Abstract
Repetitive DNA sequences are abundant in the human genome and can adopt alternative (i.e., non-B) DNA structures. These sequences contribute to diverse biological functions, including genomic instability. Previously, we found that Z-DNA-, H-DNA- and cruciform DNA-forming sequences are mutagenic, implicating them in cancer etiology. These sequences can stimulate the formation of DNA double-strand breaks (DSBs), causing deletions via cleavage by the endonuclease ERCC1-XPF. Interestingly, the activity of ERCC1-XPF in H-DNA-induced mutagenesis is nucleotide excision repair (NER)-dependent, but its role in Z-DNA-induced mutagenesis is NER-independent. Instead, Z-DNA is processed by ERCC1-XPF in a mechanism dependent on the mismatch repair (MMR) complex, MSH2-MSH3. These observations indicate distinct mechanisms of non-B-induced genomic instability. However, the roles of NER and MMR proteins, as well as additional nucleases (CtIP and MRE11), in the processing of cruciform DNA remain unknown. Here, we present data on the processing of cruciform-forming short inverted repeats (IRs) by DNA repair proteins using mammalian cell-based systems. From this pilot study, we show that, in contrast to H-DNA and Z-DNA, short IRs are processed in a NER- and MMR-independent manner, and the nucleases CtIP and MRE11 suppress short IR-induced genomic instability in mammalian cells.
1. Introduction
Over a dozen alternative or non-B DNA structures have been identified since the 1950s [1,2,3,4]. These structure-forming sequences have documented roles in physiological processes, such as transcription, translation, replication, chromatin structure and evolution; however, they also play roles in pathophysiological processes such as genomic instability, which can contribute to disease etiology [2,5,6,7,8,9,10,11]. Non-B DNA-forming sequences are abundant in the human genome, enriched at ‘hotspots’ of genomic instability in human cancer, and have been shown to be associated with human disease [12,13,14,15,16]. An overrepresentation of such sequences in frequently mutated loci also suggests that they are determinants of mutagenesis [10]. Hence, the abundance of repetitive DNA sequences that can adopt non-B DNA structures and their roles in normal physiological and pathophysiological conditions warrants further study, particularly in regard to the mechanisms underlying their function and mutagenic processing. In this pilot study, we focus on determining the proteins involved in the mutagenic processing of cruciform DNA relative to our published mechanisms on the mutagenic processing of Z-DNA and H-DNA (Figure 1).
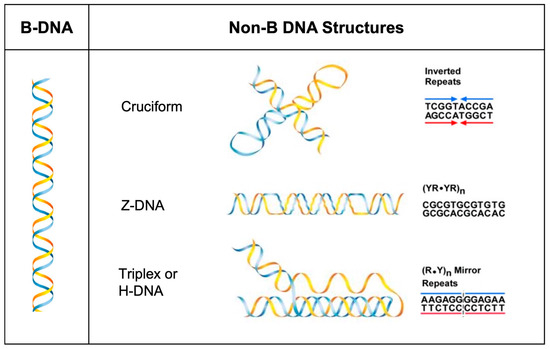
Figure 1.
Schematic of non-B DNA structures. Left panel: canonical B-DNA duplex. Right panel: non-B DNA structures with the characteristic repetitive motifs at which they form. Listed here are cruciform DNA (inverted repeats), Z-DNA [alternating purines and pyrimidines; (YR-YR)n], triplex or H-DNA [mirror repeats; (R.Y)n]. The blue and yellow color indicate the complementary strands of DNA (Adapted from [5]).
We have previously shown that Z-DNA-forming sequences induce large deletions and complex rearrangements in yeast, mammalian cells and mouse chromosomes via the stimulation of DNA double-strand break (DSB) formation. This Z-DNA-induced genomic instability is facilitated by the endonuclease ERCC1-XPF, the recruitment of which is dependent on the mismatch repair (MMR) protein complex, MSH2-MSH3.
In addition, we have previously shown that H-DNA-forming sequences from the human c-MYC promoter can also stimulate the formation of DSBs, leading to large deletions in yeast, mammalian cells, and mouse chromosomes. Microhomologies at the deletion breakpoints indicate microhomology-mediated end-joining (MMEJ) mechanisms in the formation of the deletions [17,18,19]. Interestingly, we have identified both replication-dependent and replication-independent mechanisms of H-DNA-induced genomic instability. We have shown that ERCC1-XPF and XPG process H-DNA-forming sequences in a nucleotide excision repair (NER)-dependent manner, leading to large deletions via the formation of DSBs. This endonuclease-based cleavage mechanism could explain the replication-independent mechanisms of H-DNA-induced genomic instability [17]. In addition, we found that the absence of the flap endonuclease 1 (FEN1) protein led to an increase in H-DNA-induced mutagenesis, suggesting that FEN1 protects the genome from H-DNA obstructions during replication by cleaving this mutagenic structure.
With regard to inverted repeats (IRs), much of the research has focused on long IRs (>100 bp), which can stimulate deletion events in prokaryotes and yeast [20,21]. Similarly, long IRs have been studied extensively with regard to their contributions to human disease. However, a study of ~20,000 translocation breakpoints in human cancer genomes revealed IRs with stem lengths between 10–30 bps (short IRs) to be enriched within a 200 bp region around the breakpoints [15,22], implicating them in cancer etiology. We have also shown that a short IR sequence of 29 bp in length can stimulate DSBs, leading to deletions containing microhomologies at the breakpoint junctions in yeast and mammalian cells [22]. The DSBs can occur in a replication-independent manner via cleavage of the cruciform structures by the enzyme ERCC1-XPF or in a replication-dependent manner via fork stalling [22].
Z-DNA, H-DNA, and hairpin/cruciform DNA are therefore biologically functional motifs and intrinsic sources of genomic instability in different organisms and cell types, and we have found that all these sequences/structures are substrates for the enzyme complex ERCC1-XPF [17,22,23,24]. Interestingly, in addition to Z-DNA, H-DNA, and hairpin/cruciform DNA, G-quadruplexes, R-loops and D-loops are also substrates for the enzyme complex ERCC1-XPF [24]; however, for this study, we focus on the roles of different DNA repair proteins in the mutagenic processing of cruciform DNA, comparing these preliminary findings with our previously published mechanisms of H-DNA and Z-DNA processing.
The role of ERCC1-XPF in H-DNA-induced genomic instability is NER dependent, while the role associated with Z-DNA-induced mutagenesis is NER independent (but dependent on MSH2-MSH3). However, the roles of NER proteins on hairpins have shown conflicting results in both bacterial and mammalian cells, as reported by different groups, where their effects on CAG repeats resulted in either an increase or a decrease in deletion events [24,25,26,27]. Additionally, MMR proteins can also bind/remove mismatch-containing small cruciform/hairpins, though not those formed by perfect IRs [28,29]. However, the role of MMR proteins in CAG repeat-mediated instability has shown conflicting results in mice and human systems, where, in the presence of MMR proteins, contraction events at CAG repeats were seen in humans [26] and expansion events were seen in mice [30]. Interestingly, experiments in different biological systems indicate that MMR proteins may play a role in trinucleotide repeat expansions by a mechanism not related to the classical strand slippage model observed during non-expandable microsatellite replication [31,32]. More recently, using in vitro assays, it was also shown that MMR protein complexes can decrease the levels of cruciform DNA [33].
We have previously shown that non-B DNA structures induce genomic instability via the stimulation of DSB formations, resulting in large deletions [17,22,23]. These large deletions show microhomologies at the breakpoint junctions, suggesting a role of MMEJ in processing the DSBs induced by non-B DNA structures. An important step involved in the initiation of this pathway is DNA end resection at the site of a DSB catalyzed by the CtIP and MRE11 proteins [34]. Interestingly, apart from their roles in homologous recombination (HR), MRE11 and CtIP have also been shown to mediate MMEJ repair mechanisms [35]. In addition to its role in resection, MRE11 can also function as an endonuclease and cleave hairpin loops in vitro [36,37,38]. Interestingly, other groups have demonstrated endonuclease-mediated cleavage of single-stranded DNA next to the hairpins by the yeast homolog of CtIP, i.e., Sae2 [38,39]. While the CtIP protein augments the function of MRE11, studies on its endonuclease functions are inconsistent [40]. Relevant to non-B DNA processing, CtIP and MRE11 are required to preserve stability at AT-rich common fragile sites and Alu-IRs [41], demonstrating that CtIP and MRE11 could have diverse roles in processing structures formed at IRs. In addition, the small loop within the cruciform structure may also provide a substrate for CtIP and MRE11. Hence, while CtIP and MRE11 may process IR-induced DSBs via HR or MMEJ, they could also be involved in cleaving the structures formed at IRs.
Together, these studies indicate that, while ERCC1-XPF can cleave a variety of non-B DNA structures, distinct DNA repair mechanisms/proteins are involved in their mutagenic processing. Since the mechanisms involved in the mutagenic processing of short IRs are not clear, we performed a preliminary analysis of different DNA repair proteins and their roles in short IR-induced genomic instability in human cells. Thus, in this pilot study, we sought to ascertain the involvement of NER and MMR proteins and additional DNA repair proteins (CtIP and MRE11) on short IR-induced genomic instability.
2. Materials and Methods
2.1. Mutagenesis Assays in Mammalian Cell Lines
We utilized mutation-reporter shuttle vectors to study non-B DNA-induced mutagenesis [42], which can replicate in both bacterial and mammalian cells. For this study, we used the pSP189-based mutation-reporters containing the SV40 origin of replication and T antigen, which allows for replication in mammalian cells, along with the pBR327 origin of replication, which allows for replication in bacterial cells. This reporter also contains the supF mutation-reporter gene, which facilitates the use of bacterial strains carrying amber mutations as indicators of the functional activity of the supF gene via blue−white screening [42,43,44]. In this study we have cloned a 29-bp IR sequence capable of forming cruciform DNA structures (IR) into the supF-containing reporter vector, pSP189, using standard cloning protocols. The supF-containing pSupFG1 vector was used as a control (B-DNA) reporter (Figure 2a).
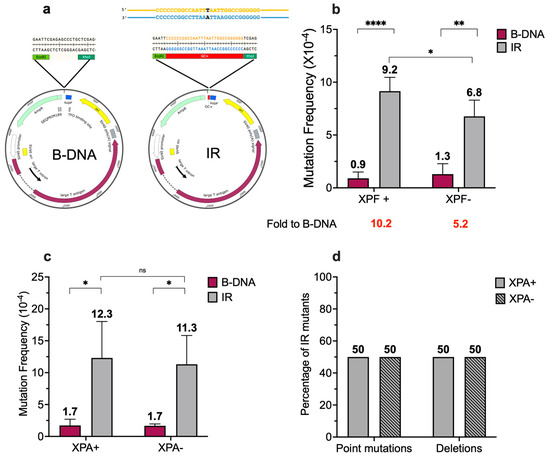
Figure 2.
Short IR-induced mutagenesis decreases in the absence of ERCC1-XPF and remains unchanged in the presence or absence of XPA. (a) Schematic of the control (B-DNA) and 29-bp GC-rich (IR) plasmids used in this study. The supF-containing pSupFG1 vector was used as a control reporter ((B-DNA) and a 29-bp IR sequence capable of forming a cruciform DNA structure (IR) was cloned into the supF-containing reporter vector, pSP189, using standard cloning protocols. There are two stretches of IRs (14-bp long) that form the stem of the cruciform structure (regions in blue and yellow) and a loop that consists of a single nucleotide (highlighted in black). (b) Mutation frequencies were measured in human XPF-proficient fibroblast cell lines in the presence of control siRNA. Mutation reporters containing control (B-DNA) sequences or short, perfect inverted repeat (IR) sequences were transfected into the human XPF-proficient cells at T48 and collected 48 h later at T96. Mutation frequencies were calculated as the ratio of white colonies to the total number of colonies. Experiments were performed in triplicate, data are expressed as mean ± SD, and a 2-way ANOVA with Šidák post hoc test, * p < 0.05, ** p < 0.01, **** p < 0.0001 was used for statistical analysis. (c) Mutation frequencies were measured in human XPA-proficient and deficient cell lines. Mutation reporters containing control (B-DNA) sequences or short inverted repeat (IR) sequences were transfected into the cells and isolated 48 h post-transfection. Mutation frequencies were calculated as the ratio of white (mutant) colonies to the total number of colonies (blue plus white). Experiments were performed in triplicate. Data are expressed as mean ± SD, and a 2-way ANOVA with Šidák posthoc test, * p < 0.05, was used for statistical analysis. (d) Percentage distribution of different types of mutants. Mutants are characterized as point mutations and deletions. Gray solid bars represent samples from XPA-proficient cells, and gray patterned bars represent samples from XPA-proficient cells. (Adapted from [45]).
2.2. Cell Lines
Human XPA-deficient (XPA2OSSV) and XPA-proficient (SV40-immortalized human fibroblasts complemented with the XPA gene) [46,47] were maintained in Dulbecco’s Modified Eagle’s Medium with supplements [DMEM + 10% heat-inactivated fetal bovine serum (HI-FBS) + 1% Penicillin-Streptomycin]. Human XPF-deficient and XPF-proficient [48] cells were maintained in Dulbecco’s Modified Eagle’s Medium with supplements [DMEM + 10% fetal bovine serum (FBS) + 1% Penicillin-Streptomycin].
2.3. siRNA-Mediated Knockdown of DNA Repair Proteins and Mutation Reporter Transfection
We performed siRNA-mediated transient knockdowns to study the effects of different proteins on short IR-induced mutagenesis. Two transfections were performed with the ON-TARGET plus siRNA SMARTpool for human (MSH2, CtIP, or MRE11) and non-targeting control SMARTpool. A reverse transfection was performed using 25–40 nM siRNA in OptiMEM Reduced-Serum Medium. The 48 h time point was selected because it gave >80% knockdown of protein levels at the time of mutation reporter transfection into the human cells. Next, a forward transfection of siRNA was performed with 2.5 µg of the IR- or B-DNA-containing reporters with the GenePORTER transfection reagent (Genlantis Inc., now AMSBIO, San Diego, CA, USA). Samples were collected at T48 and T96 (48 h after the second transfection) to verify protein knockdown by Western blotting using the following antibodies: anti-MSH2, anti-CtIP, anti-MRE11 and anti-beta actin as loading control (Table 1). Transfected reporters were collected at T96 by the alkaline lysis method using a QIAPrep Spin Miniprep kit (Qiagen, Germantown, MD, USA) with slight modifications to the protocol. The extracted mutation reporters were DpnI digested and purified, then used for the transformation of electrocompetent MBM7070 cells to determine mutation frequency via blue−white screening.

Table 1.
Antibodies used in this study.
2.4. Mutation Frequency and Mutation Spectra Analyses
Mutation frequencies were determined by dividing the total number of white (mutant) colonies by the total number of blue (wild-type), and white (mutant) colonies counted on X-Gal, IPTG and carbenicillin agar plates. Experiments were performed in triplicate, with at least 20,000 colonies being counted for each replicate. Statistical significance was calculated using a two-way ANOVA test.
Mutation spectra were evaluated by sequencing randomly selected white mutant colonies and a control blue wild-type colony. Mutation reporter isolation was performed using the QIAPrep Spin Miniprep kit. The isolated reporters were sequenced by Sanger’s sequencing method using primers specific to a region around the mutation-reporter gene and the IR or control B-DNA sequence (seqPriim189 CAAAAAAGGGAATAAGGGCG). The obtained sequences were used to characterize the types of mutations around the IR sequence and a similar region in the control B-DNA reporter.
3. Results and Discussion
Based on substrate preferences, short IRs may be recognized and processed by NER, MMR, MMEJ, and/or HR repair proteins/pathways. Studies to determine which proteins are involved are ongoing, and the initial findings are presented below.
It is known that NER is required for the repair of helix-distorting bulky adducts [49,50], thereby validating its potential role in processing non-B DNA structures, as seen with the mutagenic processing of H-DNA [17]. We have previously shown that ERCC1-XPF (a key nuclease in the NER pathway) is involved in short IR-induced genomic instability in yeast and mammalian systems [22]. In line with our previous observation in mammalian cells, we found that short IRs are mutagenic in human cells, with a 10.2-fold increase in short IR-induced mutagenesis over that of control B-DNA in wild-type human XPF-proficient fibroblasts (Figure 2b). Further, we observed a decrease in the fold induction (5.2-fold) of short IR-induced mutagenesis over that of control B-DNA in the human XPF-deficient fibroblasts, accompanied by a decrease in mutation frequency in the absence of XPF (9.2 × 10−4 vs. 6.8 × 10−4, p < 0.1) (Figure 2b, gray bars), indicating a role for ERCC1-XPF in short IR-induced mutagenesis in human cells. Interestingly, while short IRs induced mutations in both the presence and absence of XPA (a critical NER recognition and verification protein), the presence (12.3 × 10−4) or absence of XPA (11.3 × 10−4) did not impact the short IR-induced mutagenesis (Figure 2c, gray bars).
We did not detect any significant differences in the mutation spectra associated with the short IR-induced genomic instability in human XPA-proficient vs. deficient cells, with ~50% deletions and ~50% point mutations observed in both cell lines (Figure 2d). Thus, it appears that, unlike H-DNA [17], functional NER is not required for the mutagenic processing of short IRs in human cells.
Since we have previously shown that the cleavage of Z-DNA by ERCC1-XPF was dependent on its interaction with MSH2-MSH3, we evaluated the effect of MSH2 (an essential MMR protein) in the presence or absence of XPF on short IR-induced genomic instability. Upon knockdown of MSH2 in XPF-proficient cells (Figure S1a,b) we found that the short IR-induced genomic instability was not different in the presence (9.5 × 10−4) or absence (9.9 × 10−4) of MSH2 (Figure 3a, gray bars). This is not unexpected, given previous reports indicating that MMR proteins bind more specifically to mismatch-containing hairpins/cruciforms rather than to perfect repeats [29]. Thus, it appears that, unlike Z-DNA [23], MSH2 is not required for the mutagenic processing of short IRs in human cells.
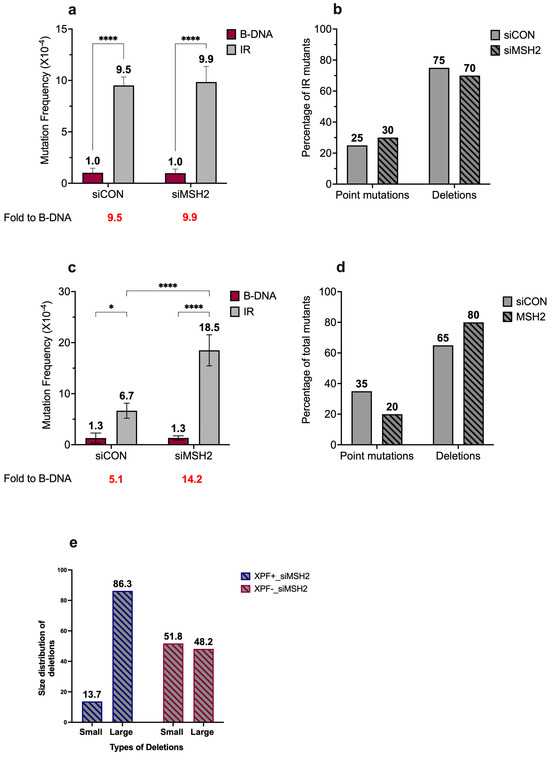
Figure 3.
Short IR-induced mutagenesis remains unchanged in the absence of MSH2 and increases in the absence of both MSH2 and XPF. (a) Mutation frequencies were measured in human XPF-proficient cell lines in the presence of siCON or siMSH2. Mutation reporters containing control (B-DNA) sequences or short inverted repeat (IR) sequences were transfected into the human XPF-proficient cells at T48 and collected 48 h later at T96. Mutation frequencies were calculated as the ratio of white colonies to the total number (blue plus white) of colonies counted. Experiments were performed in triplicate, data are expressed as mean ± SD, and a 2-way ANOVA with Šidák posthoc test, **** p < 0.0001 was used for statistical analysis. (b) Percentage distribution of different types of mutants in human XPF-proficient cell lines following treatment with siCON or siMSH2. Mutants are characterized as point mutations and deletions. Gray solid bars represent siCON-treated samples, while gray patterned bars represent siMSH2-treated samples. (c) Mutation frequencies were measured in human XPF-deficient cell lines in the presence of siCON or siMSH2. Mutation reporters containing control (B-DNA) sequences or short inverted repeat (IR) sequences were transfected into the human XPF-deficient cells at T48 and collected 48 h later at T96. Mutation frequencies were calculated as the ratio of white colonies to the total number (blue plus white) of colonies counted. Experiments were performed in triplicate, data are expressed as mean ± SD, and a 2-way ANOVA with Šidák posthoc test, * p < 0.05, **** p < 0.0001 was used for statistical analysis. (d) Percentage distribution of different types of mutants in human XPF-deficient cell lines following treatment with siCON or siMSH2. Mutants are characterized as point mutations and deletions. Gray solid bars represent siCON-treated samples, and gray patterned bars represent siMSH2-treated samples. (e) Comparative analysis of short IR-induced deletions from human XPF-proficient and XPF-deficient cells treated with siMSH2. The deletions were categorized as small deletions (<100 bp) and large deletions (>100 bp). Blue patterned bars correspond to XPF-proficient cells treated with siMSH2, and red patterned bars represent human XPF-deficient cells treated with siMSH2. (Adapted from [45]).
Upon analysis of the mutation spectra, we did not observe any considerable differences in the distribution of mutation types associated with the short IR-induced genomic instability in the presence or absence of MSH2, with ~25% point mutations and ~75% deletions observed in mutants from both the control and MSH2-depleted cells (Figure 3b). However, a difference was seen in the position of the breakpoint junctions around the IR sequence in the presence of MSH2 compared to those detected in the absence of MSH2. In the presence of MSH2, most of the breakpoint junctions were located in the tip of the loop region within the IR sequence, resulting in partial loss of the IR sequence. In contrast, in the absence of MSH2, ~50% of the breakpoint junctions were mapped outside the IR sequence, resulting in the complete loss of the IR sequence (Figure S2).
MSH2 has a variety of functions in several DNA repair pathways, such as MMR, HR, single-strand annealing (SSA), etc. [51,52,53,54], interacting, stimulating or inhibiting the activity of different proteins [52]. For example, MutSβ (MSH2-MSH3) can stimulate the activity of the SMX trinuclease in Holliday junction (HJ) resolution [55]. MutSα (MSH2-MSH6) can interact with the BLM helicase and promote dissolution of HJs [56]. Because HJs are structurally similar to cruciform DNA, we speculate that MSH2, via its interacting partners MSH3, MSH6, or other interacting proteins, may regulate the processing of structures formed at short IRs. Therefore, the presence or absence of MSH2 could lead to differential processing around the IR sequence, leading to alterations in the mutation spectra.
Surprisingly, upon the knockdown of MSH2 in XPF-deficient cells (Figure S1a,c), we observed that the absence of both MSH2 and XPF led to an increase in short IR-induced genomic instability (6.7 × 10−4 vs. 18.5 × 10−4) (Figure 3c, gray bars). We also observed a minor shift in the mutation spectra associated with short IR-induced genomic instability in the presence or absence of MSH2, with ~65% deletions and ~35% point mutations in mutants from the wild-type cells, while there were ~80% deletions and ~20% point mutations in mutants from the MSH2-depleted cells (Figure 3d). Interestingly, we also observed an alteration in the size of the deletions in MSH2-depleted samples in the presence or absence of XPF. The deletions were grouped as >100 bp or <100 bp, and, in the presence of XPF, ~14% of deletions were <100 bp while the remaining ~86% were >100 bp (blue dashed bars). In contrast, ~52% of deletions were <100 bp and the remaining ~48% were >100 bp in the absence of XPF (red dashed bars) (Figure 3e). Mutants from the control B-DNA samples predominantly contained point mutations in the presence or absence of MSH2.
The differences in the sizes of the deletions generated in the human XPF-proficient or deficient cells in the absence of MSH2 could be attributed to the different mechanisms that might be involved in processing the short IRs as a result of the interplay between ERCC1-XPF and MSH2. Therefore, under these conditions, one might speculate on the roles of additional proteins/nucleases in the mutagenic processing of short IRs. Because IRs can adopt cruciform structures, they may be susceptible to processing by enzymes that cleave around the single-stranded regions (loops), as seen with ERCC1-XPF-mediated cleavage of the 29-bp IR substrate around the loop region indicating a ‘center break’ mechanism [22,57]. In addition, the presence of four-way junctions in the cruciform structure, which are remarkably similar to an HJ intermediate, could make these potential substrates for HJ resolvases (e.g., GEN1, SLX1, MUS81) [58,59,60] in a ‘resolution’-type mechanism. Interestingly, Inagaki et al. (2013) have shown that palindrome-mediated translocations associated with the Palindromic AT-rich repeat (PATRR) entail two sequential cleavage reactions by the GEN1 and Artemis proteins, suggesting a mechanism that involves the resolution of the four-way junction of the cruciform structure [61]. Additionally, Kaushal et al. (2019) have shown that the fragility linked to the Flex1 long AT-rich region of the common fragile site FRA16D could be attributed to its capacity to adopt a cruciform structure. The fragility involves the yeast Mus81-Mms4 and Slx1-4/Rad1-10 protein complexes [62]. Since these proteins play a role in HJ resolution and the processing of cruciform structures formed at long IRs, additional studies must be undertaken to determine their roles and those of other proteins in short IR-induced genomic instability.
Previously, we demonstrated a role for the ERCC1-XPF nuclease in the mutagenic processing of short IRs [22]. Here, we extended our study to include the CtIP and MRE11 nucleases (critical in DSB repair processing) [63,64]. Upon knockdown of CtIP and MRE11 in XPF-proficient cells (Figure S3a,b), we found that the short IR-induced genomic instability increased in the absence of CtIP (9.9 × 10−4 vs. 27.5 × 10−4) and MRE11 (9.9 × 10−4 vs. 26.2 × 10−4) (Figure 4a,c). This suggests a role for CtIP and MRE11 in suppressing short IR-induced genomic instability. Upon analysis of the mutation spectra, we observed a shift in the mutation types between the wild-type cells and the CtIP- or MRE11-depleted cells. When CtIP or MRE11 were depleted, 100% of the mutants contained deletion events, whereas the wild-type cells had ~75% deletion events and ~25% point mutations (Figure 4b,d).
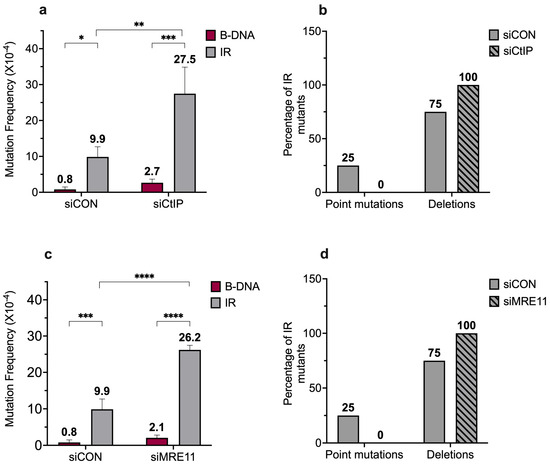
Figure 4.
Short IR-induced mutagenesis increases in the absence of CtIP and MRE11. (a) Mutation frequencies were measured in human XPF-proficient cell lines in the presence of siCON or siCtIP. Mutation reporters containing control (B-DNA) sequences or short inverted repeat (IR) sequences were transfected into the human XPF-proficient cells at T48, being collected 48 h later at T96. Mutation frequencies were calculated as the ratio of white colonies to the total number of colonies. Experiments were performed in triplicate, data are expressed as mean ± SD, and a 2-way ANOVA with Šidák post hoc test, * p < 0.05, ** p < 0.01, *** p < 0.001 was used for statistical analysis. (b) Percentage distribution of different types of mutants in human XPF-proficient cells following treatment with siCON or siCtIP. Mutants are characterized as point mutations and deletions. Gray solid bars represent siCON-treated samples and gray patterned bars represent siCtIP-treated samples. (c) Mutation frequencies were measured in human XPF-proficient cell lines in the presence of siCON or siMRE11. Mutation reporters containing control (B-DNA) sequences or short inverted repeat (IR) sequences were transfected into the human XPF-proficient cells at T48 and collected 48 h later at T96. Mutation frequencies were calculated as the ratio of white colonies to the total number of colonies. Experiments were performed in triplicate, data are expressed as mean ± SD and a 2-way ANOVA with Šidák post hoc test, *** p < 0.001, **** p < 0.0001 was used for statistical analysis. (d) Percentage distribution of different types of mutants in human XPF-proficient cells following treatment with siCON or siMRE11. Mutants are characterized as point mutations and deletions. Gray solid bars represent siCON-treated samples, and gray patterned bars represent siMRE11-treated samples. (Adapted from [45]).
As mentioned previously, both CtIP and MRE11 have varied roles in the DNA damage repair processes; in this case, CtIP and MRE11 could favor the repair of the short IR-induced DSBs by the error-free HR pathway. Therefore, these proteins could potentially restore stability in short IR-induced DSBs. Because these events are error-free, they will not be captured by our blue−white screening assay. However, the presence of short IR-induced deletions in the absence of CtIP and MRE11 is intriguing. Since both CtIP and MRE11 are regulators of the key step of 5′-3′ resection, the depletion of these proteins could affect resection at the short IR-induced DSBs. Since deletion events may require resection at the DSBs, it is possible that other nucleases are able to compensate, in part, for the resection function of MRE11. One such protein is EXO1, a 5′-3′ exonuclease involved in revealing long tracts of ssDNA after initial resection by MRE11/CtIP [65,66].
Interestingly, it has been shown that the yeast homologs of MRE11 and EXO1, i.e., Mre11 and Exo1, have overlapping functions, suggesting redundancy in the resection functions and defects in Mre11 being compensated for by overexpression of ExoI [67,68]. Studies in yeast have also found roles for ExoI in repair via MMEJ mechanisms [69]. Perhaps in the absence of CtIP/MRE11, EXO1 could, to a limited extent, facilitate resection in regard to the short IR-induced DSBs and promote the more mutagenic MMEJ pathway, leading to deletions observed in the absence of CtIP and MRE11.
Our study provides evidence to suggest that short IRs may be processed in a manner distinct from that of H-DNA and Z-DNA-forming sequences, i.e., the mutagenic processing of short IRs appears to be independent of both a functional NER pathway and MSH2, which is required for functional MMR.
4. Concluding Remarks
Previously, we demonstrated that the mutagenic processing of different non-B DNA structures involves distinct mechanisms, with H-DNA being processed by NER and Z-DNA being processed by the NER nuclease ERCC1-XPF, which requires the MMR complex MSH2-MSH3 for the recruitment of ERCC1-XPF to the Z-DNA region [17,23,70]. This study provides preliminary evidence of the roles of different DNA repair proteins in the mutagenic processing of cruciform-forming short IRs. Due to the complex interplay between different DNA repair proteins on this structure, several protein/pathways are likely involved in its mutagenic processing. Future studies are necessary to define the exact mechanism(s) involved in short IR-induced genomic instability. Understanding the mechanisms responsible for DNA structure-induced genomic instability and their roles in disease etiology will allow for the development of therapeutics to treat and/or prevent diseases of genetic instability, such as cancer and neurodegenerative disorders.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/dna4020008/s1, Figure S1: Expression of XPF in XPF proficient and deficient cell lines and knockdown of MSH2 in XPF-proficient cells, Figure S2: Schematic distribution of short IR-induced deletions from human XPF-proficient cells treated with: (a) siCON (b) siMSH2, Figure S3: Knockdown of CtIP and MRE11 in XPF-proficient cells.
Author Contributions
P.M.: co-conceptualization, writing—original draft preparation, writing—review and editing. K.M.V.: co-conceptualization, writing—review and editing, funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by an NIH/NCI grant, R01CA093729 to K.M.V.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Acknowledgments
We would like to thank Richard Wood for providing the cell lines and Nathan Subramaniam for help with the mutation frequency data collection.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Sinden, R.R. DNA Structure and Function; Academic Press: San Diego, CA, USA, 1994. [Google Scholar]
- Wang, G.; Vasquez, K.M. Non-B DNA structure-induced genetic instability. Mutat. Res. 2006, 598, 103–119. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, M.; Kaushik, S.; Roy, K.; Singh, A.; Mahendru, S.; Kumar, M.; Chaudhary, S.; Ahmed, S.; Kukreti, S. A bouquet of DNA structures: Emerging diversity. Biochem. Biophys. Rep. 2016, 5, 388–395. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Choi, J.; Majima, T. Conformational changes of non-B DNA. Chem. Soc. Rev. 2011, 40, 5893–5909. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Bacolla, A.; Wang, G.; Vasquez, K.M. Non-B DNA structure-induced genetic instability and evolution. Cell Mol. Life Sci. 2010, 67, 43–62. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Vasquez, K.M. Effects of Replication and Transcription on DNA Structure-Related Genetic Instability. Genes 2017, 8, 17. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Vasquez, K.M. Impact of alternative DNA structures on DNA damage, DNA repair, and genetic instability. DNA Repair 2014, 19, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Vasquez, K.M. Dynamic alternative DNA structures in biology and disease. Nat. Rev. Genet. 2023, 24, 211–234. [Google Scholar] [CrossRef] [PubMed]
- Bacolla, A.; Wells, R.D. Non-B DNA conformations, genomic rearrangements, and human disease. J. Biol. Chem. 2004, 279, 47411–47414. [Google Scholar] [CrossRef] [PubMed]
- Georgakopoulos-Soares, I.; Morganella, S.; Jain, N.; Hemberg, M.; Nik-Zainal, S. Noncanonical secondary structures arising from non-B DNA motifs are determinants of mutagenesis. Genome Res. 2018, 28, 1264–1271. [Google Scholar] [CrossRef] [PubMed]
- Georgakopoulos-Soares, I.; Chan, C.S.Y.; Ahituv, N.; Hemberg, M. High-throughput techniques enable advances in the roles of DNA and RNA secondary structures in transcriptional and post-transcriptional gene regulation. Genome Biol. 2022, 23, 159. [Google Scholar] [CrossRef]
- Bacolla, A.; Wojciechowska, M.; Kosmider, B.; Larson, J.E.; Wells, R.D. The involvement of non-B DNA structures in gross chromosomal rearrangements. DNA Repair 2006, 5, 1161–1170. [Google Scholar] [CrossRef]
- Wells, R.D. Non-B DNA conformations, mutagenesis and disease. Trends Biochem. Sci. 2007, 32, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Bacolla, A.; Wells, R.D. Non-B DNA conformations as determinants of mutagenesis and human disease. Mol. Carcinog. 2009, 48, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Bacolla, A.; Tainer, J.A.; Vasquez, K.M.; Cooper, D.N. Translocation and deletion breakpoints in cancer genomes are associated with potential non-B DNA-forming sequences. Nucleic. Acids. Res. 2016, 44, 5673–5688. [Google Scholar] [CrossRef] [PubMed]
- Mirkin, S.M. DNA structures, repeat expansions and human hereditary disorders. Curr. Opin. Struct. Biol. 2006, 16, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wang, G.; Del Mundo, I.M.; McKinney, J.A.; Lu, X.; Bacolla, A.; Boulware, S.B.; Zhang, C.; Zhang, H.; Ren, P.; et al. Distinct Mechanisms of Nuclease-Directed DNA-Structure-Induced Genetic Instability in Cancer Genomes. Cell Rep. 2018, 22, 1200–1210. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Carbajal, S.; Vijg, J.; DiGiovanni, J.; Vasquez, K.M. DNA structure-induced genomic instability in vivo. J. Natl. Cancer Inst. 2008, 100, 1815–1817. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Vasquez, K.M. Naturally occurring H-DNA-forming sequences are mutagenic in mammalian cells. Proc. Natl. Acad. Sci. USA 2004, 101, 13448–13453. [Google Scholar] [CrossRef] [PubMed]
- Weston-Hafer, K.; Berg, D.E. Limits to the role of palindromy in deletion formation. J. Bacteriol. 1991, 173, 315–318. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nasar, F.; Jankowski, C.; Nag, D.K. Long palindromic sequences induce double-strand breaks during meiosis in yeast. Mol. Cell Biol. 2000, 20, 3449–3458. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, G.; Bacolla, A.; Zhao, J.; Spitser, S.; Vasquez, K.M. Short Inverted Repeats Are Hotspots for Genetic Instability: Relevance to Cancer Genomes. Cell Rep. 2015, 10, 1674–1680. [Google Scholar] [CrossRef] [PubMed]
- McKinney, J.A.; Wang, G.; Mukherjee, A.; Christensen, L.; Subramanian, S.H.S.; Zhao, J.; Vasquez, K.M. Distinct DNA repair pathways cause genomic instability at alternative DNA structures. Nat. Commun. 2020, 11, 236. [Google Scholar] [CrossRef] [PubMed]
- Li, T.T.; Vasquez, K.M. Multi-Faceted Roles of ERCC1-XPF Nuclease in Processing Non-B DNA Structures. DNA 2022, 2, 231–247. [Google Scholar] [CrossRef]
- Parniewski, P.; Bacolla, A.; Jaworski, A.; Wells, R.D. Nucleotide excision repair affects the stability of long transcribed (CTG*CAG) tracts in an orientation-dependent manner in Escherichia coli. Nucleic. Acids. Res. 1999, 27, 616–623. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lin, Y.; Dion, V.; Wilson, J.H. Transcription promotes contraction of CAG repeat tracts in human cells. Nat. Struct. Mol. Biol. 2006, 13, 179–180. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Wilson, J.H. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol. Cell Biol. 2007, 27, 6209–6217. [Google Scholar] [CrossRef] [PubMed]
- Taghian, D.G.; Hough, H.; Nickoloff, J.A. Biased short tract repair of palindromic loop mismatches in mammalian cells. Genetics 1998, 148, 1257–1268. [Google Scholar] [CrossRef] [PubMed]
- Owen, B.A.; Yang, Z.; Lai, M.; Gajec, M.; Badger, J.D., 2nd; Hayes, J.J.; Edelmann, W.; Kucherlapati, R.; Wilson, T.M.; McMurray, C.T. (CAG)(n)-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat. Struct. Mol. Biol. 2005, 12, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Manley, K.; Shirley, T.L.; Flaherty, L.; Messer, A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat. Genet. 1999, 23, 471–473. [Google Scholar] [CrossRef] [PubMed]
- Richard, G.F. The Startling Role of Mismatch Repair in Trinucleotide Repeat Expansions. Cells 2021, 10, 1019. [Google Scholar] [CrossRef] [PubMed]
- Lahue, R.S. New developments in Huntington’s disease and other triplet repeat diseases: DNA repair turns to the dark side. Neuronal Signal. 2020, 4, NS20200010. [Google Scholar] [CrossRef] [PubMed]
- Mengoli, V.; Ceppi, I.; Sanchez, A.; Cannavo, E.; Halder, S.; Scaglione, S.; Gaillard, P.H.; McHugh, P.J.; Riesen, N.; Pettazzoni, P.; et al. WRN helicase and mismatch repair complexes independently and synergistically disrupt cruciform DNA structures. EMBO J. 2023, 42, e111998. [Google Scholar] [CrossRef] [PubMed]
- Truong, L.N.; Li, Y.; Shi, L.Z.; Hwang, P.Y.; He, J.; Wang, H.; Razavian, N.; Berns, M.W.; Wu, X. Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl. Acad. Sci. USA 2013, 110, 7720–7725. [Google Scholar] [CrossRef] [PubMed]
- Yun, M.H.; Hiom, K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 2009, 459, 460–463. [Google Scholar] [CrossRef] [PubMed]
- Paull, T.T.; Gellert, M. The 3’ to 5’ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell 1998, 1, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, K.M.; Yuan, S.S.; Lee, E.Y.; Sung, P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 1998, 273, 21447–21450. [Google Scholar] [CrossRef] [PubMed]
- Paull, T.T.; Gellert, M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999, 13, 1276–1288. [Google Scholar] [CrossRef] [PubMed]
- Lengsfeld, B.M.; Rattray, A.J.; Bhaskara, V.; Ghirlando, R.; Paull, T.T. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol. Cell 2007, 28, 638–651. [Google Scholar] [CrossRef] [PubMed]
- Andres, S.N.; Williams, R.S. CtIP/Ctp1/Sae2, molecular form fit for function. DNA Repair 2017, 56, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, Y.; Truong, L.N.; Shi, L.Z.; Hwang, P.Y.; He, J.; Do, J.; Cho, M.J.; Li, H.; Negrete, A.; et al. CtIP maintains stability at common fragile sites and inverted repeats by end resection-independent endonuclease activity. Mol. Cell 2014, 54, 1012–1021. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhao, J.; Vasquez, K.M. Methods to determine DNA structural alterations and genetic instability. Methods 2009, 48, 54–62. [Google Scholar] [CrossRef]
- Kraemer, K.H.; Seidman, M.M. Use of supF, an Escherichia coli tyrosine suppressor tRNA gene, as a mutagenic target in shuttle-vector plasmids. Mutat. Res. 1989, 220, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Ariza, R.R.; Roldan-Arjona, T.; Hera, C.; Pueyo, C. A method for selection of forward mutations in supF gene carried by shuttle-vector plasmids. Carcinogenesis 1993, 14, 303–305. [Google Scholar] [CrossRef] [PubMed]
- Mandke, P. Elucidation of factors affecting short inverted repeat-induced genomic instability in eukaryotic systems. PhD Thesis, University of Texas at Austin, Austin, TX, USA, 2022. [Google Scholar]
- Levy, D.D.; Saijo, M.; Tanaka, K.; Kraemer, K.H. Expression of a transfected DNA repair gene (XPA) in xeroderma pigmentosum group A cells restores normal DNA repair and mutagenesis of UV-treated plasmids. Carcinogenesis 1995, 16, 1557–1563. [Google Scholar] [CrossRef] [PubMed]
- Manandhar, M.; Lowery, M.G.; Boulware, K.S.; Lin, K.H.; Lu, Y.; Wood, R.D. Transcriptional consequences of XPA disruption in human cell lines. DNA Repair 2017, 57, 76–90. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zacal, N.J.; Rainbow, A.J.; Zhu, X.D. XPF with mutations in its conserved nuclease domain is defective in DNA repair but functions in TRF2-mediated telomere shortening. DNA Repair 2007, 6, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, C.M.; Sancar, C. Mechanisms and Maps of Nucleotide Excision Repair. In DNA Damage, DNA Repair and Disease; Royal Society of Chemistry: London, UK, 2020; Volume 2, pp. 1–23. [Google Scholar] [CrossRef]
- Wood, R.D. Nucleotide excision repair in mammalian cells. J. Biol. Chem. 1997, 272, 23465–23468. [Google Scholar] [CrossRef]
- Zhao, J.; Jain, A.; Iyer, R.R.; Modrich, P.L.; Vasquez, K.M. Mismatch repair and nucleotide excision repair proteins cooperate in the recognition of DNA interstrand crosslinks. Nucleic. Acids. Res. 2009, 37, 4420–4429. [Google Scholar] [CrossRef] [PubMed]
- Young, S.J.; West, S.C. Coordinated roles of SLX4 and MutSbeta in DNA repair and the maintenance of genome stability. Crit. Rev. Biochem. Mol. Biol. 2021, 56, 157–177. [Google Scholar] [CrossRef] [PubMed]
- Jiricny, J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006, 7, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Iyer, R.R.; Pluciennik, A.; Burdett, V.; Modrich, P.L. DNA mismatch repair: Functions and mechanisms. Chem. Rev. 2006, 106, 302–323. [Google Scholar] [CrossRef]
- Young, S.J.; Sebald, M.; Shah Punatar, R.; Larin, M.; Masino, L.; Rodrigo-Brenni, M.C.; Liang, C.C.; West, S.C. MutSbeta Stimulates Holliday Junction Resolution by the SMX Complex. Cell Rep. 2020, 33, 108289. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhang, R.; Wang, X.W.; Linke, S.P.; Sengupta, S.; Hickson, I.D.; Pedrazzi, G.; Perrera, C.; Stagljar, I.; Littman, S.J.; et al. The mismatch DNA repair heterodimer, hMSH2/6, regulates BLM helicase. Oncogene 2004, 23, 3749–3756. [Google Scholar] [CrossRef] [PubMed]
- Lobachev, K.S.; Rattray, A.; Narayanan, V. Hairpin- and cruciform-mediated chromosome breakage: Causes and consequences in eukaryotic cells. Front. Biosci. 2007, 12, 4208–4220. [Google Scholar] [CrossRef] [PubMed]
- Svendsen, J.M.; Harper, J.W. GEN1/Yen1 and the SLX4 complex: Solutions to the problem of Holliday junction resolution. Genes Dev. 2010, 24, 521–536. [Google Scholar] [CrossRef] [PubMed]
- Garner, E.; Kim, Y.; Lach, F.P.; Kottemann, M.C.; Smogorzewska, A. Human GEN1 and the SLX4-associated nucleases MUS81 and SLX1 are essential for the resolution of replication-induced Holliday junctions. Cell Rep. 2013, 5, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Brazda, V.; Laister, R.C.; Jagelska, E.B.; Arrowsmith, C. Cruciform structures are a common DNA feature important for regulating biological processes. BMC Mol. Biol. 2011, 12, 33. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, H.; Ohye, T.; Kogo, H.; Tsutsumi, M.; Kato, T.; Tong, M.; Emanuel, B.S.; Kurahashi, H. Two sequential cleavage reactions on cruciform DNA structures cause palindrome-mediated chromosomal translocations. Nat. Commun. 2013, 4, 1592. [Google Scholar] [CrossRef] [PubMed]
- Kaushal, S.; Wollmuth, C.E.; Das, K.; Hile, S.E.; Regan, S.B.; Barnes, R.P.; Haouzi, A.; Lee, S.M.; House, N.C.M.; Guyumdzhyan, M.; et al. Sequence and Nuclease Requirements for Breakage and Healing of a Structure-Forming (AT)n Sequence within Fragile Site FRA16D. Cell Rep. 2019, 27, 1151–1164.e1155. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Kanagaraj, R.; Mihaljevic, B.; Schwendener, S.; Sartori, A.A.; Gerrits, B.; Shevelev, I.; Janscak, P. MRE11 complex links RECQ5 helicase to sites of DNA damage. Nucleic. Acids. Res. 2009, 37, 2645–2657. [Google Scholar] [CrossRef] [PubMed]
- Sartori, A.A.; Lukas, C.; Coates, J.; Mistrik, M.; Fu, S.; Bartek, J.; Baer, R.; Lukas, J.; Jackson, S.P. Human CtIP promotes DNA end resection. Nature 2007, 450, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Mimitou, E.P.; Symington, L.S. DNA end resection: Many nucleases make light work. DNA Repair 2009, 8, 983–995. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Caron, P.; Legube, G.; Paull, T.T. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic. Acids. Res. 2014, 42, e19. [Google Scholar] [CrossRef] [PubMed]
- Moreau, S.; Morgan, E.A.; Symington, L.S. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics 2001, 159, 1423–1433. [Google Scholar] [CrossRef]
- Lam, A.F.; Krogh, B.O.; Symington, L.S. Unique and overlapping functions of the Exo1, Mre11 and Pso2 nucleases in DNA repair. DNA Repair 2008, 7, 655–662. [Google Scholar] [CrossRef] [PubMed]
- McVey, M.; Lee, S.E. MMEJ repair of double-strand breaks (director’s cut): Deleted sequences and alternative endings. Trends Genet. 2008, 24, 529–538. [Google Scholar] [CrossRef] [PubMed]
- McKinney, J.A.; Wang, G.; Vasquez, K.M. Distinct mechanisms of mutagenic processing of alternative DNA structures by repair proteins. Mol. Cell Oncol. 2020, 7, 1743807. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).