul, L.; Saleh, M.C.; van Rij, R.P. Histone-derived piRNA biogenesis depends on the ping-pong partners Piwi5 and Ago3 in Aedes aegypti. Nucleic Acids Res. 2017, 45, 4881–4892. [Google Scholar] [CrossRef] [PubMed]
- Varjak, M.; Maringer, K.; Watson, M.; Sreenu, V.B.; Fredericks, A.C.; Pondeville, E.; Donald, C.L.; Sterk, J.; Kean, J.; Vazeille, M.; et al. Aedes aegypti Piwi4 Is a Noncanonical PIWI Protein Involved in Antiviral Responses. mSphere 2017, 2, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Tassetto, M.; Kunitomi, M.; Whitfield, Z.J.; Dolan, P.T.; Sanchez-Vargas, I.; Garcia-Knight, M.; Ribiero, I.; Chen, T.; Olson, K.E.; Andino, R. Control of RNA viruses in mosquito cells through the acquisition of vDNA and endogenous viral elements. eLife 2019, 8, e41244. [Google Scholar] [CrossRef] [PubMed]
- Joosten, J.; Taskopru, E.; Jansen, P.; Pennings, B.; Vermeulen, M.; Van Rij, R.P. PIWI proteomics identifies Atari and Pasilla as piRNA biogenesis factors in Aedes mosquitoes. Cell Rep. 2021, 35, 109073. [Google Scholar] [CrossRef] [PubMed]
- Halbach, R.; Miesen, P.; Joosten, J.; Taşköprü, E.; Rondeel, I.; Pennings, B.; Vogels, C.B.F.; Merkling, S.H.; Koenraadt, C.J.; Lambrechts, L.; et al. A satellite repeat-derived piRNA controls embryonic development of Aedes. Nature 2020, 580, 274–277. [Google Scholar] [CrossRef]
- Betting, V.; Joosten, J.; Halbach, R.; Thaler, M.; Miesen, P.; Van Rij, R.P. A piRNA-lncRNA regulatory network initiates responder and trailer piRNA formation during mosquito embryonic development. RNA 2021, 27, 1155–1172. [Google Scholar] [CrossRef]
- Ma, Q.; Srivastav, S.P.; Gamez, S.; Dayama, G.; Feitosa-Suntheimer, F.; Patterson, E.I.; Johnson, R.M.; Matson, E.M.; Gold, A.; Brackney, D.E.; et al. A mosquito small RNA genomics resource reveals dynamic evolution and host responses to viruses and transposons. Genome Res. 2021, 31, 512–528. [Google Scholar] [CrossRef] [PubMed]
- Morazzani, E.M.; Wiley, M.R.; Murreddu, M.G.; Adelman, Z.N.; Myles, K.M. Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma. PLoS Pathog. 2012, 8, e1002470. [Google Scholar] [CrossRef]
- Vodovar, N.; Bronkhorst, A.W.; van Cleef, K.W.; Miesen, P.; Blanc, H.; van Rij, R.P.; Saleh, M.-C. Arbovirus-derived piRNAs exhibit a ping-pong signature in mosquito cells. PLoS ONE 2012, 7, e30861. [Google Scholar] [CrossRef] [PubMed]
- Macias, V.; Coleman, J.; Bonizzoni, M.; James, A.A. piRNA pathway gene expression in the malaria vector mosquito Anopheles stephensi. Insect Mol. Biol. 2014, 23, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Hess, A.M.; Prasad, A.N.; Ptitsyn, A.; Ebel, G.D.; Olson, K.E.; Barbacioru, C.; Monighetti, C.; Campbell, C.L. Small RNA profiling of Dengue virus-mosquito interactions implicates the PIWI RNA pathway in anti-viral defense. BMC Microbiol. 2011, 11, 45. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.C.; Brackney, D.E.; Campbell, C.L.; Bondu-Hawkins, V.; Hjelle, B.; Ebel, G.D.; Olson, K.E.; Blair, C.D. Comparison of dengue virus type 2-specific small RNAs from RNA interference-competent and -incompetent mosquito cells. PLoS Negl. Trop. Dis. 2010, 4, e848. [Google Scholar] [CrossRef] [PubMed]
- Palatini, U.; Masri, R.A.; Cosme, L.V.; Koren, S.; Thibaud-Nissen, F.; Biedler, J.K.; Krsticevic, F.; Johnston, J.S.; Halbach, R.; Crawford, J.E.; et al. Improved reference genome of the arboviral vector Aedes albopictus. Genome Biol. 2020, 21, 215. [Google Scholar] [CrossRef]
- Whitfield, Z.J.; Dolan, P.T.; Kunitomi, M.; Tassetto, M.; Seetin, M.G.; Oh, S.; Heiner, C.; Paxinos, E.; Andino, R. The Diversity, Structure, and Function of Heritable Adaptive Immunity Sequences in the Aedes aegypti Genome. Curr. Biol. 2017, 27, 3511–3519.e7. [Google Scholar] [CrossRef]
- Suzuki, Y.; Frangeul, L.; Dickson, L.B.; Blanc, H.; Verdier, Y.; Vinh, J.; Lambrechts, L.; Saleh, M.-C. Uncovering the Repertoire of Endogenous Flaviviral Elements in Aedes Mosquito Genomes. J. Virol. 2017, 91, 10–1128. [Google Scholar] [CrossRef]
- Qu, J.; Betting, V.; van Iterson, R.; Kwaschik, F.M.; van Rij, R.P. Chromatin profiling identifies transcriptional readthrough as a conserved mechanism for piRNA biogenesis in mosquitoes. Cell Rep. 2023, 42, 112257. [Google Scholar] [CrossRef]
- Marconcini, M.; Pischedda, E.; Houe, V.; Palatini, U.; Lozada-Chavez, N.; Sogliani, D.; Failloux, A.-B.; Bonizzoni, M. Profile of Small RNAs, vDNA Forms and Viral Integrations in Late Chikungunya Virus Infection of Aedes albopictus Mosquitoes. Viruses 2021, 13, 553. [Google Scholar] [CrossRef] [PubMed]
- Lewis, S.H.; Quarles, K.A.; Yang, Y.; Tanguy, M.; Frezal, L.; Smith, S.A.; Sharma, P.P.; Cordaux, R.; Gilbert, C.; Giraud, I.; et al. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat. Ecol. Evol. 2018, 2, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Akbari, O.S.; Antoshechkin, I.; Amrhein, H.; Williams, B.; Diloreto, R.; Sandler, J.; Hay, B.A. The developmental transcriptome of the mosquito Aedes aegypti, an invasive species and major arbovirus vector. G3 2013, 3, 1493–1509. [Google Scholar] [CrossRef] [PubMed]
- Crava, C.M.; Varghese, F.S.; Pischedda, E.; Halbach, R.; Palatini, U.; Marconcini, M.; Gasmi, L.; Redmond, S.; Afrane, Y.; Ayala, D.; et al. Population genomics in the arboviral vector Aedes aegypti reveals the genomic architecture and evolution of endogenous viral elements. Mol. Ecol. 2021, 30, 1594–1611. [Google Scholar] [CrossRef]
- Campbell, C.L.; Black, W.C.; Hess, A.M.; Foy, B.D. Comparative genomics of small RNA regulatory pathway components in vector mosquitoes. BMC Genom. 2008, 9, 425. [Google Scholar] [CrossRef] [PubMed]
- Miesen, P.; Ivens, A.; Buck, A.H.; van Rij, R.P. Small RNA Profiling in Dengue Virus 2-Infected Aedes Mosquito Cells Reveals Viral piRNAs and Novel Host miRNAs. PLoS Negl. Trop. Dis. 2016, 10, e0004452. [Google Scholar] [CrossRef] [PubMed]
- Walsh, E.; Torres, T.Z.B.; Ruckert, C. Culex Mosquito Piwi4 Is Antiviral against Two Negative-Sense RNA Viruses. Viruses 2022, 14, 2758. [Google Scholar] [CrossRef] [PubMed]
- Joosten, J.; Overheul, G.J.; Van Rij, R.P.; Miesen, P. Endogenous piRNA-guided slicing triggers responder and trailer piRNA production from viral RNA in Aedes aegypti mosquitoes. Nucleic Acids Res. 2021, 49, 8886–8899. [Google Scholar] [CrossRef]
- Joosten, J.; Miesen, P.; Taskopru, E.; Pennings, B.; Jansen, P.; Huynen, M.A.; Vermeulen, M.; Van Rij, R.P. The Tudor protein Veneno assembles the ping-pong amplification complex that produces viral piRNAs in Aedes mosquitoes. Nucleic Acids Res. 2019, 47, 2546–2559. [Google Scholar] [CrossRef]
- Xu, M.; You, Y.; Hunsicker, P.; Hori, T.; Small, C.; Griswold, M.D.; Hecht, N.B. Mice deficient for a small cluster of Piwi-interacting RNAs implicate Piwi-interacting RNAs in transposon control. Biol. Reprod. 2008, 79, 51–57. [Google Scholar] [CrossRef]
- Homolka, D.; Pandey, R.R.; Goriaux, C.; Brasset, E.; Vaury, C.; Sachidanandam, R.; Fauvarque, M.-O.; Pillai, R.S. PIWI Slicing and RNA Elements in Precursors Instruct Directional Primary piRNA Biogenesis. Cell Rep. 2015, 12, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.H.; Fu, Y.; Cecchini, K.; Ozata, D.M.; Arif, A.; Yu, T.; Colpan, C.; Gainetdinov, I.; Weng, Z.; Zamore, P.D. The evolutionarily conserved piRNA-producing locus pi6 is required for male mouse fertility. Nat. Genet. 2020, 52, 728–739. [Google Scholar] [CrossRef] [PubMed]
- Gebert, D.; Neubert, L.K.; Lloyd, C.; Gui, J.; Lehmann, R.; Teixeira, F.K. Large Drosophila germline piRNA clusters are evolutionarily labile and dispensable for transposon regulation. Mol. Cell 2021, 81, 3965–3978.e5. [Google Scholar] [CrossRef] [PubMed]
- Ge, D.T.; Wang, W.; Tipping, C.; Gainetdinov, I.; Weng, Z.; Zamore, P.D. The RNA-Binding ATPase, Armitage, Couples piRNA Amplification in Nuage to Phased piRNA Production on Mitochondria. Mol. Cell 2019, 74, 982–995.e6. [Google Scholar] [CrossRef]
- Lim, A.K.; Kai, T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2007, 104, 6714–6719. [Google Scholar] [CrossRef] [PubMed]
- Webster, A.; Li, S.; Hur, J.K.; Wachsmuth, M.; Bois, J.S.; Perkins, E.M.; Patel, D.J.; Aravin, A.A. Aub and Ago3 Are Recruited to Nuage through Two Mechanisms to Form a Ping-Pong Complex Assembled by Krimper. Mol. Cell 2015, 59, 564–575. [Google Scholar] [CrossRef] [PubMed]
- Sienski, G.; Donertas, D.; Brennecke, J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 2012, 151, 964–980. [Google Scholar] [CrossRef]
- Le Thomas, A.; Rogers, A.K.; Webster, A.; Marinov, G.K.; Liao, S.E.; Perkins, E.M.; Hur, J.K.; Aravin, A.A.; Fejes Tóth, K. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013, 27, 390–399. [Google Scholar] [CrossRef] [PubMed]
- Sienski, G.; Batki, J.; Senti, K.A.; Donertas, D.; Tirian, L.; Meixner, K.; Brennecke, J. Silencio/CG9754 connects the Piwi-piRNA complex to the cellular heterochromatin machinery. Genes Dev. 2015, 29, 2258–2271. [Google Scholar] [CrossRef]
- Akkouche, A.; Mugat, B.; Barckmann, B.; Varela-Chavez, C.; Li, B.; Raffel, R.; Pélisson, A.; Chambeyron, S. Piwi Is Required during Drosophila Embryogenesis to License Dual-Strand piRNA Clusters for Transposon Repression in Adult Ovaries. Mol. Cell 2017, 66, 411–419.e4. [Google Scholar] [CrossRef]
- Andersen, P.R.; Tirian, L.; Vunjak, M.; Brennecke, J. A heterochromatin-dependent transcription machinery drives piRNA expression. Nature 2017, 549, 54–59. [Google Scholar] [CrossRef] [PubMed]
- ElMaghraby, M.F.; Andersen, P.R.; Puhringer, F.; Hohmann, U.; Meixner, K.; Lendl, T.; Tirian, L.; Brennecke, J. A Heterochromatin-Specific RNA Export Pathway Facilitates piRNA Production. Cell 2019, 178, 964–979.e20. [Google Scholar] [CrossRef] [PubMed]
- Duempelmann, L.; Skribbe, M.; Buhler, M. Small RNAs in the Transgenerational Inheritance of Epigenetic Information. Trends Genet. 2020, 36, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; He, P.; Kanrar, N.; Fejes Toth, K.; Aravin, A.A. Maternally inherited siRNAs initiate piRNA cluster formation. Mol. Cell 2023, 83, 3835–3851.e7. [Google Scholar] [CrossRef] [PubMed]
- Barckmann, B.; Pierson, S.; Dufourt, J.; Papin, C.; Armenise, C.; Port, F.; Grentzinger, T.; Chambeyron, S.; Baronian, G.; Desvignes, J.-P.; et al. Aubergine iCLIP Reveals piRNA-Dependent Decay of mRNAs Involved in Germ Cell Development in the Early Embryo. Cell Rep. 2015, 12, 1205–1216. [Google Scholar] [CrossRef] [PubMed]
- Rozhkov, N.V.; Schostak, N.G.; Zelentsova, E.S.; Yushenova, I.A.; Zatsepina, O.G.; Evgen’ev, M.B. Evolution and dynamics of small RNA response to a retroelement invasion in Drosophila. Mol. Biol. Evol. 2013, 30, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Akkouche, A.; Grentzinger, T.; Fablet, M.; Armenise, C.; Burlet, N.; Braman, V.; Chambeyron, S.; Vieira, C. Maternally deposited germline piRNAs silence the tirant retrotransposon in somatic cells. EMBO Rep. 2013, 14, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Grentzinger, T.; Armenise, C.; Brun, C.; Mugat, B.; Serrano, V.; Pelisson, A.; Chambeyron, S. piRNA-mediated transgenerational inheritance of an acquired trait. Genome Res. 2012, 22, 1877–1888. [Google Scholar] [CrossRef] [PubMed]
- Senti, K.A.; Jurczak, D.; Sachidanandam, R.; Brennecke, J. piRNA-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear piRNA repertoire. Genes Dev. 2015, 29, 1747–1762. [Google Scholar] [CrossRef]
- Desset, S.; Buchon, N.; Meignin, C.; Coiffet, M.; Vaury, C. In Drosophila melanogaster the COM locus directs the somatic silencing of two retrotransposons through both Piwi-dependent and -independent pathways. PLoS ONE 2008, 3, e1526. [Google Scholar] [CrossRef]
- Desset, S.; Meignin, C.; Dastugue, B.; Vaury, C. COM, a heterochromatic locus governing the control of independent endogenous retroviruses from Drosophila melanogaster. Genetics 2003, 164, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Duc, C.; Yoth, M.; Jensen, S.; Mouniee, N.; Bergman, C.M.; Vaury, C.; Brasset, E. Trapping a somatic endogenous retrovirus into a germline piRNA cluster immunizes the germline against further invasion. Genome Biol. 2019, 20, 127. [Google Scholar] [CrossRef] [PubMed]
- Meignin, C.; Dastugue, B.; Vaury, C. Intercellular communication between germ line and somatic line is utilized to control the transcription of ZAM, an endogenous retrovirus from Drosophila melanogaster. Nucleic Acids Res. 2004, 32, 3799–3806. [Google Scholar] [CrossRef] [PubMed]
- Mevel-Ninio, M.; Pelisson, A.; Kinder, J.; Campos, A.R.; Bucheton, A. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics 2007, 175, 1615–1624. [Google Scholar] [CrossRef] [PubMed]
- Ninova, M.; Holmes, H.; Lomenick, B.; Fejes Toth, K.; Aravin, A.A. Pervasive SUMOylation of heterochromatin and piRNA pathway proteins. Cell Genom. 2023, 3, 100329. [Google Scholar] [CrossRef] [PubMed]
- Godneeva, B.; Ninova, M.; Fejes-Toth, K.; Aravin, A. SUMOylation of Bonus, the Drosophila homolog of Transcription Intermediary Factor 1, safeguards germline identity by recruiting repressive chromatin complexes to silence tissue-specific genes. eLife 2023, 12, e89493. [Google Scholar] [CrossRef]
- Andreev, V.I.; Yu, C.; Wang, J.; Schnabl, J.; Tirian, L.; Gehre, M.; Handler, D.; Duchek, P.; Novatchkova, M.; Baumgartner, L.; et al. Panoramix SUMOylation on chromatin connects the piRNA pathway to the cellular heterochromatin machinery. Nat. Struct. Mol. Biol. 2022, 29, 130–142. [Google Scholar] [CrossRef] [PubMed]
- Batki, J.; Schnabl, J.; Wang, J.; Handler, D.; Andreev, V.I.; Stieger, C.E.; Novatchkova, M.; Lampersberger, L.; Kauneckaite, K.; Xie, W.; et al. The nascent RNA binding complex SFiNX licenses piRNA-guided heterochromatin formation. Nat. Struct. Mol. Biol. 2019, 26, 720–731. [Google Scholar] [CrossRef]
- Zhao, K.; Cheng, S.; Miao, N.; Xu, P.; Lu, X.; Zhang, Y.; Wang, M.; Ouyang, X.; Yuan, X.; Liu, W.; et al. A Pandas complex adapted for piRNA-guided transcriptional silencing and heterochromatin formation. Nat. Cell Biol. 2019, 21, 1261–1272. [Google Scholar] [CrossRef]
- Fabry, M.H.; Ciabrelli, F.; Munafo, M.; Eastwood, E.L.; Kneuss, E.; Falciatori, I.; Falconio, F.A.; J Hannon, G.J.; Czech, B. piRNA-guided co-transcriptional silencing coopts nuclear export factors. eLife 2019, 8, e47999. [Google Scholar] [CrossRef]
- Murano, K.; Iwasaki, Y.W.; Ishizu, H.; Mashiko, A.; Shibuya, A.; Kondo, S.; Adachi, S.; Suzuki, S.; Saito, K.; Natsume, T.; et al. Nuclear RNA export factor variant initiates piRNA-guided co-transcriptional silencing. EMBO J. 2019, 38, e102870. [Google Scholar] [CrossRef] [PubMed]
- Ninova, M.; Chen, Y.A.; Godneeva, B.; Rogers, A.K.; Luo, Y.; Fejes Toth, K.; Aravin, A.A. Su(var)2-10 and the SUMO Pathway Link piRNA-Guided Target Recognition to Chromatin Silencing. Mol. Cell 2020, 77, 556–570.e6. [Google Scholar] [CrossRef] [PubMed]
- Ninova, M.; Godneeva, B.; Chen, Y.A.; Luo, Y.; Prakash, S.J.; Jankovics, F.; Erdélyi, M.; Aravin, A.A.; Fejes Tóth, K. The SUMO Ligase Su(var)2-10 Controls Hetero- and Euchromatic Gene Expression via Establishing H3K9 Trimethylation and Negative Feedback Regulation. Mol. Cell 2020, 77, 571–585.e4. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Stuwe, E.; Luo, Y.; Ninova, M.; Le Thomas, A.; Rozhavskaya, E.; Li, S.; Vempati, S.; Laver, J.D.; Patel, D.J.; et al. Cutoff Suppresses RNA Polymerase II Termination to Ensure Expression of piRNA Precursors. Mol. Cell 2016, 63, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Mohn, F.; Sienski, G.; Handler, D.; Brennecke, J. The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell 2014, 157, 1364–1379. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, J.; Schultz, N.; Zhang, F.; Parhad, S.S.; Tu, S.; Vreven, T.; Zamore, P.D.; Weng, Z.; Theurkauf, W.E. The HP1 Homolog Rhino Anchors a Nuclear Complex that Suppresses piRNA Precursor Splicing. Cell 2014, 157, 1353–1363. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.E.; Shrivastava, G.; Gittis, A.G.; Ganesan, S.; Martin-Martin, I.; Valenzuela Leon, P.C.; Olson, K.E.; Calvo, E. Aedes aegypti Piwi4 Structural Features Are Necessary for RNA Binding and Nuclear Localization. Int. J. Mol. Sci. 2021, 22, 12733. [Google Scholar] [CrossRef] [PubMed]
- Aravin, A.A.; van der Heijden, G.W.; Castaneda, J.; Vagin, V.V.; Hannon, G.J.; Bortvin, A. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet. 2009, 5, e1000764. [Google Scholar] [CrossRef] [PubMed]
- Murota, Y.; Ishizu, H.; Nakagawa, S.; Iwasaki, Y.W.; Shibata, S.; Kamatani, M.K.; Saito, K.; Okano, H.; Siomi, H.; Siomi, M.C. Yb integrates piRNA intermediates and processing factors into perinuclear bodies to enhance piRISC assembly. Cell Rep. 2014, 8, 103–113. [Google Scholar] [CrossRef]
- Qi, H.; Watanabe, T.; Ku, H.Y.; Liu, N.; Zhong, M.; Lin, H. The Yb body, a major site for Piwi-associated RNA biogenesis and a gateway for Piwi expression and transport to the nucleus in somatic cells. J. Biol. Chem. 2011, 286, 3789–3797. [Google Scholar] [CrossRef]
- Saito, K.; Ishizu, H.; Komai, M.; Kotani, H.; Kawamura, Y.; Nishida, K.M.; Siomi, H.; Siomi, M.C. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 2010, 24, 2493–2498. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Hu, H.; Webster, A.; Zou, F.; Du, J.; Patel, D.J.; Sachidanandam, R.; Fejes Toth, K.; Aravin, A.A.; Li, S. Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex. Nat. Commun. 2021, 12, 4061. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Iwasaki, Y.W.; Shibuya, A.; Carninci, P.; Tsuchizawa, Y.; Ishizu, H.; Siomi, M.C.; Siomi, H. Krimper Enforces an Antisense Bias on piRNA Pools by Binding AGO3 in the Drosophila Germline. Mol. Cell 2015, 59, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Post, C.; Clark, J.P.; Sytnikova, Y.A.; Chirn, G.W.; Lau, N.C. The capacity of target silencing by Drosophila PIWI and piRNAs. RNA 2014, 20, 1977–1986. [Google Scholar] [CrossRef] [PubMed]
- Sytnikova, Y.A.; Rahman, R.; Chirn, G.W.; Clark, J.P.; Lau, N.C. Transposable element dynamics and PIWI regulation impacts lncRNA and gene expression diversity in Drosophila ovarian cell cultures. Genome Res. 2014, 24, 1977–1990. [Google Scholar] [CrossRef] [PubMed]
- Genzor, P.; Konstantinidou, P.; Stoyko, D.; Manzourolajdad, A.; Marlin Andrews, C.; Elchert, A.R.; Stathopoulos, C.; D Haase, A.D. Cellular abundance shapes function in piRNA-guided genome defense. Genome Res. 2021, 31, 2058–2068. [Google Scholar] [CrossRef] [PubMed]
- Gainetdinov, I.; Colpan, C.; Arif, A.; Cecchini, K.; Zamore, P.D. A Single Mechanism of Biogenesis, Initiated and Directed by PIWI Proteins, Explains piRNA Production in Most Animals. Mol. Cell 2018, 71, 775–790.e5. [Google Scholar] [CrossRef] [PubMed]
- Han, B.W.; Wang, W.; Li, C.; Weng, Z.; Zamore, P.D. Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 2015, 348, 817–821. [Google Scholar] [CrossRef] [PubMed]
- Mohn, F.; Handler, D.; Brennecke, J. Noncoding RNA. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 2015, 348, 812–817. [Google Scholar] [CrossRef]
- Wang, W.; Han, B.W.; Tipping, C.; Ge, D.T.; Zhang, Z.; Weng, Z.; Zamore, P.D. Slicing and Binding by Ago3 or Aub Trigger Piwi-Bound piRNA Production by Distinct Mechanisms. Mol. Cell 2015, 59, 819–830. [Google Scholar] [CrossRef]
- Wenda, J.M.; Homolka, D.; Yang, Z.; Spinelli, P.; Sachidanandam, R.; Pandey, R.R.; Pillai, R.S. Distinct Roles of RNA Helicases MVH and TDRD9 in PIWI Slicing-Triggered Mammalian piRNA Biogenesis and Function. Dev. Cell 2017, 41, 623–637.e9. [Google Scholar] [CrossRef] [PubMed]
- Henderson, C.; Brustolin, M.; Hegde, S.; Dayama, G.; Lau, N.; Hughes, G.L.; Bergey, C.; Rasgon, J.L. Transcriptomic and small RNA response to Mayaro virus infection in Anopheles stephensi mosquitoes. PLoS Negl. Trop. Dis. 2022, 16, e0010507. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, F.K.; Okuniewska, M.; Malone, C.D.; Coux, R.X.; Rio, D.C.; Lehmann, R. piRNA-mediated regulation of transposon alternative splicing in the soma and germ line. Nature 2017, 552, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Richards, G.; Cassab, A.; Bourouis, M.; Jarry, B.; Dissous, C. The normal developmental regulation of a cloned sgs3 ‘glue’ gene chromosomally integrated in Drosophila melanogaster by P element transformation. EMBO J. 1983, 2, 2137–2142. [Google Scholar] [CrossRef] [PubMed]
- Scholnick, S.B.; Morgan, B.A.; Hirsh, J. The cloned dopa decarboxylase gene is developmentally regulated when reintegrated into the Drosophila genome. Cell 1983, 34, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Spradling, A.C.; Rubin, G.M. The effect of chromosomal position on the expression of the Drosophila xanthine dehydrogenase gene. Cell 1983, 34, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Lozovsky, E.R.; Nurminsky, D.; Wimmer, E.A.; Hartl, D.L. Unexpected stability of mariner transgenes in Drosophila. Genetics 2002, 160, 527–535. [Google Scholar] [CrossRef]
- Lidholm, D.A.; Lohe, A.R.; Hartl, D.L. The transposable element mariner mediates germline transformation in Drosophila melanogaster. Genetics 1993, 134, 859–868. [Google Scholar] [CrossRef]
- DeLuca, S.Z.; Spradling, A.C. Efficient Expression of Genes in the Drosophila Germline Using a UAS Promoter Free of Interference by Hsp70 piRNAs. Genetics 2018, 209, 381–387. [Google Scholar] [CrossRef]
- Poyhonen, M.; de Vanssay, A.; Delmarre, V.; Hermant, C.; Todeschini, A.L.; Teysset, L.; Ronsseray, S. Homology-dependent silencing by an exogenous sequence in the Drosophila germline. G3 2012, 2, 331–338. [Google Scholar] [CrossRef]
- Giordano, E.; Rendina, R.; Peluso, I.; Furia, M. RNAi triggered by symmetrically transcribed transgenes in Drosophila melanogaster. Genetics 2002, 160, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Fanti, L.; Dorer, D.R.; Berloco, M.; Henikoff, S.; Pimpinelli, S. Heterochromatin protein 1 binds transgene arrays. Chromosoma 1998, 107, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Dayama, G.; Bulekova, K.; Lau, N.C. Extending and Running the Mosquito Small RNA Genomics Resource Pipeline. Methods Mol. Biol. 2022, 2509, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Palatini, U.; Contreras, C.A.; Gasmi, L.; Bonizzoni, M. Endogenous viral elements in mosquito genomes: Current knowledge and outstanding questions. Curr. Opin. Insect Sci. 2022, 49, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Baidaliuk, A.; Miesen, P.; Frangeul, L.; Crist, A.B.; Merkling, S.H.; Fontaine, A.; Lequime, S.; Moltini-Conclois, I.; Blanc, H. Non-retroviral Endogenous Viral Element Limits Cognate Virus Replication in Aedes aegypti Ovaries. Curr. Biol. 2020, 30, 3495–3506.e6. [Google Scholar] [CrossRef]
- Dezordi, F.Z.; Vasconcelos, C.; Rezende, A.M.; Wallau, G.L. In and Outs of Chuviridae Endogenous Viral Elements: Origin of a Potentially New Retrovirus and Signature of Ancient and Ongoing Arms Race in Mosquito Genomes. Front. Genet. 2020, 11, 542437. [Google Scholar] [CrossRef]
- Chirn, G.W.; Rahman, R.; Sytnikova, Y.A.; Matts, J.A.; Zeng, M.; Gerlach, D.; Yu, M.; Berger, B.; Naramura, M.; Kile, B.T.; et al. Conserved piRNA Expression from a Distinct Set of piRNA Cluster Loci in Eutherian Mammals. PLoS Genet. 2015, 11, e1005652. [Google Scholar] [CrossRef]
- Robine, N.; Lau, N.C.; Balla, S.; Jin, Z.; Okamura, K.; Kuramochi-Miyagawa, S.; Blower, M.D.; Lai, E.C. A broadly conserved pathway generates 3′UTR-directed primary piRNAs. Curr. Biol. 2009, 19, 2066–2076. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Inagaki, S.; Mituyama, T.; Kawamura, Y.; Ono, Y.; Sakota, E.; Kotani, H.; Asai, K.; Siomi, H.; Siomi, M.C. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 2009, 461, 1296–1299. [Google Scholar] [CrossRef]
- Li, X.Z.; Roy, C.K.; Dong, X.; Bolcun-Filas, E.; Wang, J.; Han, B.W.; Xu, J.; Moore, M.J.; Schimenti, J.C.; Weng, Z.; et al. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol. Cell 2013, 50, 67–81. [Google Scholar] [CrossRef]
- de Vanssay, A.; Bouge, A.L.; Boivin, A.; Hermant, C.; Teysset, L.; Delmarre, V.; Antoniewski, C.; Ronsseray, S. Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature 2012, 490, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Hermant, C.; Boivin, A.; Teysset, L.; Delmarre, V.; Asif-Laidin, A.; van den Beek, M.; Antoniewski, C.; Ronsseray, S. Paramutation in Drosophila Requires Both Nuclear and Cytoplasmic Actors of the piRNA Pathway and Induces Cis-spreading of piRNA Production. Genetics 2015, 201, 1381–1396. [Google Scholar] [CrossRef] [PubMed]
- Le Thomas, A.; Stuwe, E.; Li, S.; Du, J.; Marinov, G.; Rozhkov, N.; Chen, Y.-C.A.; Luo, Y.; Sachidanandam, R.; Fejes Toth, K.; et al. Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes Dev. 2014, 28, 1667–1680. [Google Scholar] [CrossRef] [PubMed]
- Okamura, K.; Lai, E.C. Endogenous small interfering RNAs in animals. Nat. Rev. Mol. Cell Biol. 2008, 9, 673–678. [Google Scholar] [CrossRef] [PubMed]
- Gamez, S.; Antoshechkin, I.; Mendez-Sanchez, S.C.; Akbari, O.S. The Developmental Transcriptome of Aedes albopictus, a Major Worldwide Human Disease Vector. G3 2020, 10, 1051–1062. [Google Scholar] [CrossRef] [PubMed]
- Okamura, K.; Balla, S.; Martin, R.; Liu, N.; Lai, E.C. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat. Struct. Mol. Biol. 2008, 15, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Okamura, K.; Chung, W.J.; Ruby, J.G.; Guo, H.; Bartel, D.P.; Lai, E.C. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 2008, 453, 803–806. [Google Scholar] [CrossRef] [PubMed]
- Rozhkov, N.V.; Aravin, A.A.; Zelentsova, E.S.; Schostak, N.G.; Sachidanandam, R.; McCombie, W.R.; Hannon, G.J.; Evgen’ev, M.B. Small RNA-based silencing strategies for transposons in the process of invading Drosophila species. RNA 2010, 16, 1634–1645. [Google Scholar] [CrossRef] [PubMed]
- Selvaraju, D.; Wierzbicki, F.; Kofler, R. Experimentally evolving Drosophila erecta populations may fail to establish an effective piRNA based host defense against invading P-elements. Genome Res. 2024, gr-278706. [Google Scholar] [CrossRef]
- Hoa, N.T.; Keene, K.M.; Olson, K.E.; Zheng, L. Characterization of RNA interference in an Anopheles gambiae cell line. Insect Biochem. Mol. Biol. 2003, 33, 949–957. [Google Scholar] [CrossRef]
- Keene, K.M.; Foy, B.D.; Sanchez-Vargas, I.; Beaty, B.J.; Blair, C.D.; Olson, K.E. RNA interference acts as a natural antiviral response to O’nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2004, 101, 17240–17245. [Google Scholar] [CrossRef] [PubMed]
- Samuel, G.H.; Pohlenz, T.; Dong, Y.; Coskun, N.; Adelman, Z.N.; Dimopoulos, G.; Myles, K.M. RNA interference is essential to modulating the pathogenesis of mosquito-borne viruses in the yellow fever mosquito Aedes aegypti. Proc. Natl. Acad. Sci. USA 2023, 120, e2213701120. [Google Scholar] [CrossRef] [PubMed]
- Myles, K.M.; Wiley, M.R.; Morazzani, E.M.; Adelman, Z.N. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proc. Natl. Acad. Sci. USA 2008, 105, 19938–19943. [Google Scholar] [CrossRef] [PubMed]
- Scherer, C.; Knowles, J.; Sreenu, V.B.; Fredericks, A.C.; Fuss, J.; Maringer, K.; Fernandez-Sesma, A.; Merits, A.; Varjak, M.; Kohl, A.; et al. An Aedes aegypti-Derived Ago2 Knockout Cell Line to Investigate Arbovirus Infections. Viruses 2021, 13, 1066. [Google Scholar] [CrossRef] [PubMed]
- Gestuveo, R.J.; Parry, R.; Dickson, L.B.; Lequime, S.; Sreenu, V.B.; Arnold, M.J.; Khromykh, A.A.; Schnettler, E.; Lambrechts, L.; Varjak, M.; et al. Mutational analysis of Aedes aegypti Dicer 2 provides insights into the biogenesis of antiviral exogenous small interfering RNAs. PLoS Pathog. 2022, 18, e1010202. [Google Scholar] [CrossRef] [PubMed]
- Palatini, U.; Miesen, P.; Carballar-Lejarazu, R.; Ometto, L.; Rizzo, E.; Tu, Z.; van Rij, R.P.; Bonizzoni, M. Comparative genomics shows that viral integrations are abundant and express piRNAs in the arboviral vectors Aedes aegypti and Aedes albopictus. BMC Genom. 2017, 18, 512. [Google Scholar] [CrossRef] [PubMed]
- Cecere, G. Small RNAs in epigenetic inheritance: From mechanisms to trait transmission. FEBS Lett. 2021, 595, 2953–2977. [Google Scholar] [CrossRef]
- McEnany, J.; Meir, Y.; Wingreen, N.S. piRNAs of Caenorhabditis elegans broadly silence nonself sequences through functionally random targeting. Nucleic Acids Res. 2022, 50, 1416–1429. [Google Scholar] [CrossRef]
- Cornes, E.; Bourdon, L.; Singh, M.; Mueller, F.; Quarato, P.; Wernersson, E.; Bienko, M.; Li, B.; Cecere, G. piRNAs initiate transcriptional silencing of spermatogenic genes during C. elegans germline development. Dev. Cell 2022, 57, 180–196.e7. [Google Scholar] [CrossRef]
- Wu, W.S.; Brown, J.S.; Shiue, S.C.; Chung, C.J.; Lee, D.E.; Zhang, D.; Lee, H.-C. Transcriptome-wide analyses of piRNA binding sites suggest distinct mechanisms regulate piRNA binding and silencing in C. elegans. RNA 2023, 29, 557–569. [Google Scholar] [CrossRef]
- Charlesworth, A.G.; Seroussi, U.; Lehrbach, N.J.; Renaud, M.S.; Sundby, A.E.; Molnar, R.I.; Lao, R.X.; Willis, A.R.; Woock, J.R.; Aber, M.J.; et al. Two isoforms of the essential C. elegans Argonaute CSR-1 differentially regulate sperm and oocyte fertility. Nucleic Acids Res. 2021, 49, 8836–8865. [Google Scholar] [CrossRef] [PubMed]
- Wedeles, C.J.; Wu, M.Z.; Claycomb, J.M. Protection of germline gene expression by the C. elegans Argonaute CSR-1. Dev. Cell 2013, 27, 664–671. [Google Scholar] [CrossRef]
- Cecere, G.; Hoersch, S.; O’Keeffe, S.; Sachidanandam, R.; Grishok, A. Global effects of the CSR-1 RNA interference pathway on the transcriptional landscape. Nat. Struct. Mol. Biol. 2014, 21, 358–365. [Google Scholar] [CrossRef]
- Seth, M.; Shirayama, M.; Tang, W.; Shen, E.Z.; Tu, S.; Lee, H.C.; Weng, Z.; Mello, C.C. The Coding Regions of Germline mRNAs Confer Sensitivity to Argonaute Regulation in C. elegans. Cell Rep. 2018, 22, 2254–2264. [Google Scholar] [CrossRef]
- Shirayama, M.; Seth, M.; Lee, H.C.; Gu, W.; Ishidate, T.; Conte, D., Jr.; Mello, C.C. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 2012, 150, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Olmo, R.P.; Todjro, Y.M.H.; Aguiar, E.; de Almeida, J.P.P.; Ferreira, F.V.; Armache, J.N.; de Faria, I.J.S.; Ferreira, A.G.A.; Amadou, S.C.G.; Silva, A.T.S.; et al. Mosquito vector competence for dengue is modulated by insect-specific viruses. Nat. Microbiol. 2023, 8, 135–149. [Google Scholar] [CrossRef]
- Barucci, G.; Cornes, E.; Singh, M.; Li, B.; Ugolini, M.; Samolygo, A.; Didier, C.; Dingli, F.; Loew, D.; Quarato, P.; et al. Small-RNA-mediated transgenerational silencing of histone genes impairs fertility in piRNA mutants. Nat. Cell Biol. 2020, 22, 235–245. [Google Scholar] [CrossRef]
- Montgomery, B.E.; Vijayasarathy, T.; Marks, T.N.; Cialek, C.A.; Reed, K.J.; Montgomery, T.A. Dual roles for piRNAs in promoting and preventing gene silencing in C. elegans. Cell Rep. 2021, 37, 110101. [Google Scholar] [CrossRef]
- Reed, K.J.; Svendsen, J.M.; Brown, K.C.; Montgomery, B.E.; Marks, T.N.; Vijayasarathy, T.; Parker, D.M.; Nishimura, E.O.; Updike, D.L.; Montgomery, T.A. Widespread roles for piRNAs and WAGO-class siRNAs in shaping the germline transcriptome of Caenorhabditis elegans. Nucleic Acids Res. 2020, 48, 1811–1827. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Barik, T.K.; Johnson, R.M.; Rasgon, J.L. In vitro and in vivo host range of Anopheles gambiae densovirus (AgDNV). Sci. Rep. 2015, 5, 12701. [Google Scholar] [CrossRef]
- Johnson, R.M.; Rasgon, J.L. Densonucleosis viruses (‘densoviruses’) for mosquito and pathogen control. Curr. Opin. Insect Sci. 2018, 28, 90–97. [Google Scholar] [CrossRef]
- Viswanatha, R.; Mameli, E.; Rodiger, J.; Merckaert, P.; Feitosa-Suntheimer, F.; Colpitts, T.M.; Mohr, S.E.; Hu, Y.; Perrimon, N. Bioinformatic and cell-based tools for pooled CRISPR knockout screening in mosquitos. Nat. Commun. 2021, 12, 6825. [Google Scholar] [CrossRef]
- Feng, X.; Lopez Del Amo, V.; Mameli, E.; Lee, M.; Bishop, A.L.; Perrimon, N.; Gantz, V.M. Optimized CRISPR tools and site-directed transgenesis towards gene drive development in Culex quinquefasciatus mosquitoes. Nat. Commun. 2021, 12, 2960. [Google Scholar] [CrossRef]
- Frokjaer-Jensen, C.; Jain, N.; Hansen, L.; Davis, M.W.; Li, Y.; Zhao, D.; Rebora, K.; Millet, J.R.M.; Liu, X.; Kim, S.K.; et al. An Abundant Class of Non-coding DNA Can Prevent Stochastic Gene Silencing in the C. elegans Germline. Cell 2016, 166, 343–357. [Google Scholar] [CrossRef]
- Zhang, D.; Tu, S.; Stubna, M.; Wu, W.S.; Huang, W.C.; Weng, Z.; Lee, H.-C. The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science 2018, 359, 587–592. [Google Scholar] [CrossRef]
- Aljohani, M.D.; El Mouridi, S.; Priyadarshini, M.; Vargas-Velazquez, A.M.; Frokjaer-Jensen, C. Engineering rules that minimize germline silencing of transgenes in simple extrachromosomal arrays in C. elegans. Nat. Commun. 2020, 11, 6300. [Google Scholar] [CrossRef]
- Makeyeva, Y.V.; Shirayama, M.; Mello, C.C. Cues from mRNA splicing prevent default Argonaute silencing in C. elegans. Dev. Cell 2021, 56, 2636–2648.e4. [Google Scholar] [CrossRef] [PubMed]
- Grishok, A.; Sinskey, J.L.; Sharp, P.A. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev. 2005, 19, 683–696. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lau, N.C.; Macias, V.M. Transposon and Transgene Tribulations in Mosquitoes: A Perspective of piRNA Proportions. DNA 2024, 4, 104-128. https://doi.org/10.3390/dna4020006
Lau NC, Macias VM. Transposon and Transgene Tribulations in Mosquitoes: A Perspective of piRNA Proportions. DNA. 2024; 4(2):104-128. https://doi.org/10.3390/dna4020006
Chicago/Turabian StyleLau, Nelson C., and Vanessa M. Macias. 2024. "Transposon and Transgene Tribulations in Mosquitoes: A Perspective of piRNA Proportions" DNA 4, no. 2: 104-128. https://doi.org/10.3390/dna4020006
APA StyleLau, N. C., & Macias, V. M. (2024). Transposon and Transgene Tribulations in Mosquitoes: A Perspective of piRNA Proportions. DNA, 4(2), 104-128. https://doi.org/10.3390/dna4020006





