Abstract
Cheese ripening involves microbial changes, with starter lactic acid bacteria (SLAB) initiating fermentation and nonstarter lactic acid bacteria (NSLAB) driving flavor and texture development. However, heat-resistant spores of Clostridium and Bacillus can survive pasteurization and cause spoilage during ripening. This study evaluated NSLAB dynamics in the presence of spores during cheese ripening. Cheddar cheese samples at pilot-scale level (110 L) with four treatments, namely control, with spores of B. licheniformis (T1), with spores of Cl. tyrobutyricum (T2), and both spores (T3) at 2.0 Log10 CFU/mL, were ripened at 7 °C for six months. SLAB declined from 8.0 to 0.2 Log10 CFU/g, while NSLAB increased from 2.0 to 8.5 Log10 CFU/g by month three and maintained their counts up to six months, unaffected by spore presence. Spore counts were ≤1.45 Log10 CFU/g in controls but reached 2.94 ± 0.02 (T2) and 2.48 ± 0.03 (T3), correlating with spoilage signs after five months. MALDI-TOF identified L. rhamnosus (up to 37%) and L. paracasei (up to 25%) as dominant NSLAB across treatments. Physicochemical parameters were not significantly affected by higher spore levels. While NSLAB dominated, they were inadequate to prevent spoilage in spore-inoculated samples exceeding 2.0 logs during cheese ripening.
1. Introduction
Cheese, a highly diverse fermented milk product, represents one of the most versatile and widely consumed dairy foods globally. The variety of cheeses, differentiated by flavor, texture, and form, is shaped by regional resources, cultural preferences, and technological advancements in cheesemaking [1]. Beyond its culinary appeal, cheese can be regarded as a complex ecosystem characterized by a diverse microbiota derived from raw milk, starter cultures, adjunct cultures, and the environment. This microbiota, often called the “cheese flora”, is critical in defining cheese’s sensory attributes and quality through biochemical and microbiological interactions during ripening. Ripening, a crucial phase in cheese production, includes a series of metabolic processes driven by LAB and NSLAB, which collectively contribute to the flavor, texture, and overall consumer acceptability of the final product [2].
Starter LAB, primarily consisting of species such as Lactococcus lactis, St. thermophilus, and L. delbrueckii, is intentionally added to milk to initiate fermentation. These bacteria rapidly ferment lactose into lactic acid, leading to pH reduction and casein coagulation, which are essential for curd formation [3]. Additionally, SLAB contributes to proteolysis and lipolysis during the first 4–5 weeks of ripening, producing peptides and free amino acids that serve as precursors for flavor compounds [4]. On the other hand, NSLAB, including species from the Lactobacillus, Leuconostoc, and Pediococcus genera, are not intentionally added but originate from raw milk [5]. They emerge as dominant populations during the later stages of cheese ripening when the starter LAB declines due to nutrient depletion and acidity stress [6]. NSLAB contributes significantly to proteolysis and lipolysis, leading to the breakdown of casein into smaller peptides, free amino acids, and volatile compounds that influence cheese flavor, texture, and aroma [3]. The dynamics of starter LAB and NSLAB populations evolve throughout ripening. Starter LAB typically reaches peak concentrations (108 to 109 CFU/g) during the first five to six weeks but gradually declines due to acid-induced stress and depletion of lactose. In contrast, NSLAB, initially present at lower levels, proliferates over time and becomes the dominant microflora (107 to 109 CFU/g) in aged cheeses [7]. Their ability to survive and grow in nutrient-limited environments allows them to influence the final cheese characteristics through their enzymatic activities [8]. Common NSLAB species found during cheddar ripening include L. rhamnosus, L. paracasei, L. plantarum, and Leuc. mesenteroides, with L. rhamnosus often dominating due to its high tolerance to acidic, low-moisture conditions, and its contribution to flavor-enhancing proteolytic activity [5]. During ripening, physicochemical changes such as a steady decline in moisture content, accumulation of FFA, and consistent acidity (pH 5.0) are typically observed. These changes are critical for the development of texture and flavor, and they influence microbial populations by selectively promoting the growth of acid-tolerant NSLAB while suppressing pathogens and spoilage microbes [9]. Cheese composition standards vary depending on the cheese variety; however, for semi-hard ripened cheeses such as cheddar, the U.S. FDA requires that the product must contain a minimum of 33% milkfat and a maximum of 39% moisture content to meet its standard [10]. These regulatory parameters support proper textural development, ensure microbial stability, and facilitate controlled ripening. Maintaining these compositional requirements is essential for preserving the balance between microbial activity and physicochemical conditions during aging, thereby providing both the sensory quality and safety of the final cheese product. Therefore, understanding the interactions between SLAB and NSLAB is essential for controlling cheese ripening and ensuring desirable sensory attributes. Studying their population dynamics is particularly important to identify dominant NSLAB strains in the presence of aerobic and anaerobic spore formers, which can impact cheese quality. Certain NSLAB strains may suppress the growth of spoilage organisms, including spore-forming bacteria such as Clostridium and Bacillus, which can lead to defects like late-blowing or off-flavors [11,12]. By characterizing NSLAB communities at different ripening stages, we aimed to develop a novel strategy to enhance beneficial microbial interactions and mitigate spoilage risks. This study investigated the population changes of NSLAB in presence of spores during cheese ripening.
2. Materials and Methods
2.1. Experimental Design
The cheese manufacturing process was first standardized at lab scale with 10 L of milk in a small double-jacketed vat with a heating system. After standardization of the process, two independent trials of each cheese namely control, spiked with aerobic spore-former B. licheniformis T1 (BL) at 2log10 CFU/g, spiked with anaerobic spore-former Cl. tyrobutyricum T2 (CT) at 2log10 CFU/g, and the mixture of both T3 (BL+CT) at 1:1, were made in the batches of 110 kg milk each at Davis Dairy Plant (DDP) at South Dakota State University, Brookings. Cheese samples were ripened at 7 °C for 6 months, and microbial and chemical analyses were performed monthly in triplicates for two independent trials.
2.2. Materials
Pasteurized standardized milk was procured from Davis Dairy Plant (DDP), SDSU, Brookings. The starter culture for the cheese manufacturing was procured from CHR Hansen (DVS 970, 500U, Milwaukee, WI, USA). The aerobic spore-former (B. licheniformis 6634) and the anaerobic spore-former (Cl. tyrobutyricum VPI 5392) isolates were procured from ATCC (Manassas, VA, USA).
2.3. Spore Preparation
2.3.1. Aerobic Spores
Spores of B. licheniformis (6634, ATCC, USA) were prepared by spreading 1 mL of actively growing broth culture of B. licheniformis onto TSA plates and incubated under aerobic conditions at 37 °C for up to 3 days to promote sporulation, and spores were harvested by flooding the plate surface with 10 mL of sterile distilled water, allowing it to soak for 2 to 3 min, and then gently scraping with a sterile spreader. The spore suspension was collected into sterile 50 mL centrifuge tubes and centrifuged at 4500× g for 30 min. The resulting pellets were washed twice by resuspending in 20 mL sterile distilled water, followed by centrifugation at 4500× g for 30 min at 20 °C. The final spore pellets were resuspended in 10 to 15 mL of sterile distilled water and heat-treated at 80 °C for 20 min to eliminate any remaining vegetative cells [13].
2.3.2. Anaerobic Spores
Spores of C. tyrobutyricum (VPI 5392, ATCC, USA) were prepared by spreading 1 mL of actively growing broth culture of Cl. tyrobutyricum onto RCA plates and incubated under anaerobic conditions at 37 °C for up to 7 days to promote sporulation, and spores were harvested by flooding the plate surface with 10 mL of sterile distilled water, allowing it to soak for 2 to 3 min, and then gently scraping with a sterile spreader. The spore suspension was collected into sterile 50 mL centrifuge tubes and then centrifuged at 4500× g for 30 min. The resulting pellets were washed twice by resuspending in 20 mL sterile distilled water, followed by centrifugation at 4500× g for 30 min at 20 °C. The final spore pellets were resuspended in 10 to 15 mL of sterile distilled water and heat-treated at 80 °C for 20 min to eliminate any remaining vegetative cells [13].
2.4. Cheddar Cheese Manufacturing
2.4.1. Lab-Scale Cheese Manufacturing Process Standardization
The cheese manufacturing process was standardized at the laboratory scale using a scale cheese vat (Vevor, Panyu, Guangzhou, China) with 10 L of pasteurized milk per batch with the method described by Hanlon et al., 2022 [14], as after adjusting milk to 32 °C, mesophilic starter culture (DVS 970, CHR Hansen, USA) was added. Once pH increased by 0.2 units, rennet was added, and the curd was set for 30 to 35 min. Curd firmness was checked using a stainless-steel rod, followed by horizontal and vertical cutting and a 5 min rest. The curd was then cooked gradually to 39 °C at a rate of 1 °C every 5 min. Whey was drained, and cheddaring (packing, cutting, piling, and re-piling) was performed until pH reached 5.5. The curd was cubed (2.5 × 2.5 cm), salted, filled into hoops, pressed overnight, vacuum-packed, and ripened at 7 °C for 6 months.
2.4.2. Pilot Scale Cheese Manufacturing
For the pilot scale trials, pasteurized standardized (casein/fat ratio of 0.7) milk was procured from the DDP (SDSU, Brookings). The cheese samples were manufactured using the method explained by Hanlon et al. (2022) in a Double O cheese vat (Kusel Equipment Co., Watertown, WI, USA) with 110 L of milk in each batch [14]. After adjusting the milk temperature to 32 °C, mesophilic starter culture (DVS 970, CHR Hansen, USA) and/or spores were added to the milk, and after a 0.2 increase in the pH, rennet was then added and allowed to set for 30 to 35 min. The setting of the curd was checked with a stainless-steel rod by inserting it vertically and lifting it with an inclination to check if any curd particles were sticking or not. After that, the curd was cut horizontally and vertically, respectively, with the cheese knives and allowed to rest for 5 min. Then, the curd was cooked slowly to 39 °C at a rate of 1 °C increase after every five minutes. After cooking, the whey was drained through drainage with a strainer to prevent curd loss. The curd then went through the process of cheddaring (packing, cutting, piling, and re-piling) until the pH reached 5.5. After that, the curd was cut into small cubes (2.5 cm × 2.5 cm), and salt (non-iodized) (Great Value, Brookings, SD, USA) was added and mixed properly. The curd was then filled into hoops and pressed overnight. The pressed curd was then vacuum packaged in a 6 × 6 × 6 inch block and kept at 7 °C for 6 months for ripening.
2.5. Microbial Analysis
2.5.1. Total Plate Counts (TPC) and Spore Counts (SC) of Pasteurized Milk Used for Cheese Manufacturing
Microbial analysis of pasteurized milk was performed using the standard method [15], as total plate counts were enumerated on TSA incubated at 37 °C for 48 h, and spore counts were enumerated on TSA after heat treatment of milk at 80 °C for 20 min to eliminate any vegetative cells and incubated at 37 °C for 48 h.
2.5.2. Sample Preparation of Cheese for Microbial Enumeration
For each analysis, 11 g of cheese was aseptically transferred into 99 mL of 2% (w/v) sodium citrate buffer to ensure proper dispersion of microbial cells. The mixture was homogenized in a high-speed stomacher (Stomacher 400 Circulator, Seward, Welwyn Garden City, Hertfordshire, UK) for 3 min. Serial dilutions were prepared using sterile PBS (Fisher Scientific, Geel, Belgium), and appropriate dilutions were plated on selective and non-selective media for microbial enumeration [16].
2.5.3. Total Plate Counts of Cheese Samples During Ripening
Following homogenization, serial dilutions of the cheese samples were prepared using sterile PBS (Fisher Scientific, Geel, Belgium). For the enumeration of total plate counts, the standard pour plate method [16] was used, where appropriate dilutions were pour-plated in triplicate on TSA (Remel, ThermoFisher, Lenexa, KS, USA) using a sterile pipette. The plates were incubated aerobically at 37 °C for 48 h. After incubation, colonies were counted manually using a standard colony counter, and results were expressed as colony-forming units per gram (CFU/g) of cheese. All samples were analyzed in triplicate, and the mean values were recorded and converted to log10 CFU/g for statistical analysis.
2.5.4. Spore Counts of Cheese Samples During Ripening
For spore enumeration, aliquots of the homogenized and diluted samples were subjected to heat treatment at 80 °C for 20 min in a shaking water bath (BS-06, JeioTech, Daejeon, Republic of Korea) to inactivate vegetative cells, ensuring that only heat-resistant spores were quantified [17]. Following heat treatment, samples were immediately cooled in an ice bath and plated in triplicate on selective media. Aerobic spores were enumerated by pour plating on TSA (Remel, ThermoFisher, KS, USA) and incubated aerobically at 37 °C for 48 h. Anaerobic spores were enumerated by pour plating on RCM agar (OXOID, Hants, Basingstoke, Hampshire, UK) and incubated anaerobically at 37 °C for 48 h using anaerobic jars with anaerobe container system sachets with the indicator (BD, Cockeysville, MD, USA). Colonies were manually counted and reported as colony-forming units per gram (CFU/g) of cheese. All counts were performed in triplicate and expressed as log10 CFU/g (Limit of Detection—1 Log). Strict aseptic techniques were followed throughout the procedure to ensure the accuracy and reproducibility of the results.
2.5.5. Starter and Non-Starter Lactic Acid Bacteria (NSLAB) Counts of Cheese Samples During Ripening
After homogenization of 11 g of cheese in 99 mL of 2% sodium citrate buffer using a high-speed stomacher (Stomacher 400 Circulator, Seward, UK) for 3 min, serial dilutions were prepared in sterile PBS (Fisher Scientific, Geel, Belgium). For starters, i.e., Lactococci, appropriate dilutions were plated on M17 agar (OXOID, Hants, UK), and for NSLAB, serial dilutions were plated on De Man, Rogosa, and Sharpe (MRS) agar (BD Difco, MD, USA) in triplicate [18]. M17 plates were incubated aerobically at 37 °C for 72 h, and MRS plates were incubated anaerobically at 37 °C for 72 h using anaerobic jars with anaerobe container system sachets with an indicator (BD, MD, USA) to ensure optimal growth conditions for obligate and facultative anaerobes. After incubation, colonies were counted manually and expressed as CFU/g of cheese [19]. All analyses were performed in triplicate, and the results are reported as mean log10 CFU/g.
2.5.6. Identification of NSLAB Isolates Using MALDI-TOF
Representative colonies from MRS plates were randomly selected based on distinct morphological characteristics and subcultured on their respective media to obtain pure isolates [20]. Species-level identification of isolates was performed using MALDI-TOF at Animal Disease Research and Diagnostic Laboratory (ADRDL), South Dakota State University (SDSU), Brookings, SD, USA.
2.6. Physicochemical Analysis
2.6.1. Moisture Content
The moisture content of cheddar cheese was determined by [21], according to which 3 g of cheese sample was grated on empty dried aluminum dishes and kept in a hot air oven (Isotemp Oven, Fisher Scientific, USA) at 102 ± 2 °C for 5 h, and then, the dishes were cooled to room temperature in a desiccator, and the dried weight was taken. The moisture content of the cheese sample was determined using Equation (1).
- where
- W3—the weight of the dish with the dried sample (g)
- W2—the weight of the empty dish (g)
- W—the weight of the sample (g)
2.6.2. Protein Content
The total protein content of cheese samples was determined by the micro Kjeldahl method [22] based on the determination of nitrogen concentration. A 0.3 to 0.5 g sample of cheese was placed into Kjeldahl tubes, and 10 mL of H2SO4 (98%, ACS Grade, VWR Chemicals, Radnor PA, USA) was added along with catalyst tablets (FOSS Kjeltec, Runcorn, Cheshire, UK). The samples were then digested in a digester (DKL heating Digester, Velp Scientifica) at a range of 350–400 °C for 4 h to convert nitrogen into NH4+. Then, the samples were distilled into a distillation unit (UDK 129, Velp Scientifica), and the nitrogen was trapped in saturated boric acid (ACS, Spectrum Chemicals, Gardena, CA, USA) with the mixed indicator (Methyl red and Bromocresol green). The boric acid with dissolved nitrogen was then titrated with 0.1 N hydrochloric acid (HCl) till the green color changed to light pink. The total nitrogen content of the cheese samples was determined using Equation (2), and protein content was determined using Equation (3).
- where
- N—normality of HCl
- Vs—volume of HCl used for sample (ml)
- Vb—volume of HCl used for blank (ml)
- W—weight of sample (g)
Protein % = N% × 6.38
2.6.3. Fat Content
The fat content of cheese was determined using the Modified Mojonnier Ether Extraction method [23]. The fat content of the cheese samples was determined using Equation (4).
- W3—the weight of the dish with the dried sample (g)
- W2—the weight of the empty dish (g)
- W—the weight of the sample (g)
2.6.4. pH
Cheese pH was determined by homogenizing 20 g of grated cheese with 12 mL of deionized water, followed by measurement using a calibrated pH meter (Orion Lab Star PH111, Thermo Scientific, Boston, MA, USA) [24].
2.6.5. Free Fatty Acids (FFA)
Free fatty acids of the cheese samples were determined by the method explained by Asif et al. 2023, according to which 50 g of sample was mixed with absolute and neutralized ethanol with 0.1 N NaOH and then titrated with NaOH, and FFA was calculated in terms of oleic acid (Equation (5)) [25].
- where
- V—volume of NaOH used for titration (in mL)
- N—normality of NaOH solution
- W—weight of the sample (in grams)
- 28.2—milliequivalent weight of oleic acid
2.6.6. Visual Inspection of Cheese Samples for Bloating/Slits/Holes
During ripening, cheese samples were routinely inspected for visual changes to monitor gas formation and the presence of slits/holes. Cheeses exhibiting signs of bloating were identified by gas accumulation within the packaging; the development of irregular eyes, cracks, and splits in the cheese matrix; and the rancid odor characteristic of butyric acid production [18].
2.7. Statistical Analysis
All data were analyzed using two-way analysis of variance (ANOVA) to determine the effects of ripening time, treatment, and their interaction on the measured parameters at p < 0.05. Time and treatment were considered fixed factors, and where significant differences were found, mean comparisons were conducted using Tukey’s post hoc analysis. The statistical analyses were performed using OriginPro 2025 (OriginLab Corporation, Northampton, MA, USA). Results are presented as means ± standard deviations of triplicate measurements unless otherwise stated.
3. Results and Discussion
3.1. Microbial Analyses
3.1.1. Total Plate Counts and Spore Counts of Pasteurized Milk Used for Cheese Manufacturing
The microbial quality of raw milk is a critical determinant of cheese safety, starter culture performance, and overall product quality. The TPC of all four milk batches used for cheese manufacture ranged between 2.75 ± 0.03 and 2.83 ± 0.02 Log10 CFU/mL, with spore counts not detected (ND) in any sample (Table 1). These values are within the optimal microbiological limits for cheese production to ensure controlled fermentation and minimize the risk of spoilage or off-flavor development [6]. Maintaining low microbial counts in milk is essential for preventing the introduction of heat-resistant spore formers such as Bacillus and Clostridium, which can persist through pasteurization and contribute to late spoilage in aged cheeses [26]. These results confirm that the milk was microbiologically suitable for controlled cheese production, ensuring reliable starter culture performance and reproducibility. Silva et al. (2023) also reported that milk with total aerobic counts exceeding 6.0 Log10 CFU/mL increases the risk of spoilage and undesirable microbial activity during cheese ripening. In contrast, the low TPC observed in this study highlights the high hygienic quality of the milk used and its suitability for producing semi-hard cheeses, where ripening and microbial population examinations are required [27]. Likewise, Franciosi et al. (2011) emphasized the importance of microbial baseline stability in milk used for cheese production to ensure reproducible fermentation dynamics and flavor development [28]. This microbiological quality allowed for a focused assessment of microbial changes driven primarily by spore inoculation and natural NSLAB development rather than variability arising from the raw material. Hence, the milk used in all treatments provided a suitable and consistent substrate for cheese manufacture and evaluating microbial changes during cheese ripening under controlled experimental conditions.

Table 1.
Total plate counts (Log10CFU/mL) and spore counts (CFU/mL) of different batches of pasteurized milk used for different cheese manufacturing.
3.1.2. Total Plate Counts (TPC) of Cheese Samples During Ripening
TPC remained relatively constant across the six-month ripening period in all cheese samples, ranging from 8.3 to 9.9 Log10 CFU/g (Table 2). The control cheese showed values increasing from 8.31 ± 0.06 Log10 CFU/g from one month to 8.92 ± 0.07 Log10 CFU/g after six months, indicating sustained microbial activity likely dominated by LAB. Similarly, T1 (BL), T2 (CT), and T3 (BL+CT) cheeses exhibited TPC values within the range of 8.35 to 9.91 Log10 CFU/g, with minor fluctuations observed across treatments but no significant decline, suggesting the maintenance of a robust and metabolically active microbiota. These findings are consistent with Hanlon et al. (2022), who reported that TPC values after 90 days of ripening in semi-hard cheeses reached approximately 8.5 Log10 CFU/g [14], aligning with the average mid-ripening counts observed in this study. Similarly, Beresford et al. (2001) and Gatti et al. (2014) emphasized that relatively constant TPC in ripening cheeses indicates a healthy LAB population and balanced microbial ecology, which are essential for flavor development and safety [6,29]. This stability suggests that the core microbial population in all samples remained consistent regardless of the presence or absence of spores and that both starter and NSLAB populations established a favorable ecological niche in the initial ripening stages. Although samples inoculated with spores exhibited greater variability in TPC at early stages, no statistically significant differences were observed between overall control and spore-inoculated cheeses during later stages of ripening. This supports that lactic acid bacteria can effectively dominate the cheese ecosystem, maintaining metabolic activity and microbial density despite potential competitive pressure from spores.

Table 2.
Total plate counts (TPC) (Log10CFU/g) on Tryptic soy agar (TSA) of different cheese samples stored at 7 °C for ripening.
3.1.3. Changes in Spore Counts of Cheese Samples and Observing Spoilage in Cheese Samples During Ripening
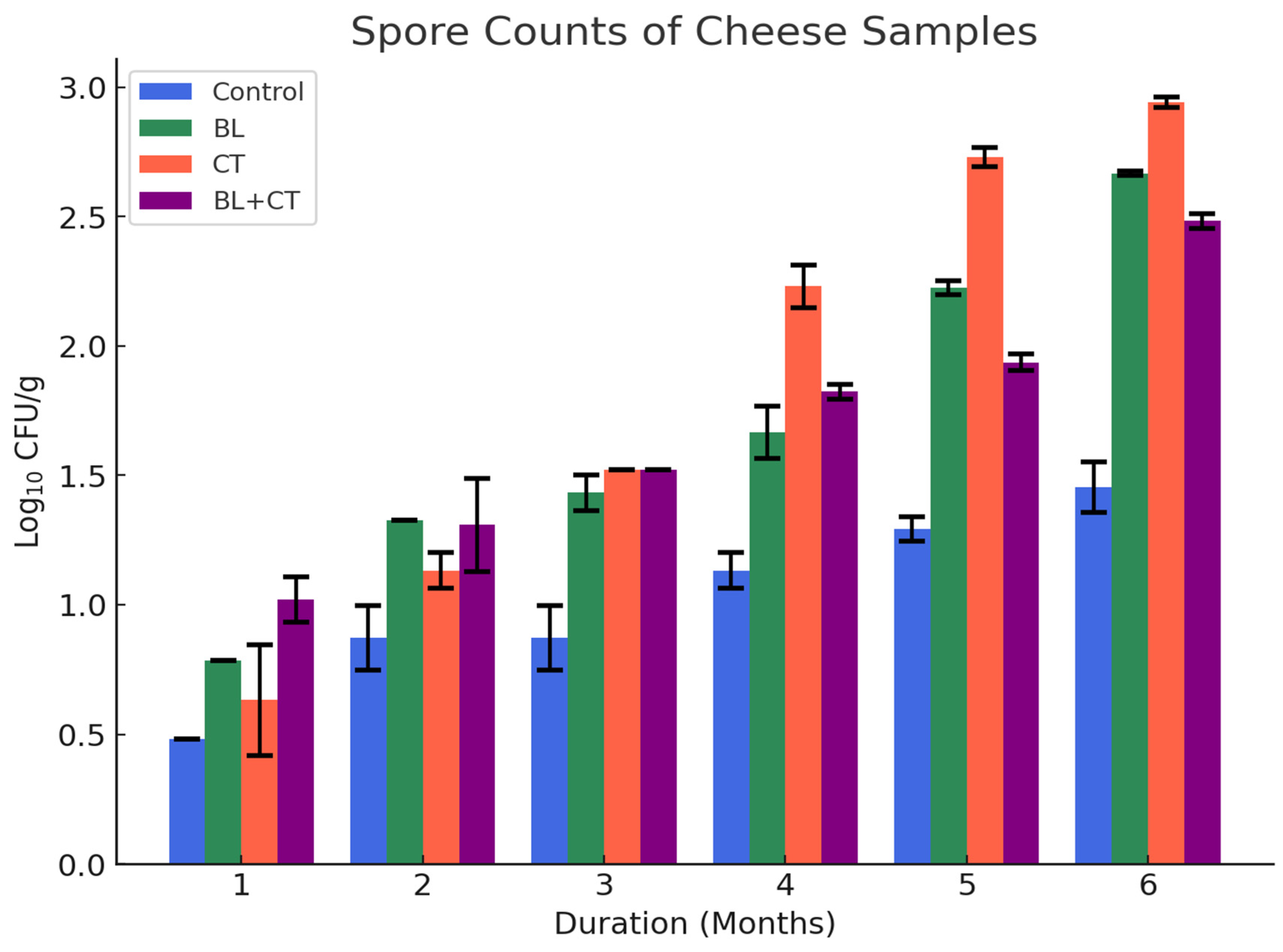
Spore counts of the cheese samples increased progressively during ripening in the spore inoculated cheese samples (T1 (BL), T2 (CT), and T3 (BL+CT)) while the control cheese sample had the lowest spore levels (1.45 ± 0.09) after six months (Figure 1). After one month, all samples, including the control, had negligible or undetectable SC (≤0.5 Log10 CFU/g). However, after three months, SC in the T2 (CT) cheese increased significantly to 1.52 ± 0.00 Log10 CFU/g after three months and continued to rise to 2.94 ± 0.02 Log10 CFU/g up to six months. Similarly, the T1 (BL) cheese gradually increased from 1.43 ± 0.07 Log10 CFU/g at three months to 2.67 ± 0.01 Log10 CFU/g after six months. The T3 (BL+CT) combination reached 2.38 ± 0.04 Log10 CFU/g at the end of ripening. The control cheese, by contrast, maintained SC below 1.45 ± 0.10 Log10 CFU/g, indicating minimal natural contamination and validating the effectiveness of processing conditions. These trends indicate that spore proliferation in cheese is largely driven by the initial inoculation load and favorable ripening conditions, such as low redox potential, moderate acidity, and nutrient diffusion in the matrix [30]. The substantial increase in spore counts, especially in the T2 (CT) and T3 (BL+CT) treatments, suggests that the ripening environment at the end of ripening favored anaerobic spore germination, particularly that of Cl. tyrobutyricum, a known cause of late-blowing defect. These defects were confirmed by visual bloating, cracks, and slits observed in the T2 (CT) and T3 (BL+CT) cheese samples after five months with spore counts of 2.23 ± 0.03 and 1.93 ± 0.03, respectively (Figure 2). This is in line with Garde López-Brea et al. (2013) and Ávila et al. (2023), who reported that anaerobic niches within the cheese core provide ideal conditions for the outgrowth of clostridial spores during extended ripening [31,32]. Notably, no visible spoilage defects such as cracks or bloating were observed in the control and BL cheese despite a slight increase in SC, indicating that visual spoilage may require a threshold of at least 2.0 Log10 CFU/g or higher [33]. This suggests that the onset of spoilage depends not only on the presence of spores but also on their population level and the microenvironment within the cheese during ripening stages. The appearance of visual spoilage after five months in the T2 (CT) and T3 (BL+CT) samples suggests that, as ripening progresses, the cheese environment becomes increasingly anaerobic, creating favorable conditions for anaerobic spores activation and growth. Collectively, these results reinforce the importance of monitoring spore dynamics during cheese maturation, as even controlled production environments can support significant outgrowth when spores are present. The elevated spore loads in inoculated cheeses and the resulting physical defects highlight the importance of strategies aimed at reducing spore presence or enhancing the growth of beneficial microbes to suppress them.
Figure 1.
Spore counts (Log10 CFU/g) of cheese samples stored at 7 °C for ripening. BL—cheese inoculated with aerobic spores (B. licheniformis); CT—cheese inoculated with anaerobic spores (Cl. tyrobutyricum); BL+CT—cheese inoculated with aerobic and anaerobic spores (B. licheniformis and Cl. tyrobutyricum). Counts below 1.0 Log10 CFU/g are near the detection limit and should be considered semi-quantitative.

Figure 2.
Visual observation of cheese samples after 5 months of ripening at 7 °C. T1 (BL)—cheese inoculated with aerobic spores (B. licheniformis); T2 (CT)—cheese inoculated with anaerobic spores (Cl. tyrobutyricum); T3 (BL+CT)—cheese inoculated with aerobic and anaerobic spores (B. licheniformis and Cl. tyrobutyricum).
3.1.4. Starter and NSLAB Counts of Cheese Samples During Ripening
The population dynamics of SLAB and NSLAB during cheese ripening in this study revealed a distinct trend consistent with expected microbial changes. SLAB counts showed a rapid decline from 7.94 ± 0.04 Log10 CFU/g at the first month of ripening to reaching 2.36 ± 0.09 Log10 CFU/g levels by month three and reaching near-zero levels after the six-month ripening (Table 3). This decline is attributed to acid accumulation, depletion of fermentable carbohydrates, and increasing environmental stress, which limit the metabolic activity and survival of starter organisms [34,35]. In contrast, NSLAB populations increased progressively, reaching 8.4 Log10 CFU/g by the third month and remaining above 8.4 Log10 CFU/g through the end of ripening (Table 4). This pattern was observed across all treatments, including those inoculated with aerobic T1 (BL) and anaerobic T2 (CT) spores.

Table 3.
Starter counts (Log10CFU/g) on M17 agar of different cheese samples stored at 7 °C for ripening.

Table 4.
Non-starter lactic acid bacteria (NSLAB) counts (Log10CFU/g) on de Man Rogosa agar (MRS) of different cheese samples stored at 7 °C for ripening.
This suggests that the presence of aerobic or anaerobic spore formers did not affect the overall NSLAB population, and spores and NSLAB were resilient and were not competitively excluded. The persistence of high spores alongside NSLAB indicates that NSLAB may not be sufficient to fully suppress spore proliferation, especially when spores were inoculated in high numbers initially. Therefore, in the absence of supportive interventions, the native NSLAB microbiota, despite their abundance, may lack the biopreservative effect required to control spore-driven spoilage under standard ripening conditions [9]. The stability of NSLAB alongside the increasing SC suggests that biocontrol strategies utilizing NSLAB to suppress spore formers may require strain-specific approaches, as not all NSLAB exhibit strong competitive exclusion or antimicrobial activity against these contaminants, or it is possible are that their numbers are low to exert a significant antimicrobial effect [31]. While some NSLAB strains produce organic acids, bacteriocins, or other antimicrobial compounds that can inhibit spore formers, their effectiveness depends on their abundance, metabolic activity, and interaction with the cheese matrix [26]. Previous studies have reported that the optimal pH range for NSLAB growth and metabolic activity lies between 4.5 and 6.0 [36]. This aligns with our observation, as the cheese pH during the active growth phase of NSLAB remained around 5.0 across treatments, supporting their sustained viability and functionality. The maintenance of this favorable pH environment likely contributed to the successful establishment and dominant populations of NSLAB during the ripening process despite the presence of competing microorganisms. In contrast, lipolytic enzymes from LAB are known to exhibit maximal activity near neutral pH, particularly around pH 7.0 [37]. This may explain the relatively low extent of lipolysis observed in our study despite a slight increase, as the ripening pH of approximately 5.0 was suboptimal for strong lipolytic activity.
3.1.5. Shifts in NSLAB Population and Emergence of Dominant NSLAB
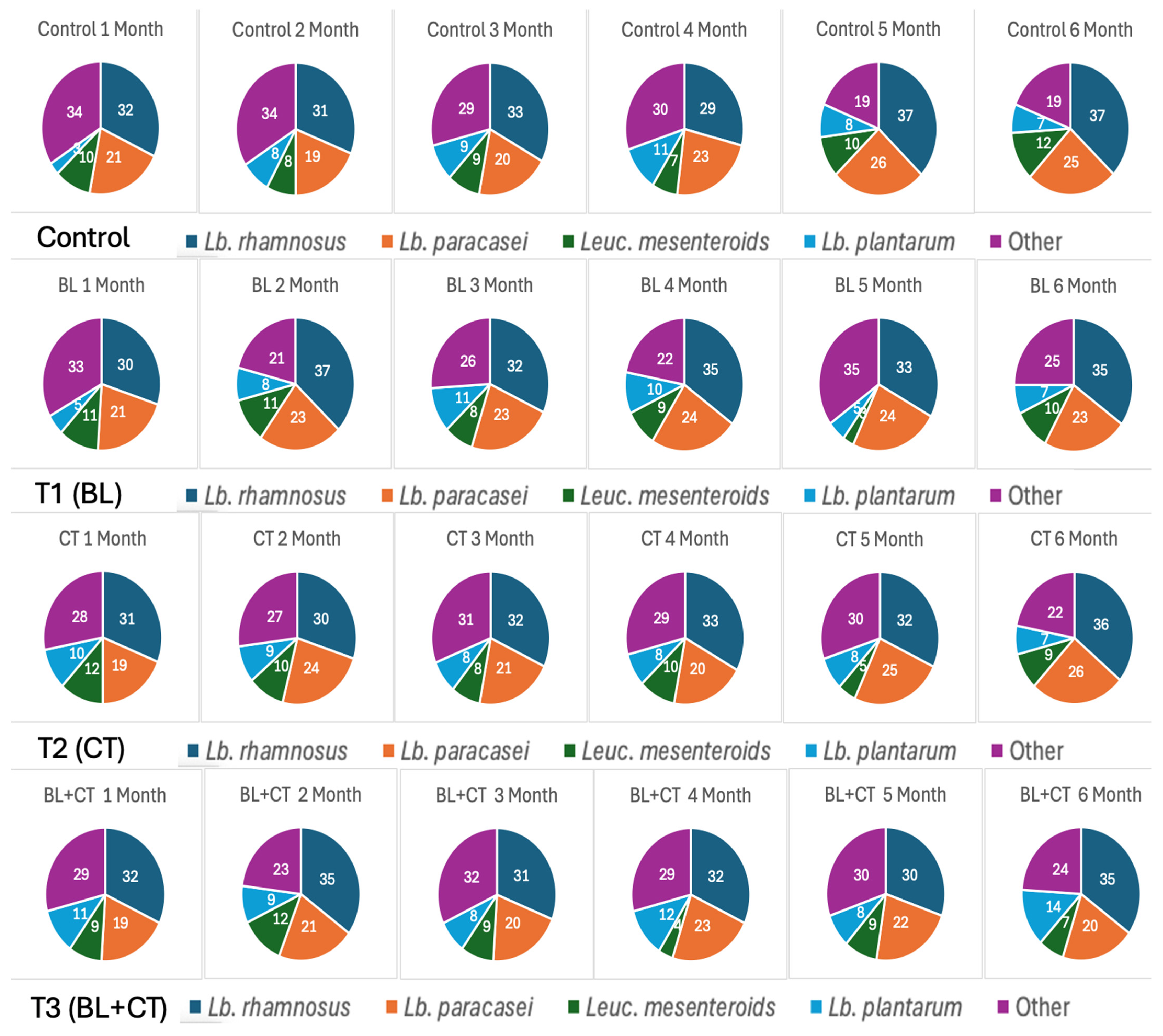
NSLAB is crucial to the development of flavor, texture, and safety in ripened cheeses, particularly cheddar [38]. Among the most commonly found NSLAB in cheddar are species of the genus Lactobacillus, including L. rhamnosus, L. paracasei, and L. plantarum, which contribute significantly to secondary proteolysis and microbial stability during aging [7]. Based on the MALDI-TOF data, across the four cheese types, L. rhamnosus consistently emerged as the most dominant NSLAB species throughout ripening (Figure 3). Its abundance increased from approximately 24% in the first month to 34–37% after six months. This dominance suggests that L. rhamnosus is well adapted to the ripening environment, thriving under low pH, reduced moisture, and increasing proteolysis, conditions that typically suppress other microbial groups [39,40]. Its ability to persist despite aerobic and anaerobic spores further underscores its resilience and ecological competitiveness.
Figure 3.
6 Dominance of L. rhamnosus and L. paracasei during cheese ripening. T1 (BL)—cheese inoculated with aerobic spores (B. licheniformis); T2 (CT)—cheese inoculated with anaerobic spores (Cl. tyrobutyricum); T3 (BL+CT)—cheese inoculated with aerobic and anaerobic spores (B. licheniformis and Cl. tyrobutyricum).
Similarly, L. paracasei showed steady growth and was the second most dominant species by the end of ripening, particularly in T1 (BL) and T2 (CT) cheeses, accounting for 20–25% of the relative population by month 6. Its growth trajectory mirrored the increase in NSLAB counts, suggesting it played an active role in secondary proteolysis and microbial balance [41]. In contrast, Leuc. mesenteroides and L. plantarum showed more variable and generally lower levels (<10%), likely due to their sensitivity to salt, pH, or competition from more robust LAB species [42,43]. Interestingly, in samples with higher spore counts (T2 (CT) and T3 (BL+CT)), L. rhamnosus and L. paracasei still maintained dominance, suggesting these species may possess inherent resistance mechanisms or competitive traits that allow them to co-exist or even inhibit spore formers indirectly. This microbial dynamic aligns closely with the trends observed in spore activity and chemical composition. Cheeses with higher spore counts T2 (CT) and T3 (BL+CT) also showed higher L. rhamnosus and L. paracasei dominance, suggesting that these NSLAB species may contribute to the competitive inhibition of spores or control their activity through acid production or nutrient competition [44]. Meanwhile, the pH (around 5.0) and rising protein and FFA levels suggest active metabolic processes [9], likely driven by NSLAB, such as L. rhamnosus, which is known for its proteolytic capacity and role in flavor development during ripening [45]. In conclusion, while the overall NSLAB population showed some variability, L. rhamnosus and L. paracasei consistently dominated across all cheese samples, including the sample spiked with spores. This highlights their potential to be selectively enhanced to help control spoilage associated with spores and provides a strong basis for the selection of specific NSLAB strains during cheese ripening. A previous study indicated the suitability of inulin, as both L. rhamnosus and L. paracasei preferentially utilized inulin [46]. This strategic choice may provide targeted enhancement of the dominant NSLAB populations, increasing their competitive and antimicrobial potential during cheese ripening.
3.2. Physicochemical Analysis
3.2.1. Moisture Content During Cheese Ripening
The moisture content of cheese samples decreased progressively during the 6-month ripening period, starting from initial values between 37.32% and 38.25% during the first month to 34.49 to 35.40% after six months across all treatments (Table 5), consistent with typical moisture loss due to evaporation and structural reorganization within the cheese matrix [47]. The results observed were similar to the previous studies, where the moisture content of cheddar cheese ranges from 37 to 40% [14,47,48]. In control cheese, moisture dropped steadily from 38.09 ± 0.64% during the first month to 34.49 ± 0.17% at six months, indicating typical dehydration during ripening. The BL cheese followed a similar trend, decreasing from 37.57 ± 1.04% to 34.21 ± 0.53%, suggesting that inoculation with B. licheniformis did not significantly affect moisture loss. The T2 (CT) cheese showed more variation, with moisture declining from 38.25 ± 0.06% to 35.40 ± 1.68%, and higher standard deviations were observed at the fourth month (35.72 ± 2.44%), likely due to gas formation and structural irregularities caused by Cl. tyrobutyricum [31]. The T3 (BL+CT) cheese showed a consistent reduction from 37.32 ± 0.25% to 34.40 ± 0.41%, with minor fluctuations. Despite some variability, especially in T2 (CT) samples, all cheeses exhibited the expected moisture decline due to evaporation under storage conditions. These results indicate that while anaerobic spore activity may influence localized moisture retention, overall moisture loss trends remained consistent across treatments [49]. The consistent moisture decline across all treatments reflects normal ripening-related dehydration. B. licheniformis had minimal impact on moisture loss, while the greater variability in T2 (CT)-spiked cheese suggests localized moisture retention due to gas formation and matrix disruption. Despite this, overall moisture trends remained within expected cheddar ripening ranges, indicating well-controlled ripening conditions and only minor microbial influence on bulk moisture dynamics.

Table 5.
Moisture content (%) of different cheese samples stored at 7 °C for ripening.
3.2.2. Protein Content During Cheese Ripening
The protein content of all cheese samples remained relatively consistent over the six-month ripening period (Table 6), with values ranging between 23.72% and 25.35%. In the control cheese, protein content increased slightly from 24.16 ± 0.17% at the first month to 25.26 ± 0.10% after six months; a similar trend was observed in the other treatment groups. The T1 (BL) cheese showed a slight rise from 24.83 ± 0.32% to 25.16 ± 0.19%, while T2 (CT) cheese exhibited a more variable increase from 23.72 ± 0.77% to 25.08 ± 1.14%, likely reflecting fluctuations due to internal structural disruption from anaerobic gas production or minor shifts in microbial proteolytic activity. The T3 (BL+CT) cheese showed a steady increase from 24.13 ± 0.76% to 25.30 ± 0.31%. These findings are consistent with previous studies indicating that increases in total protein percentage during ripening are primarily due to the reduction in moisture rather than the synthesis of new protein or extensive proteolysis [14,47]. Although NSLAB and residual rennet may contribute to secondary proteolysis, which releases peptides and amino acids, this is not reflected in gross protein percentage but rather in nitrogen solubility or free amino acid assays. The stability of gross protein content, despite ongoing microbial activity, suggests that the effects of proteolysis were likely masked by concurrent moisture evaporation, which concentrates solids. Variability in the CT cheese may indicate localized proteolytic effects or matrix disruption, but overall, protein changes align with typical ripening behavior and do not directly quantify NSLAB-driven proteolysis.

Table 6.
Protein content (%) of different cheese samples stored at 7 °C for ripening.
3.2.3. Fat Content During Cheese Ripening
The fat content of the cheese sample did not vary significantly (p < 0.05) throughout the six months of ripening, with values ranging from 33.25% to 35.02%, showing no significant trends of increase or decrease over time (Table 7). In control cheese, the fat content varied slightly from 33.90 ± 0.15% in the first month to 33.72 ± 0.21% in six months, reflecting minor fluctuations likely due to moisture changes. Similarly, BL cheese showed slight variability, peaking at 35.02 ± 0.99% after three months but decreasing to 33.36 ± 0.17% by six months, with no consistent trend. T2 (CT) cheese exhibited a slight decline from 34.49 ± 0.40% to 34.36 ± 0.82%, with a temporary drop to 33.25 ± 0.01% after three months, possibly due to structural disruption caused by spore germination or internal gas formation. The T3 (BL+CT) cheese followed a similar pattern, with values ranging from 34.48 ± 0.62% to 33.73 ± 0.82%, suggesting that fat content remained largely unaffected by inoculation. These modest changes are consistent with previous findings that fat remains relatively the same during ripening, with no significant loss, as microbial lipolysis primarily affects triglyceride breakdown into free fatty acids rather than altering bulk fat content [25,49]. The slight variations observed may be attributed to moisture loss and natural variation in curd structure rather than lipolytic activity, as confirmed by other studies showing that fat content changes minimally during the maturation of semi-hard cheeses [50]. However, the overall fat content was within standards in all cheese samples.

Table 7.
Fat content (%) of different cheese samples stored at 7 °C for ripening.
3.2.4. Free Fatty Acids and pH During Cheese Ripening
Monitoring FFA levels and pH during cheese ripening is essential for understanding lipolysis, microbial activity, and flavor development. FFAs contribute significantly to the sensory profile of cheese, imparting desirable notes when balanced but leading to rancidity if they accumulate excessively. pH, on the other hand, is a critical physicochemical parameter that influences microbial population, enzymatic activity, and textural stability. FFA content (% oleic acid) increased progressively in all cheese samples over the six-month ripening period (Table 8) and coincided with steady microbial counts of NSLAB, particularly L. rhamnosus and L. paracasei, which are known to contribute to lipolysis through strain-dependent lipase and esterase activities [51]. The control cheese showed a gradual rise from 0.66 ± 0.01% to 0.84 ± 0.07%, while higher final values were observed in spore-inoculated cheeses: T1 (BL) (0.91 ± 0.01%), T2 (CT) (0.93 ± 0.05%), and T3 (BL+CT) (0.92 ± 0.08%). The most notable increase was observed in the T2 (CT) cheese, likely due to the lipase activity of Cl. tyrobutyricum, which is known to hydrolyze milk triglycerides into short- and medium-chain FFAs [52]. NSLAB may have contributed synergistically to lipolysis as well, as they possess strain-dependent esterase and lipase activities that are typically enhanced in the late-ripening phase [53]. The control cheese maintained a pH of around 5.00 ± 0.04, with similar values observed in T1 (BL), T2 (CT), and T3 (BL+CT) cheeses, showing no significant variation due to spore inoculation (Table 9). This pH stability reflects a balanced interplay between acidification by starter lactic acid bacteria and buffering from proteolysis and microbial metabolism during ripening [8]. A consistent pH environment around 5.0 is optimal for promoting NSLAB activity while simultaneously inhibiting the growth of spoilage and pathogenic organisms. The absence of pH fluctuations suggests that despite increased FFA release and potential microbial competition, the cheese matrix retained its buffering capacity and compositional integrity, supporting microbial balance and safe maturation [54]. Sensory evaluation of the samples was not conducted due to the intentional addition of spores; however, no off odors were detected during handling. This may be due to the free fatty acid (FFA) levels that remained below the threshold (1.5–2.0%) typically associated with developing off odors [25]. The observed microbial distribution and metabolic outputs, including FFA release, indicate that lipolysis during ripening was an outcome of NSLAB persistence, with pH buffering enabling these processes without compromising the cheese microbial or structural integrity.

Table 8.
Free fatty acid (FFA) content (% oleic acid) of different cheese samples stored at 7 °C for ripening.

Table 9.
pH content of different cheese samples stored at 7 °C for ripening.
Overall, the ripening process was marked by consistent microbial counts, with NSLAB—particularly L. rhamnosus and L. paracasei—dominating across all cheese samples. These bacteria thrived under consistent pH conditions (5.0) and contributed to key ripening activities such as proteolysis and lipolysis, which help develop cheese flavor. The addition of aerobic (B. licheniformis) and anaerobic (Cl. tyrobutyricum) spores did not significantly disrupt total microbial balance or affect major physicochemical parameters such as moisture, protein, and fat content. However, gas formation and visible defects like slits were observed in T2 (CT) and T3 (BL+CT) samples after 5 months of ripening, indicating spoilage in cheese samples inoculated with spore counts exceeding 2.0 logs. Although NSLAB remained dominant, and pH stayed consistent, these bacteria were not able to prevent spoilage in cheese samples spiked with higher numbers of spores. This suggests that while NSLAB contributes to microbial resilience and overall balance during ripening, their natural presence alone may not be sufficient to suppress spore formers under high spore loads. Therefore, there is a need to selectively enhance NSLAB strains with strong antagonistic activity against spoilage organisms during cheese ripening.
4. Conclusions
This study provides valuable insights into the microbial and physicochemical changes during the ripening of cheddar cheese inoculated with spores. Among the native NSLAB, L. rhamnosus emerged as the most dominant and resilient species, consistently present across all treatments throughout ripening. Its persistence under low pH, moisture loss, and competitive microbial pressure highlights its metabolic adaptability and ecological competitiveness, especially in cheeses challenged with spore formers. L. paracasei also demonstrated good persistence, particularly in spore-inoculated cheeses, while Leuc. mesenteroides and L. plantarum were less consistent, suggesting sensitivity to environmental stress or competition. Cheeses inoculated with Cl. tyrobutyricum and a mixture of B. licheniformis and Cl. tyrobutyricum showed visible late-blowing defects, such as slits and bloating after five months of ripening, corresponding to spore counts exceeding 2.0 logs. Despite the constant dominance of NSLAB like L. rhamnosus and L. paracasei, these species were inadequate to prevent spoilage, suggesting that higher initial spore loads can overcome natural microbial defenses presented by NSLAB. These findings suggest that targeted enhancement of NSLAB strains, either through adjunct culture addition or nutritional support such as prebiotics, may help tip the microbial balance toward beneficial populations that can outcompete or inhibit spore formers.
In this study, physicochemical changes supported the microbial trends. Moisture content declined steadily due to syneresis and evaporation, while protein and fat contents remained relatively consistent, with slight increases largely attributed to moisture loss rather than metabolic breakdown. FFA levels rose during ripening, particularly in spore-inoculated cheeses, reflecting both spores and NSLAB lipolytic activity. The pH remained steady (5.0), supporting NSLAB growth while limiting spoilage bacteria. Based on the observed abundance and persistence of L. rhamnosus, future work could focus on developing optimized adjunct cultures from robust NSLAB strains or selectively enhancing the NSLAB population capable of suppressing spore formers and enhancing cheese quality without disrupting the native microbial balance. While NSLAB offers competitive advantages under ripening conditions, spoilage can still occur unexpectedly, especially when the initial spore load is high.
Author Contributions
Conceptualization, R.K. and S.A.; methodology, R.K.; validation, R.K., and S.A.; formal analysis, R.K.; investigation, S.A.; resources, R.K., and S.A.; data curation, R.K.; writing—original draft preparation, R.K.; writing—review and editing, R.K. and S.A.; visualization, S.A.; supervision, S.A.; project administration, S.A.; funding acquisition, S.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Midwest Dairy Association, grant number 3X3252.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author.
Acknowledgments
The authors would like to acknowledge the Department of Dairy and Food Science, SDSU, Davis Dairy Plant, SDSU, and Agricultural Experimental Station SDSU. All authors have consented to the acknowledgment.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| SLAB | Starter lactic acid bacteria |
| NSLAB | Nonstarter lactic acid bacteria |
| LBD | Late-blowing defect |
| TPC | Total plate counts |
References
- Khattab, A.R.; Guirguis, H.A.; Tawfik, S.M.; Farag, M.A. Cheese ripening: A review on modern technologies towards flavor enhancement, process acceleration and improved quality assessment. Trends Food Sci. Technol. 2019, 88, 343–360. [Google Scholar] [CrossRef]
- Forde, A.; Fitzgerald, G.F. Biotechnological approaches to the understanding and improvement of mature cheese flavour. Curr. Opin. Biotechnol. 2000, 11, 484–489. [Google Scholar] [CrossRef]
- Blaya, J.; Barzideh, Z.; LaPointe, G. Symposium review: Interaction of starter cultures and nonstarter lactic acid bacteria in the cheese environment. J. Dairy Sci. 2018, 101, 3611–3629. [Google Scholar] [CrossRef]
- Coelho, M.C.; Malcata, F.X.; Silva, C.C. Lactic acid bacteria in raw-milk cheeses: From starter cultures to probiotic functions. Foods 2022, 11, 2276. [Google Scholar] [CrossRef] [PubMed]
- Levante, A.; De Filippis, F.; La Storia, A.; Gatti, M.; Neviani, E.; Ercolini, D.; Lazzi, C. Metabolic gene-targeted monitoring of non-starter lactic acid bacteria during cheese ripening. Int. J. Food Microbiol. 2017, 257, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Gatti, M.; Bottari, B.; Lazzi, C.; Neviani, E.; Mucchetti, G. Invited review: Microbial evolution in raw-milk, long-ripened cheeses produced using undefined natural whey starters. J. Dairy Sci. 2014, 97, 573–591. [Google Scholar] [CrossRef]
- Guo, L.; Liu, B. Nonstarter Lactic Acid Bacteria in Cheese. In Handbook of Cheese Chemistry; Royal Society of Chemistry (RSC): Cambridge, UK, 2023; Volume 40, p. 48. [Google Scholar]
- Gobbetti, M.; Di Cagno, R.; Calasso, M.; Neviani, E.; Fox, P.F.; De Angelis, M. Drivers that establish and assemble the lactic acid bacteria biota in cheeses. Trends Food Sci. Technol. 2018, 78, 244–254. [Google Scholar] [CrossRef]
- Gobbetti, M.; De Angelis, M.; Di Cagno, R.; Mancini, L.; Fox, P.F. Pros and cons for using non-starter lactic acid bacteria (NSLAB) as secondary/adjunct starters for cheese ripening. Trends Food Sci. Technol. 2015, 45, 167–178. [Google Scholar] [CrossRef]
- FDA. US Cheddar Cheese, Code of Federal Regulation, Food and Drug Administration. 2024. Available online: https://www.ecfr.gov/current/title-21/chapter-I/subchapter-B/part-133/subpart-B/section-133.113 (accessed on 12 April 2025).
- Rosa, M.C.; Carmo, M.R.; Balthazar, C.F.; Guimarães, J.T.; Esmerino, E.A.; Freitas, M.Q.; Silva, M.C.; Pimentel, T.C.; Cruz, A.G. Dairy products with prebiotics: An overview of the health benefits, technological and sensory properties. Int. Dairy J. 2021, 117, 105009. [Google Scholar] [CrossRef]
- Sharma, S.; Kanwar, S.S. Effect of prebiotics on growth behavior of Lactobacillus plantarum and their impact on adherence of strict anaerobic pathogens to intestinal cell lines. J. Food Saf. 2018, 38, e12384. [Google Scholar] [CrossRef]
- Khanal, S.N.; Anand, S.; Muthukumarappan, K. Evaluation of high-intensity ultrasonication for the inactivation of endospores of 3 bacillus species in nonfat milk. J. Dairy Sci. 2014, 97, 5952–5963. [Google Scholar] [CrossRef] [PubMed]
- Hanlon, M.; Choi, J.; Goddik, L.; Park, S.H. Microbial and chemical composition of Cheddar cheese supplemented with prebiotics from pasteurized milk to aging. J. Dairy Sci. 2022, 105, 2058–2068. [Google Scholar] [CrossRef] [PubMed]
- Ryser, E.T.; Kornacki, J.L. Standard Methods. In Standard Methods for the Examination of Dairy Products, 18th ed.; American Public Health Association: Washington, DC, USA, 2024. [Google Scholar]
- Kornacki, J.L.; Ryser, E.T.; Mangione, C.M.; Wehr, H.M. Standard Methods for the Examination of Dairy Products, 18th ed.; American Public Health Association: Washington, DC, USA, 2024. [Google Scholar]
- Gómez-Torres, N.; Garde, S.; Peirotén, Á.; Ávila, M. Impact of Clostridium spp. on cheese characteristics: Microbiology, color, formation of volatile compounds and off-flavors. Food Control 2015, 56, 186–194. [Google Scholar] [CrossRef]
- Gómez-Torres, N.; Ávila, M.; Gaya, P.; Garde, S. Prevention of late blowing defect by reuterin produced in cheese by a Lactobacillus reuteri adjunct. Food Microbiol. 2014, 42, 82–88. [Google Scholar] [CrossRef]
- Barzideh, Z.; Siddiqi, M.; Mohamed, H.M.; LaPointe, G. Dynamics of starter and non-starter lactic acid bacteria populations in long-ripened cheddar cheese using propidium monoazide (PMA) treatment. Microorganisms 2022, 10, 1669. [Google Scholar] [CrossRef] [PubMed]
- Lay, J.O., Jr. MALDI-TOF mass spectrometry of bacteria. Mass Spectrom. Rev. 2001, 20, 172–194. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analysis of AOAC International; Oxford University Press: Oxford, UK, 2023. [Google Scholar]
- ISO 8968-1, IDF 20-1; IDF Milk: Determination of Nitrogen Content (Kjeldahal Method). IDF Standard 20-A; IDF: Brussels, Belgium, 2001.
- AOAC. AOAC Official Method 989.05Fat in Milk: Modified MojonnierEther Extraction Method. In Official Methods of Analysis of AOAC International; Latimer, G.W., Jr., Ed.; Oxford University Press: Oxford, UK, 2023. [Google Scholar]
- British Standard 770-5:1976; Chemical Analysis of Cheese. Part 5. Determination of pH Value, Methods for Chemical Analysis of Cheese. British Standards Institution: London, UK, 1976.
- Asif, M.; Nadeem, M.; Imran, M.; Ullah, R.; Tayyab, M.; Khan, F.A.; Al-Asmari, F.; Rahim, M.A.; Rocha, J.M.; Korma, S.A. Effect of fat contents of buttermilk on fatty acid composition, lipolysis, vitamins and sensory properties of cheddar-type cheese. Front. Microbiol. 2023, 14, 1209509. [Google Scholar] [CrossRef]
- Gopal, N.; Hill, C.; Ross, P.R.; Beresford, T.P.; Fenelon, M.A.; Cotter, P.D. The prevalence and control of Bacillus and related spore-forming bacteria in the dairy industry. Front. Microbiol. 2015, 6, 1418. [Google Scholar] [CrossRef]
- Silva, C.B.; Ferreira, L.M.; Lima, A.R.; Araújo, K.G.; Souza, R.M.; Fonseca, A.B.M.; Gonzalez, A.G. Microbiological quality and cultivable bacterial community of fresh and ripened Minas cheeses made from raw and pasteurised milk. Int. Dairy J. 2023, 143, 105662. [Google Scholar] [CrossRef]
- Franciosi, E.; Settanni, L.; Cologna, N.; Cavazza, A.; Poznanski, E. Microbial analysis of raw cows’ milk used for cheese-making: Influence of storage treatments on microbial composition and other technological traits. World J. Microbiol. Biotechnol. 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Beresford, T.P.; Fitzsimons, N.A.; Brennan, N.L.; Cogan, T.M. Recent advances in cheese microbiology. Int. Dairy J. 2001, 11, 259–274. [Google Scholar] [CrossRef]
- Fox, P.F.; Guinee, T.P.; Cogan, T.M.; McSweeney, P.L.; Fox, P.F.; Guinee, T.P.; Cogan, T.M.; McSweeney, P.L. Microbiology of cheese ripening. In Fundamentals of Cheese Science; Springer: Berlin/Heidelberg, Germany, 2017; pp. 333–390. [Google Scholar] [CrossRef]
- Ávila, M.; Sánchez, C.; Calzada, J.; Mayer, M.J.; Berruga, M.I.; López-Díaz, T.M.; Narbad, A.; Garde, S. Isolation and characterization of new bacteriophages active against Clostridium tyrobutyricum and their role in preventing the late blowing defect of cheese. Food Res. Int. 2023, 163, 112222. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Brea, S.G.; Gómez-Torres, N.; Arribas, M.Á. Spore-forming bacteria in dairy products. In Microbiology in Dairy Processing: Challenges and Opportunities; Wiley: Hoboken, NJ, USA, 2017; pp. 11–36. [Google Scholar] [CrossRef]
- Carminati, D.; Bonvini, B.; Francolino, S.; Ghiglietti, R.; Locci, F.; Tidona, F.; Mariut, M.; Abeni, F.; Zago, M.; Giraffa, G. Low-Level Clostridial Spores’ Milk to Limit the Onset of Late Blowing Defect in Lysozyme-Free, Grana-Type Cheese. Foods 2023, 12, 1880. [Google Scholar] [CrossRef] [PubMed]
- Bezie, A.; Regasa, H. The role of starter culture and enzymes/rennet for fermented dairy products manufacture—A Review. Nutr. Food Sci. Int. J. 2019, 9, 21–27. [Google Scholar]
- Williams, A.G.; Withers, S.E.; Banks, J.M. Energy sources of non-starter lactic acid bacteria isolated from Cheddar cheese. Int. Dairy J. 2000, 10, 17–23. [Google Scholar] [CrossRef]
- Bansal, V.; Veena, N. Understanding the role of pH in cheese manufacturing: General aspects of cheese quality and safety. J. Food Sci. Technol. 2024, 61, 16–26. [Google Scholar] [CrossRef]
- Thierry, A.; Collins, Y.F.; Mukdsi, M.A.; McSweeney, P.L.; Wilkinson, M.G.; Spinnler, H.E. Lipolysis and metabolism of fatty acids in cheese. In Cheese; Elsevier: Amsterdam, The Netherlands, 2017; pp. 423–444. [Google Scholar]
- Coolbear, T.; Crow, V.; Harnett, J.; Harvey, S.; Holland, R.; Martley, F. Developments in cheese microbiology in New Zealand—Use of starter and non-starter lactic acid bacteria and their enzymes in determining flavour. Int. Dairy J. 2008, 18, 705–713. [Google Scholar] [CrossRef]
- Bove, C.G.; Angelis, M.D.; Gatti, M.; Calasso, M.; Neviani, E.; Gobbetti, M. Metabolic and proteomic adaptation of L actobacillus rhamnosus strains during growth under cheese-like environmental conditions compared to de M an, R ogosa, and S harpe medium. Proteomics 2012, 12, 3206–3218. [Google Scholar] [CrossRef]
- Decadt, H.; De Vuyst, L. Insights into the microbiota and defects of present-day Gouda cheese productions. Curr. Opin. Food Sci. 2023, 52, 101044. [Google Scholar] [CrossRef]
- Banks, J.M.; Williams, A. The role of the nonstarter lactic acid bacteria in Cheddar cheese ripening. Int. J. Dairy Technol. 2004, 57, 145–152. [Google Scholar] [CrossRef]
- Wouters, J.T.; Ayad, E.H.; Hugenholtz, J.; Smit, G. Microbes from raw milk for fermented dairy products. Int. Dairy J. 2002, 12, 91–109. [Google Scholar] [CrossRef]
- Leroy, F.; De Vuyst, L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Technol. 2004, 15, 67–78. [Google Scholar] [CrossRef]
- Minervini, F.; Conte, A.; Del Nobile, M.A.; Gobbetti, M.; De Angelis, M. Dietary fibers and protective lactobacilli drive burrata cheese microbiome. Appl. Environ. Microbiol. 2017, 83, e01494-17. [Google Scholar] [CrossRef] [PubMed]
- de Souza Oliveira, R.P.; Perego, P.; de Oliveira, M.N.; Converti, A. Effect of inulin on the growth and metabolism of a probiotic strain of Lactobacillus rhamnosus in co-culture with Streptococcus thermophilus. LWT 2012, 47, 358–363. [Google Scholar] [CrossRef]
- Abed, S.M.; Ali, A.H.; Noman, A.; Niazi, S.; Ammar, A.; Bakry, A. Inulin as prebiotics and its applications in food industry and human health; a review. Int. J. Agric. Innov. Res. 2016, 5, 88–97. [Google Scholar]
- Fox, P.F.; Guinee, T.P.; Cogan, T.M.; McSweeney, P.L. Fundamentals of Cheese Science; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Tabla, R.; Gómez, A.; Rebollo, J.E.; Molina, F.; Roa, I. Effectiveness of a bacteriophage cocktail in reducing cheese early blowing caused by Escherichia coli. LWT 2022, 153, 112430. [Google Scholar] [CrossRef]
- Murtaza, M.S.; Sameen, A.; Rehman, A.; Huma, N.; Hussain, F.; Hussain, S.; Cacciotti, I.; Korma, S.A.; Ibrahim, S.A.; Ma, Y.K. Physicochemical, techno-functional, and proteolytic effects of various hydrocolloids as fat replacers in low-fat cheddar cheese. Front. Sustain. Food Syst. 2024, 8, 1440310. [Google Scholar] [CrossRef]
- Ávila, M.; Gómez-Torres, N.; Delgado, D.; Gaya, P.; Garde, S. Application of high pressure processing for controlling Clostridium tyrobutyricum and late blowing defect on semi-hard cheese. Food Microbiol. 2016, 60, 165–173. [Google Scholar] [CrossRef]
- Falentin, H.; Postollec, F.; Parayre, S.; Henaff, N.; Le Bivic, P.; Richoux, R.; Thierry, A.; Sohier, D. Specific metabolic activity of ripening bacteria quantified by real-time reverse transcription PCR throughout Emmental cheese manufacture. Int. J. Food Microbiol. 2010, 144, 10–19. [Google Scholar] [CrossRef]
- Doyle, C.J.; Gleeson, D.; Jordan, K.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. Anaerobic sporeformers and their significance with respect to milk and dairy products. Int. J. Food Microbiol. 2015, 197, 77–87. [Google Scholar] [CrossRef]
- Collins, Y.F.; McSweeney, P.L.; Wilkinson, M.G. Lipolysis and free fatty acid catabolism in cheese: A review of current knowledge. Int. Dairy J. 2003, 13, 841–866. [Google Scholar] [CrossRef]
- Dreier, M.; Meola, M.; Berthoud, H.; Shani, N.; Wechsler, D.; Junier, P. High-throughput qPCR and 16S rRNA gene amplicon sequencing as complementary methods for the investigation of the cheese microbiota. BMC Microbiol. 2022, 22, 48. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).