Single-Cell Screening through Cell Encapsulation in Photopolymerized Gelatin Methacryloyl
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioink and Cells
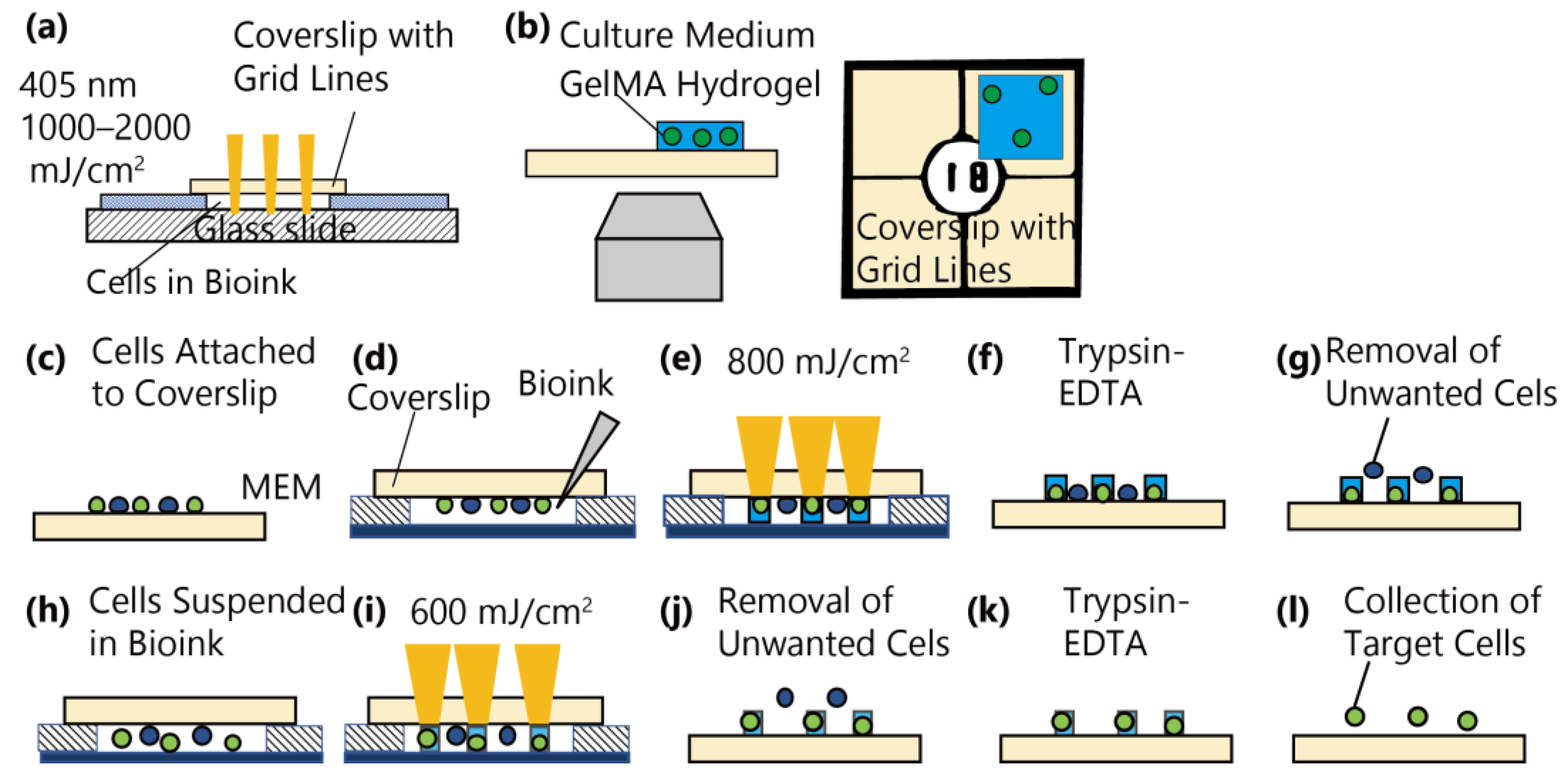
2.2. Irradiation Setup
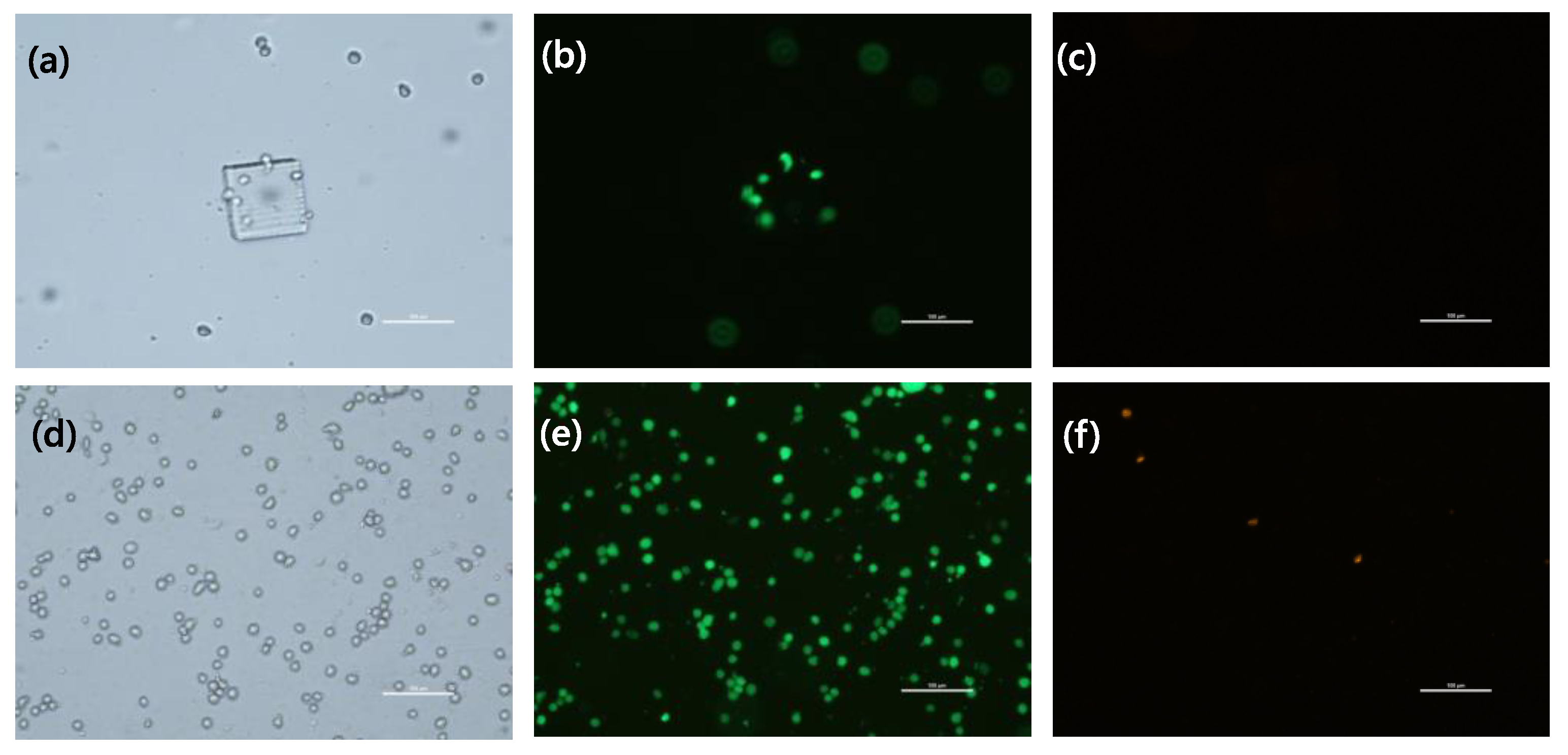
2.3. Live/Dead Assay
2.4. Observation of Cells Encapsualted in GelMA
2.5. Encapsulation of Attached and Suspended Cells in GelMA
2.6. Collection of Cells with Trypsin
3. Results and Discussion
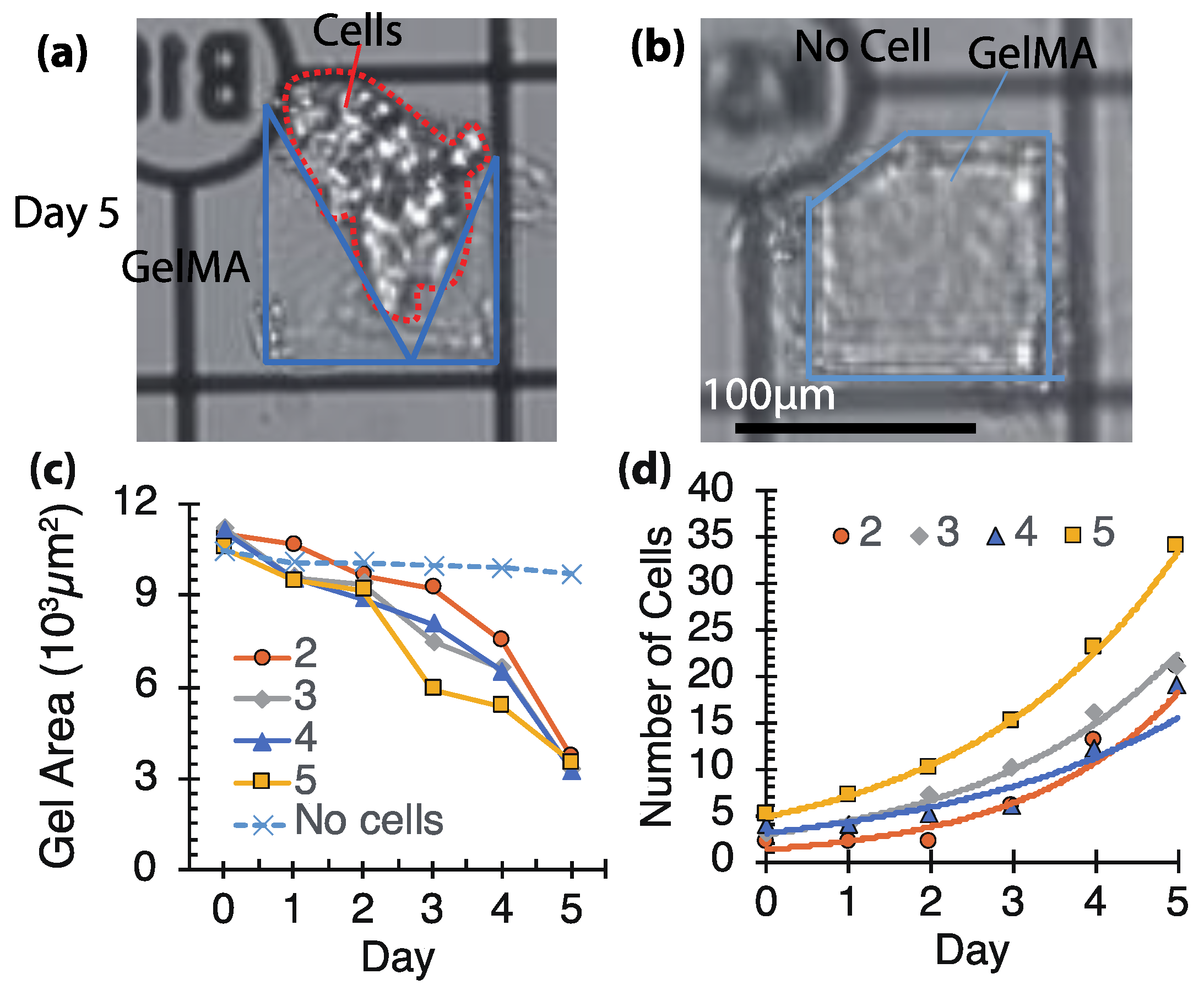
3.1. Basic Characterization with GelMA
3.2. Biodegradability of GelMA
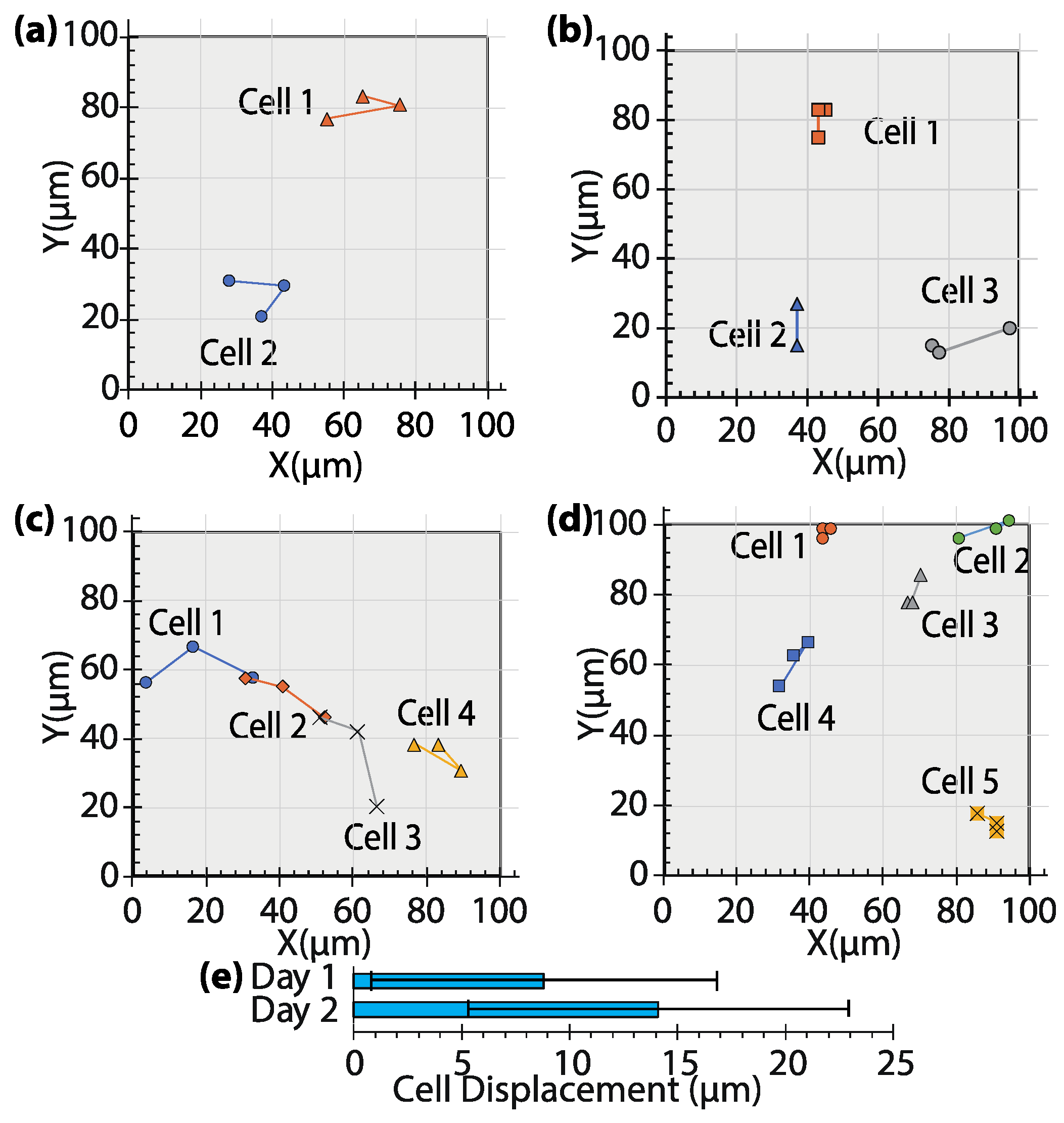
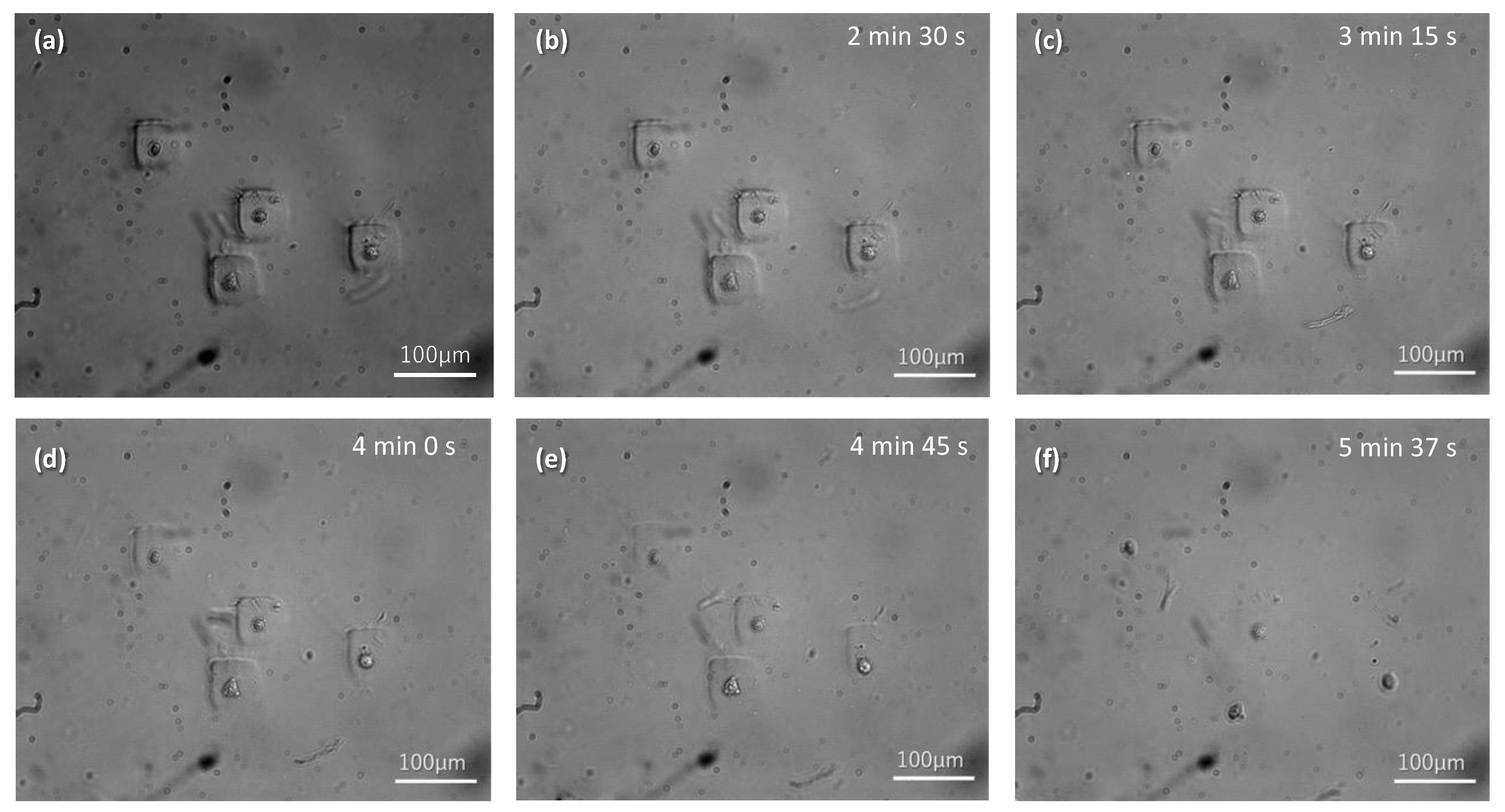
3.3. Behavior of Cells Encapsulated in the GelMA Hydrogel
3.4. Recovery of Cells Encapsulated in GelMA
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Soo Kim, H.; Devarenne, T.P.; Han, A. A High-Throughput Microfluidic Single-Cell Screening Platform Capable of Selective Cell Extraction. Lab. A Chip 2015, 15, 2467–2475. [Google Scholar] [CrossRef] [PubMed]
- Terekhov, S.S.; Smirnov, I.V.; Stepanova, A.V.; Bobik, T.V.; Mokrushina, Y.A.; Ponomarenko, N.A.; Belogurov, A.A.; Rubtsova, M.P.; Kartseva, O.V.; Gomzikova, M.O.; et al. Microfluidic Droplet Platform for Ultrahigh-Throughput Single-Cell Screening of Biodiversity. Proc. Natl. Acad. Sci. USA 2017, 114, 2550–2555. [Google Scholar] [CrossRef] [PubMed]
- Segaliny, A.I.; Li, G.; Kong, L.; Ren, C.; Chen, X.; Wang, J.K.; Baltimore, D.; Wu, G.; Zhao, W. Functional TCR T Cell Screening Using Single-Cell Droplet Microfluidics. Lab. A Chip 2018, 18, 3733–3749. [Google Scholar] [CrossRef] [PubMed]
- Müller, S.; Nebe-von-Caron, G. Functional Single-Cell Analyses: Flow Cytometry and Cell Sorting of Microbial Populations and Communities. FEMS Microbiol. Rev. 2010, 34, 554–587. [Google Scholar] [CrossRef] [PubMed]
- Wilkerson, M.J. Principles and Applications of Flow Cytometry and Cell Sorting in Companion Animal Medicine. Vet. Clin. Small Anim. Pract. 2012, 42, 53–71. [Google Scholar] [CrossRef] [PubMed]
- Adan, A.; Alizada, G.; Kiraz, Y.; Baran, Y.; Nalbant, A. Flow Cytometry: Basic Principles and Applications. Crit. Rev. Biotechnol. 2017, 37, 163–176. [Google Scholar] [CrossRef]
- McKinnon, K.M. Flow Cytometry: An Overview. Curr. Protoc. Immunol. 2018, 120, 5.1.1–5.1.11. [Google Scholar] [CrossRef] [PubMed]
- LaBelle, C.A.; Massaro, A.; Cortés-Llanos, B.; Sims, C.E.; Allbritton, N.L. Image-Based Live Cell Sorting. Trends Biotechnol. 2021, 39, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Moraes, C.; Ye, G.; Simmons, C.A.; Sun, Y. Single Cell Deposition and Patterning with a Robotic System. PLoS ONE 2010, 5, e13542. [Google Scholar] [CrossRef]
- Nagai, M.; Kato, K.; Oohara, K.; Shibata, T. Pick-and-Place Operation of Single Cell Using Optical and Electrical Measurements for Robust Manipulation. Micromachines 2017, 8, 350. [Google Scholar] [CrossRef]
- Chen, B.; Lim, S.; Kannan, A.; Alford, S.C.; Sunden, F.; Herschlag, D.; Dimov, I.K.; Baer, T.M.; Cochran, J.R. High-Throughput Analysis and Protein Engineering Using Microcapillary Arrays. Nat. Chem. Biol. 2016, 12, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Negishi, R.; Takai, K.; Tanaka, T.; Matsunaga, T.; Yoshino, T. High-Throughput Manipulation of Circulating Tumor Cells Using a Multiple Single-Cell Encapsulation System with a Digital Micromirror Device. Anal. Chem. 2018, 90, 9734–9741. [Google Scholar] [CrossRef] [PubMed]
- Negishi, R.; Iwata, R.; Tanaka, T.; Kisailus, D.; Maeda, Y.; Matsunaga, T.; Yoshino, T. Gel-Based Cell Manipulation Method for Isolation and Genotyping of Single-Adherent Cells. Analyst 2019, 144, 990–996. [Google Scholar] [CrossRef] [PubMed]
- Negishi, R.; Saito, H.; Iwata, R.; Tanaka, T.; Yoshino, T. Performance Evaluation of a High-Throughput Separation System for Circulating Tumor Cells Based on Microcavity Array. Eng. Life Sci. 2020, 20, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Kovac, J.; Voldman, J. Image-Based Single-Cell Sorting via Dual-Photopolymerized Microwell Arrays. Anal. Chem. 2014, 86, 977–981. [Google Scholar] [CrossRef] [PubMed]
- Browning, M.B.; Cereceres, S.N.; Luong, P.T.; Cosgriff-Hernandez, E.M. Determination of the in Vivo Degradation Mechanism of PEGDA Hydrogels. J. Biomed. Mater. Res. Part A 2014, 102, 4244–4251. [Google Scholar] [CrossRef]
- Perera, D.; Medini, M.; Seethamraju, D.; Falkowski, R.; White, K.; Olabisi, R.M. The Effect of Polymer Molecular Weight and Cell Seeding Density on Viability of Cells Entrapped within PEGDA Hydrogel Microspheres. J. Microencapsul. 2018, 35, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Nagai, M.; Kato, K.; Shibata, T. Integration of Microorganism Vorticella Convallaria and Poly (Ethylene Glycol) Diacrylate Hydrogel for Biohybrid Systems. Mech. Eng. Lett. 2016, 2, 16–00445. [Google Scholar] [CrossRef]
- Yue, K.; Trujillo-de Santiago, G.; Alvarez, M.M.; Tamayol, A.; Annabi, N.; Khademhosseini, A. Synthesis, Properties, and Biomedical Applications of Gelatin Methacryloyl (GelMA) Hydrogels. Biomaterials 2015, 73, 254–271. [Google Scholar] [CrossRef]
- Piao, Y.; You, H.; Xu, T.; Bei, H.-P.; Piwko, I.Z.; Kwan, Y.Y.; Zhao, X. Biomedical Applications of Gelatin Methacryloyl Hydrogels. Eng. Regen. 2021, 2, 47–56. [Google Scholar] [CrossRef]
- Ghosh, R.N.; Thomas, J.; Vaidehi, B.R.; Devi, N.G.; Janardanan, A.; Namboothiri, P.K.; Peter, M. An Insight into Synthesis, Properties and Applications of Gelatin Methacryloyl Hydrogel for 3D Bioprinting. Mater. Adv. 2023, 4, 5496–5529. [Google Scholar] [CrossRef]
- Cui, J.; Wang, H.; Shi, Q.; Sun, T.; Huang, Q.; Fukuda, T. Multicellular Co-Culture in Three-Dimensional Gelatin Methacryloyl Hydrogels for Liver Tissue Engineering. Molecules 2019, 24, 1762. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, S.; Yildirimer, L.; Zhao, H.; Ding, R.; Wang, H.; Cui, W.; Weitz, D. Injectable Stem Cell-Laden Photocrosslinkable Microspheres Fabricated Using Microfluidics for Rapid Generation of Osteogenic Tissue Constructs. Adv. Funct. Mater. 2016, 26, 2809–2819. [Google Scholar] [CrossRef]
- Wang, H.; Liu, H.; Liu, H.; Su, W.; Chen, W.; Qin, J. One-Step Generation of Core–Shell Gelatin Methacrylate (GelMA) Microgels Using a Droplet Microfluidic System. Adv. Mater. Technol. 2019, 4, 1800632. [Google Scholar] [CrossRef]
- Gao, X.; Hu, X.; Yang, D.; Hu, Q.; Zheng, J.; Zhao, S.; Zhu, C.; Xiao, X.; Yang, Y. Acoustic Quasi-Periodic Bioassembly Based Diverse Stem Cell Arrangements for Differentiation Guidance. Lab. Chip 2023, 23, 4413–4421. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Hu, X.; Shi, Y.; Liang, L.; Zhu, J.; Zhao, S.; Wang, Y.; Wu, Z.; Wang, F.; Zhou, F.; et al. Heterogeneous Tissue Construction by On-Demand Bubble-Assisted Acoustic Patterning. Lab. Chip 2023, 23, 2206–2216. [Google Scholar] [CrossRef]
- Hu, X.; Zhao, S.; Luo, Z.; Zuo, Y.; Wang, F.; Zhu, J.; Chen, L.; Yang, D.; Zheng, Y.; Zheng, Y.; et al. On-Chip Hydrogel Arrays Individually Encapsulating Acoustic Formed Multicellular Aggregates for High Throughput Drug Testing. Lab. Chip 2020, 20, 2228–2236. [Google Scholar] [CrossRef]
- Chansoria, P.; Asif, S.; Polkoff, K.; Chung, J.; Piedrahita, J.A.; Shirwaiker, R.A. Characterizing the Effects of Synergistic Thermal and Photo-Cross-Linking during Biofabrication on the Structural and Functional Properties of Gelatin Methacryloyl (GelMA) Hydrogels. ACS Biomater. Sci. Eng. 2021, 7, 5175–5188. [Google Scholar] [CrossRef]
- Kulkarni, N.S.; Chauhan, G.; Goyal, M.; Sarvepalli, S.; Gupta, V. Development of Gelatin Methacrylate (GelMa) Hydrogels for Versatile Intracavitary Applications. Biomater. Sci. 2022, 10, 4492–4507. [Google Scholar] [CrossRef]
- Allen, N.B.; Abar, B.; Johnson, L.; Burbano, J.; Danilkowicz, R.M.; Adams, S.B. 3D-Bioprinted GelMA-Gelatin-Hydroxyapatite Osteoblast-Laden Composite Hydrogels for Bone Tissue Engineering. Bioprinting 2022, 26, e00196. [Google Scholar] [CrossRef]
- Selvam, V.K.P.; Kamaludin, M.L.A.B.; Murtaza, G.; Chowdhury, R.H.; Debnath, T.; Okamoto, S.; Shibata, T.; Santra, T.S.; Nagai, M. Image-Based Gel Encapsulation of Suspended Single Cells for Parallel Single-Cell Screening. J. Robot. Mechatron. 2023, 35, 1177–1184. [Google Scholar] [CrossRef]
- Yin, J.; Yan, M.; Wang, Y.; Fu, J.; Suo, H. 3D Bioprinting of Low-Concentration Cell-Laden Gelatin Methacrylate (GelMA) Bioinks with a Two-Step Cross-Linking Strategy. ACS Appl. Mater. Interfaces 2018, 10, 6849–6857. [Google Scholar] [CrossRef] [PubMed]
- Pahoff, S.; Meinert, C.; Bas, O.; Nguyen, L.; Klein, T.J.; Hutmacher, D.W. Effect of Gelatin Source and Photoinitiator Type on Chondrocyte Redifferentiation in Gelatin Methacryloyl-Based Tissue-Engineered Cartilage Constructs. J. Mater. Chem. B 2019, 7, 1761–1772. [Google Scholar] [CrossRef] [PubMed]
- Nagai, M.; Sato, S.; Hiratsuka, S.; Kawaharada, S.; Okamoto, S.; Santra, T.S.; Shibata, T. Parallel Photothermal Coalescence of Biocompatible Photocurable PEGDA Droplets. IEEJ Trans. Sens. Micromachines 2023, 143, 49–54. [Google Scholar] [CrossRef]
- Verma, A.; Verma, M.; Singh, A. Chapter 14-Animal Tissue Culture Principles and Applications. In Animal Biotechnology, 2nd ed.; Verma, A.S., Singh, A., Eds.; Academic Press: Boston, UK, 2020; pp. 269–293. ISBN 978-0-12-811710-1. [Google Scholar]
- Vasudevan, J.; Lim, C.T.; Fernandez, J.G. Cell Migration and Breast Cancer Metastasis in Biomimetic Extracellular Matrices with Independently Tunable Stiffness. Adv. Funct. Mater. 2020, 30, 2005383. [Google Scholar] [CrossRef]
- Stillman, Z.; Jarai, B.M.; Raman, N.; Patel, P.; Fromen, C.A. Degradation Profiles of Poly(Ethylene Glycol)Diacrylate (PEGDA)-Based Hydrogel Nanoparticles. Polym. Chem. 2020, 11, 568–580. [Google Scholar] [CrossRef]
- Debnath, T.; Hattori, R.; Okamoto, S.; Shibata, T.; Santra, T.S.; Nagai, M. Automated Detection of Patterned Single-Cells within Hydrogel Using Deep Learning. Sci. Rep. 2022, 12, 18343. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panneer Selvam, V.K.; Fukunaga, T.; Suzuki, Y.; Okamoto, S.; Shibata, T.; Santra, T.S.; Nagai, M. Single-Cell Screening through Cell Encapsulation in Photopolymerized Gelatin Methacryloyl. Micro 2024, 4, 295-304. https://doi.org/10.3390/micro4020018
Panneer Selvam VK, Fukunaga T, Suzuki Y, Okamoto S, Shibata T, Santra TS, Nagai M. Single-Cell Screening through Cell Encapsulation in Photopolymerized Gelatin Methacryloyl. Micro. 2024; 4(2):295-304. https://doi.org/10.3390/micro4020018
Chicago/Turabian StylePanneer Selvam, Venkatesh Kumar, Takeru Fukunaga, Yuya Suzuki, Shunya Okamoto, Takayuki Shibata, Tuhin Subhra Santra, and Moeto Nagai. 2024. "Single-Cell Screening through Cell Encapsulation in Photopolymerized Gelatin Methacryloyl" Micro 4, no. 2: 295-304. https://doi.org/10.3390/micro4020018
APA StylePanneer Selvam, V. K., Fukunaga, T., Suzuki, Y., Okamoto, S., Shibata, T., Santra, T. S., & Nagai, M. (2024). Single-Cell Screening through Cell Encapsulation in Photopolymerized Gelatin Methacryloyl. Micro, 4(2), 295-304. https://doi.org/10.3390/micro4020018







