Functional Versatility of Vibrio cholerae Outer Membrane Proteins
Abstract
1. Introduction
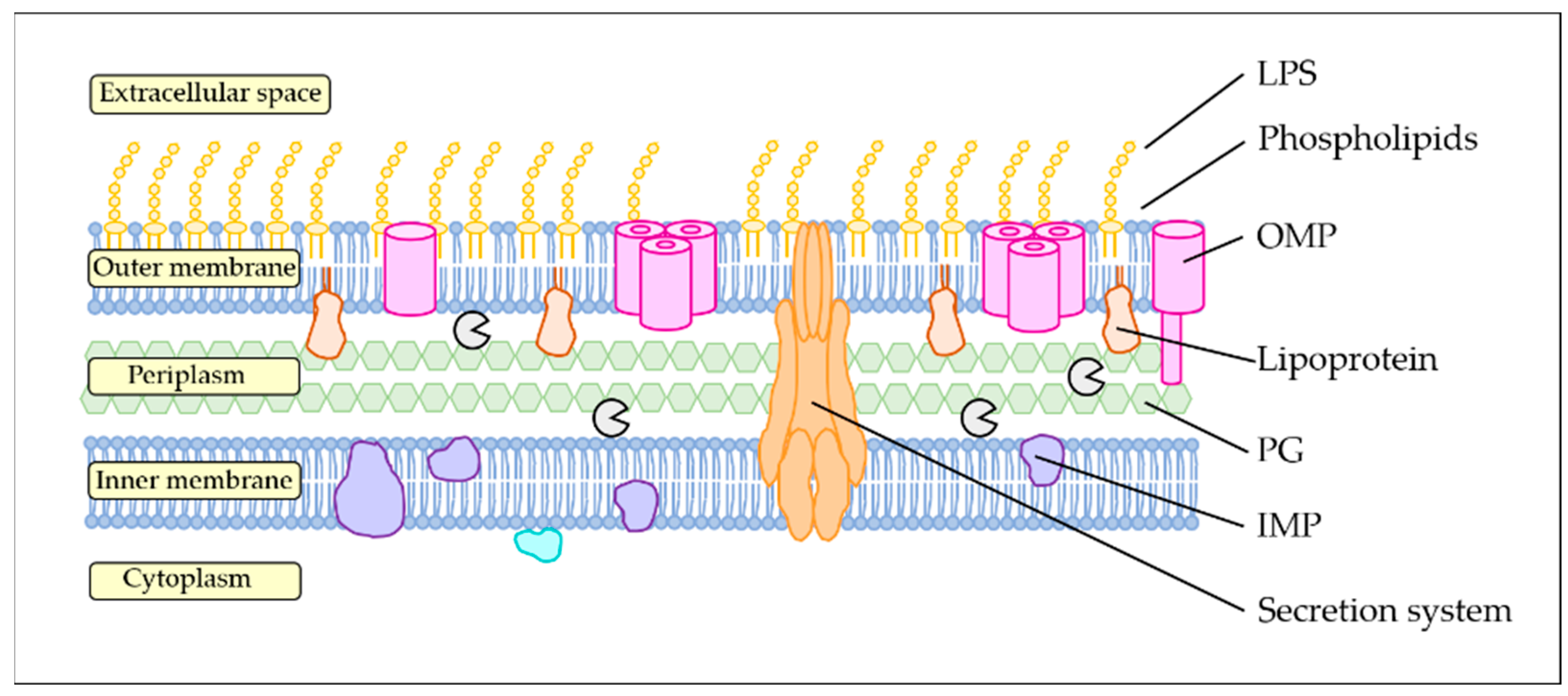
2. The Outer Membrane, a Quick Overview
3. The Outer Membrane Proteins: Structure and Transport to the Outer Membrane
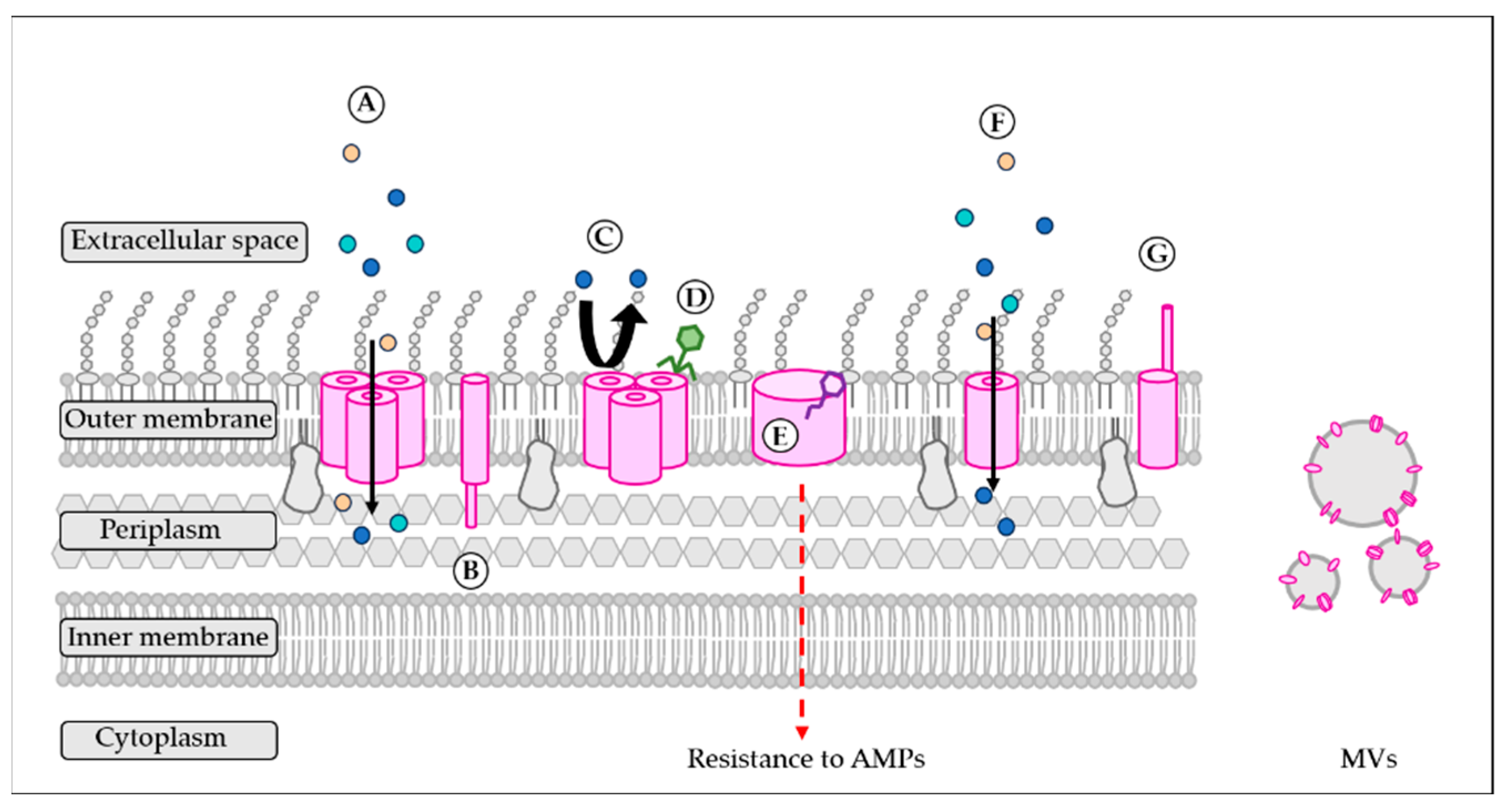
4. A Wide Variety of Functions for Adaptation to Hostile Environments
5. The Outer Membrane Proteins of V. cholerae
5.1. OmpV (VC1318)
5.2. OmpW (VCA0867)
5.3. OmpU (VC0633)
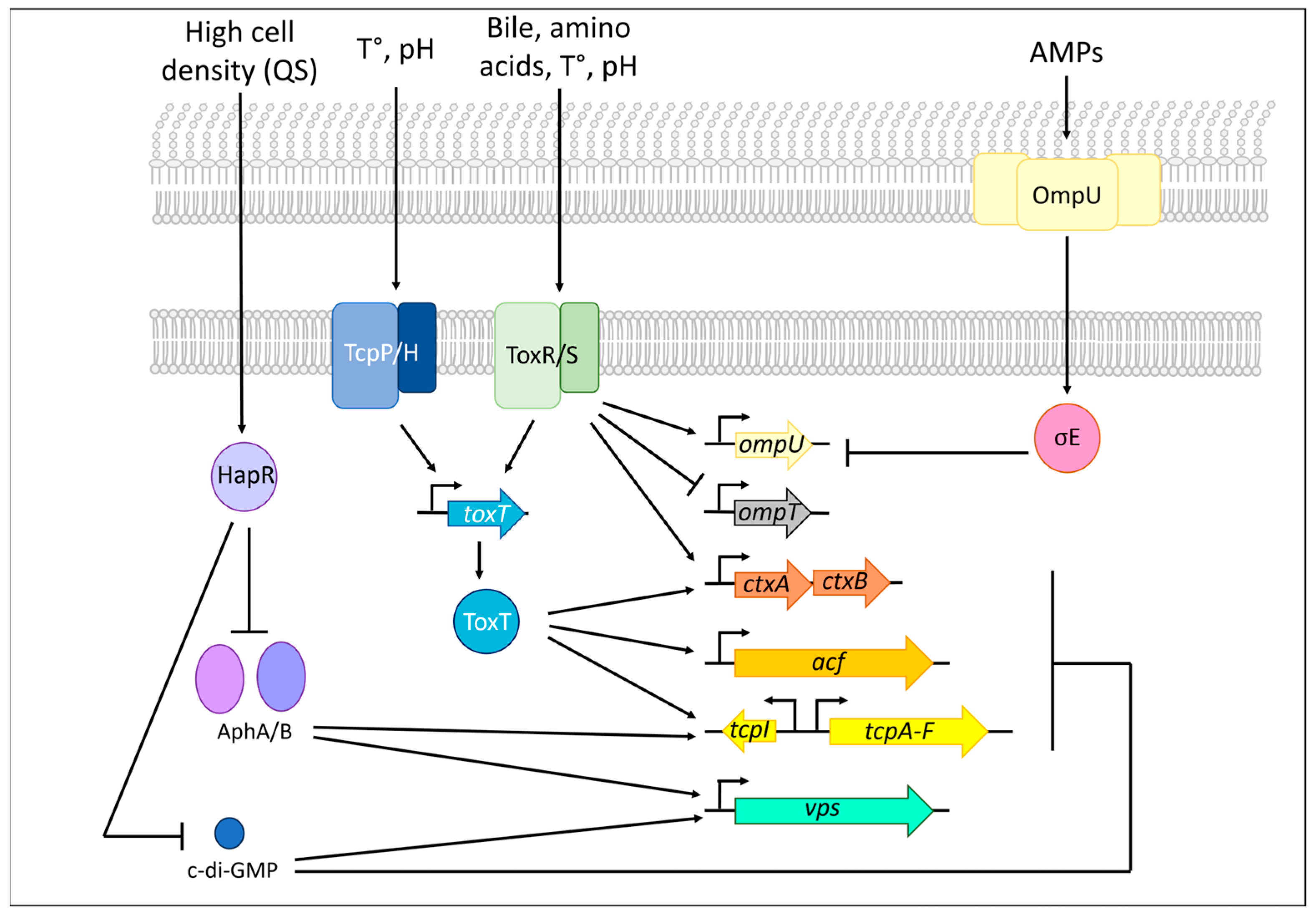
5.4. OmpT (VC1854)
5.5. Maltoporin OmpS (LamB) (VCA1028)
5.6. OmpX
5.7. OmpA (VC2213)
5.8. Chitoporin ChiP (VC0972)
5.9. LptD (VC0446)
5.10. Putative Phosphoporin PhoE (VCA1008)
5.11. TonB-Dependent Siderophore Receptors
5.12. BamA (VC2252)
5.13. ObfA (VC1154)
5.14. Uncharacterized OMPs
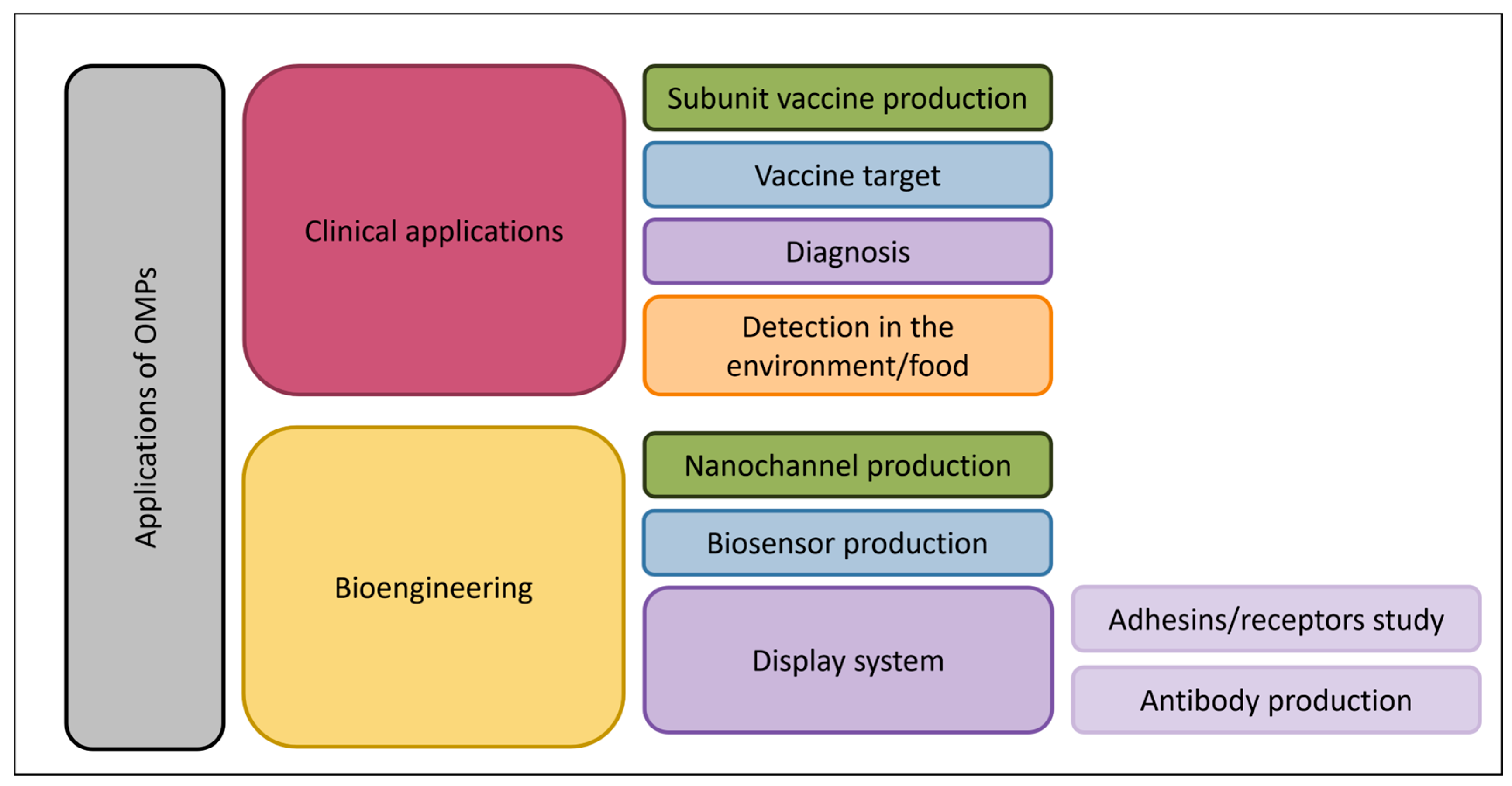
6. Discussion and Future Directions
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Rossolini, G.M.; Arena, F.; Pecile, P.; Pollini, S. Update on the antibiotic resistance crisis. Curr. Opin. Pharmacol. 2014, 18, 56–60. [Google Scholar] [CrossRef] [PubMed]
- Santajit, S.; Indrawattana, N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef] [PubMed]
- Delcour, A.H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta 2009, 1794, 808–816. [Google Scholar] [CrossRef]
- Guest, R.L.; Silhavy, T.J. Cracking outer membrane biogenesis. Biochim. Biophys. Acta-Mol. Cell Res. 2023, 1870, 119405. [Google Scholar] [CrossRef] [PubMed]
- Henderson, J.C.; Zimmerman, S.M.; Crofts, A.A.; Boll, J.M.; Kuhns, L.G.; Herrera, C.M.; Trent, M.S. The power of asymmetry: Architecture and assembly of the Gram-negative outer membrane lipid bilayer. Annu. Rev. Microbiol. 2016, 70, 255–278. [Google Scholar] [CrossRef]
- Narita, S.I.; Tokuda, H. Bacterial lipoproteins; biogenesis, sorting and quality control. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 1414–1423. [Google Scholar] [CrossRef]
- Koebnik, R.; Locher, K.P.; Van Gelder, P. Structure and function of bacterial outer membrane proteins: Barrels in a nutshell. Mol. Microbiol. 2000, 37, 239–253. [Google Scholar] [CrossRef]
- Guerrero-Mandujano, A.; Hernandez-Cortez, C.; Ibarra, J.A.; Castro-Escarpulli, G. The outer membrane vesicles: Secretion system type zero. Traffic 2017, 18, 425–432. [Google Scholar] [CrossRef]
- Reyes-Robles, T.; Dillard, R.S.; Cairns, L.S.; Silva-Valenzuela, C.A.; Housman, M.; Ali, A.; Wright, E.R.; Camilli, A. Vibrio cholerae outer membrane vesicles inhibit bacteriophage infection. J. Bacteriol. 2018, 200. [Google Scholar] [CrossRef]
- Zingl, F.G.; Thapa, H.B.; Scharf, M.; Kohl, P.; Muller, A.M.; Schild, S. Outer membrane vesicles of Vibrio cholerae protect and deliver active cholera toxin to host cells via porin-dependent uptake. mBio 2021, 12, e0053421. [Google Scholar] [CrossRef]
- Bitar, A.; Aung, K.M.; Wai, S.N.; Hammarstrom, M.L. Vibrio cholerae derived outer membrane vesicles modulate the inflammatory response of human intestinal epithelial cells by inducing microRNA-146a. Sci. Rep. 2019, 9, 7212. [Google Scholar] [CrossRef]
- Duperthuy, M.; Sjostrom, A.E.; Sabharwal, D.; Damghani, F.; Uhlin, B.E.; Wai, S.N. Role of the Vibrio cholerae matrix protein Bap1 in cross-resistance to antimicrobial peptides. PLoS Pathog. 2013, 9, e1003620. [Google Scholar] [CrossRef] [PubMed]
- Vanhove, A.S.; Duperthuy, M.; Charriere, G.M.; Le Roux, F.; Goudenege, D.; Gourbal, B.; Kieffer-Jaquinod, S.; Coute, Y.; Wai, S.N.; Destoumieux-Garzon, D. Outer membrane vesicles are vehicles for the delivery of Vibrio tasmaniensis virulence factors to oyster immune cells. Environ. Microbiol. 2015, 17, 1152–1165. [Google Scholar] [CrossRef] [PubMed]
- Giacomucci, S.; Mathieu-Denoncourt, A.; Vincent, A.T.; Jannadi, H.; Duperthuy, M. Experimental evolution of Vibrio cholerae identifies hypervesiculation as a way to increase motility in the presence of polymyxin B. Front. Microbiol. 2022, 13, 932165. [Google Scholar] [CrossRef] [PubMed]
- Clemens, J.D.; Nair, G.B.; Ahmed, T.; Qadri, F.; Holmgren, J. Cholera. Lancet 2017, 390, 1539–1549. [Google Scholar] [CrossRef]
- Halpern, M.; Izhaki, I. Fish as hosts of Vibrio cholerae. Front. Microbiol. 2017, 8, 282. [Google Scholar] [CrossRef]
- Kaper, J.B.; Morris, J.G., Jr.; Levine, M.M. Cholera. Clin. Microbiol. Rev. 1995, 8, 48–86. [Google Scholar] [CrossRef]
- Deen, J.; Mengel, M.A.; Clemens, J.D. Epidemiology of cholera. Vaccine 2020, 38 (Suppl. 1), A31–A40. [Google Scholar] [CrossRef]
- Johnson, J.A.; Salles, C.A.; Panigrahi, P.; Albert, M.J.; Wright, A.C.; Johnson, R.J.; Morris, J.G., Jr. Vibrio cholerae O139 synonym bengal is closely related to Vibrio cholerae El Tor but has important differences. Infect. Immun. 1994, 62, 2108–2110. [Google Scholar] [CrossRef]
- Li, M.; Shimada, T.; Morris, J.G., Jr.; Sulakvelidze, A.; Sozhamannan, S. Evidence for the emergence of non-O1 and non-O139 Vibrio cholerae strains with pathogenic potential by exchange of O-antigen biosynthesis regions. Infect. Immun. 2002, 70, 2441–2453. [Google Scholar] [CrossRef]
- Tobin-D’Angelo, M.; Smith, A.R.; Bulens, S.N.; Thomas, S.; Hodel, M.; Izumiya, H.; Arakawa, E.; Morita, M.; Watanabe, H.; Marin, C.; et al. Severe diarrhea caused by cholera toxin-producing vibrio cholerae serogroup O75 infections acquired in the southeastern United States. Clin. Infect. Dis. 2008, 47, 1035–1040. [Google Scholar] [CrossRef] [PubMed]
- Haley, B.J.; Choi, S.Y.; Grim, C.J.; Onifade, T.J.; Cinar, H.N.; Tall, B.D.; Taviani, E.; Hasan, N.A.; Abdullah, A.H.; Carter, L.; et al. Genomic and phenotypic characterization of Vibrio cholerae non-O1 isolates from a US Gulf Coast cholera outbreak. PLoS ONE 2014, 9, e86264. [Google Scholar] [CrossRef] [PubMed]
- Crowe, S.J.; Newton, A.E.; Gould, L.H.; Parsons, M.B.; Stroika, S.; Bopp, C.A.; Freeman, M.; Greene, K.; Mahon, B.E. Vibriosis, not cholera: Toxigenic Vibrio cholerae non-O1, non-O139 infections in the United States, 1984–2014. Epidemiol. Infect. 2016, 144, 3335–3341. [Google Scholar] [CrossRef] [PubMed]
- Ke, B.; Pang, B.; He, D.; Xu, J.; Chen, Q.; Liang, J.; Chen, J.; Li, Z.; Zhou, H.; Deng, X.; et al. Phylogenetic analysis of serogroup O5 Vibrio cholerae that caused successive cholera outbreaks—Guangdong province, China, 2020–2021. China CDC Wkly. 2022, 4, 238–241. [Google Scholar] [CrossRef]
- Mathieu-Denoncourt, A.; Giacomucci, S.; Duperthuy, M. The secretome of Vibrio cholerae. In Infections and Sepsis Development; Huang, L., Li, J., Eds.; IntechOpen: London, UK, 2021. [Google Scholar]
- Echazarreta, M.A.; Klose, K.E. Vibrio Flagellar Synthesis. Front. Cell. Infect. Microbiol. 2019, 9, 131. [Google Scholar] [CrossRef]
- Oki, H.; Kawahara, K.; Iimori, M.; Imoto, Y.; Nishiumi, H.; Maruno, T.; Uchiyama, S.; Muroga, Y.; Yoshida, A.; Yoshida, T.; et al. Structural basis for the toxin-coregulated pilus–dependent secretion of Vibrio cholerae colonization factor. Sci. Adv. 2022, 8, eabo3013. [Google Scholar] [CrossRef]
- Jaroslawski, S.; Duquesne, K.; Sturgis, J.N.; Scheuring, S. High-resolution architecture of the outer membrane of the Gram-negative bacteria Roseobacter denitrificans. Mol. Microbiol. 2009, 74, 1211–1222. [Google Scholar] [CrossRef]
- Wimley, W.C. Toward genomic identification of beta-barrel membrane proteins: Composition and architecture of known structures. Protein Sci. 2002, 11, 301–312. [Google Scholar] [CrossRef]
- Silhavy, T.J.; Kahne, D.; Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Kamio, Y.; Nikaido, H. Outer membrane of Salmonella Typhimurium: Accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry 1976, 15, 2561–2570. [Google Scholar] [CrossRef]
- Bertani, B.; Ruiz, N. Function and Biogenesis of Lipopolysaccharides. EcoSal Plus 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Malinverni, J.C.; Silhavy, T.J. An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc. Natl. Acad. Sci. USA 2009, 106, 8009–8014. [Google Scholar] [CrossRef]
- Hughes, G.W.; Hall, S.C.L.; Laxton, C.S.; Sridhar, P.; Mahadi, A.H.; Hatton, C.; Piggot, T.J.; Wotherspoon, P.J.; Leney, A.C.; Ward, D.G.; et al. Evidence for phospholipid export from the bacterial inner membrane by the Mla ABC transport system. Nat. Microbiol. 2019, 4, 1692–1705. [Google Scholar] [CrossRef]
- Sun, J.; Rutherford, S.T.; Silhavy, T.J.; Huang, K.C. Physical properties of the bacterial outer membrane. Nat. Rev. Microbiol. 2022, 20, 236–248. [Google Scholar] [CrossRef]
- Nikaido, H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef]
- Nikaido, H.; Vaara, M. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 1985, 49, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, T.S.; Parkin, J.; Khalid, S. The free energy of small solute permeation through the Escherichia coli outer membrane has a distinctly asymmetric profile. J. Phys. Chem. Lett. 2016, 7, 3446–3451. [Google Scholar] [CrossRef]
- Beveridge, T.J. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 1999, 181, 4725–4733. [Google Scholar] [CrossRef] [PubMed]
- Toyofuku, M.; Nomura, N.; Eberl, L. Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 2019, 17, 13–24. [Google Scholar] [CrossRef]
- Magana, G.; Harvey, C.; Taggart, C.C.; Rodgers, A.M. Bacterial outer membrane vesicles: Role in pathogenesis and host-cell interactions. Antibiotics 2023, 13, 32. [Google Scholar] [CrossRef]
- Toyofuku, M.; Schild, S.; Kaparakis-Liaskos, M.; Eberl, L. Composition and functions of bacterial membrane vesicles. Nat. Rev. Microbiol. 2023, 21, 415–430. [Google Scholar] [CrossRef]
- Marinacci, B.; Krzyzek, P.; Pellegrini, B.; Turacchio, G.; Grande, R. Latest update on outer membrane vesicles and their role in horizontal gene transfer: A mini-review. Membranes 2023, 13, 860. [Google Scholar] [CrossRef] [PubMed]
- Rompikuntal, P.K.; Vdovikova, S.; Duperthuy, M.; Johnson, T.L.; Ahlund, M.; Lundmark, R.; Oscarsson, J.; Sandkvist, M.; Uhlin, B.E.; Wai, S.N. Outer membrane vesicle-mediated export of processed PrtV protease from Vibrio cholerae. PLoS ONE 2015, 10, e0134098. [Google Scholar] [CrossRef]
- Zingl, F.G.; Kohl, P.; Cakar, F.; Leitner, D.R.; Mitterer, F.; Bonnington, K.E.; Rechberger, G.N.; Kuehn, M.J.; Guan, Z.; Reidl, J.; et al. Outer membrane vesiculation facilitates surface exchange and in vivo adaptation of Vibrio cholerae. Cell Host Microbe 2020, 27, 225–237.e228. [Google Scholar] [CrossRef] [PubMed]
- Jugder, B.-E.; Watnick, P.I. Vibrio cholerae sheds its coat to make itself comfortable in the gut. Cell Host Microbe 2020, 27, 161–163. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Chaudhuri, K.; Chatterjee, A.N.; Das, J. Presence of exposed phospholipids in the outer membrane of Vibrio cholerae. J Gen. Microbiol. 1992, 138, 755–761. [Google Scholar] [CrossRef]
- Chatterjee, A.; Chaudhuri, S.; Saha, G.; Gupta, S.; Chowdhury, R. Effect of bile on the cell surface permeability barrier and efflux system of Vibrio cholerae. J. Bacteriol. 2004, 186, 6809–6814. [Google Scholar] [CrossRef]
- Schulz, G.E. The structure of bacterial outer membrane proteins. Biochim. Biophys. Acta 2002, 1565, 308–317. [Google Scholar] [CrossRef]
- Dhar, R.; Slusky, J.S. Outer membrane protein evolution. Curr. Opin. Struct. Biol. 2021, 68, 122–128. [Google Scholar] [CrossRef]
- Hartojo, A.; Doyle, M.T. β-barrel membrane proteins fold via hybrid-barrel intermediate states. Curr. Opin. Struct. Biol. 2024, 87, 102830. [Google Scholar] [CrossRef]
- Schulz, G.E. β-Barrel membrane proteins. Curr. Opin. Struct. Biol. 2000, 10, 443–447. [Google Scholar] [CrossRef] [PubMed]
- Samsudin, F.; Ortiz-Suarez, M.L.; Piggot, T.J.; Bond, P.J.; Khalid, S. OmpA: A flexible clamp for bacterial cell wall attachment. Structure 2016, 24, 2227–2235. [Google Scholar] [CrossRef] [PubMed]
- Tsirigotaki, A.; De Geyter, J.; Šoštaric’, N.; Economou, A.; Karamanou, S. Protein export through the bacterial Sec pathway. Nat. Rev. Microbiol. 2017, 15, 21–36. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, B.; Clemons, W.M., Jr.; Collinson, I.; Modis, Y.; Hartmann, E.; Harrison, S.C.; Rapoport, T.A. X-ray structure of a protein-conducting channel. Nature 2004, 427, 36–44. [Google Scholar] [CrossRef]
- Mas, G.; Thoma, J.; Hiller, S. The periplasmic chaperones Skp and SurA. Subcell Biochem. 2019, 92, 169–186. [Google Scholar] [CrossRef]
- Schafer, U.; Beck, K.; Muller, M. Skp, a molecular chaperone of Gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J. Biol. Chem. 1999, 274, 24567–24574. [Google Scholar] [CrossRef]
- Chen, R.; Henning, U. A periplasmic protein (Skp) of Escherichia coli selectively binds a class of outer membrane proteins. Mol. Microbiol. 1996, 19, 1287–1294. [Google Scholar] [CrossRef]
- Rouviere, P.E.; Gross, C.A. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev. 1996, 10, 3170–3182. [Google Scholar] [CrossRef]
- Sklar, J.G.; Wu, T.; Kahne, D.; Silhavy, T.J. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007, 21, 2473–2484. [Google Scholar] [CrossRef]
- Struyvé, M.; Moons, M.; Tommassen, J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J. Mol. Biol. 1991, 218, 141–148. [Google Scholar] [CrossRef]
- Wu, T.; Malinverni, J.; Ruiz, N.; Kim, S.; Silhavy, T.J.; Kahne, D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 2005, 121, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Voulhoux, R.; Bos, M.P.; Geurtsen, J.; Mols, M.; Tommassen, J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 2003, 299, 262–265. [Google Scholar] [CrossRef]
- Tomasek, D.; Kahne, D. The assembly of β-barrel outer membrane proteins. Curr. Opin. Microbiol. 2021, 60, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Rollauer, S.E.; Sooreshjani, M.A.; Noinaj, N.; Buchanan, S.K. Outer membrane protein biogenesis in Gram-negative bacteria. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20150023. [Google Scholar] [CrossRef]
- Wibbenmeyer, J.A.; Provenzano, D.; Landry, C.F.; Klose, K.E.; Delcour, A.H. Vibrio cholerae OmpU and OmpT porins are differentially affected by bile. Infect. Immun. 2002, 70, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Lång, H.; Palva, E.T. The ompS gene of Vibrio cholerae encodes a growth-phase-dependent maltoporin. Mol. Microbiol. 1993, 10, 891–901. [Google Scholar] [CrossRef]
- Anderson, G.J.; Frazer, D.M. Current understanding of iron homeostasis. Am. J. Clin. Nutr. 2017, 106, 1559S–1566S. [Google Scholar] [CrossRef]
- Silale, A.; van den Berg, B. TonB-dependent transport across the bacterial outer membrane. Annu. Rev. Microbiol. 2023, 77, 67–88. [Google Scholar] [CrossRef]
- Park, J.S.; Lee, W.C.; Yeo, K.J.; Ryu, K.-S.; Kumarasiri, M.; Hesek, D.; Lee, M.; Mobashery, S.; Song, J.H.; Kim, S.I. Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane. FASEB J. 2012, 26, 219. [Google Scholar] [CrossRef]
- Sonntag, I.; Schwarz, H.; Hirota, Y.; Henning, U. Cell envelope and shape of Escherichia coli: Multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J. Bacteriol. 1978, 136, 280–285. [Google Scholar] [CrossRef]
- Song, T.; Mika, F.; Lindmark, B.; Liu, Z.; Schild, S.; Bishop, A.; Zhu, J.; Camilli, A.; Johansson, J.; Vogel, J.; et al. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol. Microbiol. 2008, 70, 100–111. [Google Scholar] [CrossRef]
- Potapova, A.; Garvey, W.; Dahl, P.; Guo, S.; Chang, Y.; Schwechheimer, C.; Trebino, M.A.; Floyd, K.A.; Phinney, B.S.; Liu, J.; et al. Outer membrane vesicles and the outer membrane protein OmpU govern Vibrio cholerae biofilm matrix assembly. mBio 2024, 15, e0330423. [Google Scholar] [CrossRef] [PubMed]
- Sperandio, V.; Giron, J.A.; Silveira, W.D.; Kaper, J.B. The OmpU outer membrane protein, a potential adherence factor of Vibrio cholerae. Infect. Immun. 1995, 63, 4433–4438. [Google Scholar] [CrossRef]
- Kaur, D.; Mukhopadhaya, A. Outer membrane protein OmpV mediates Salmonella enterica serovar Typhimurium adhesion to intestinal epithelial cells via fibronectin and alpha1beta1 integrin. Cell. Microbiol. 2020, 22, e13172. [Google Scholar] [CrossRef] [PubMed]
- Mathur, J.; Waldor, M.K. The Vibrio cholerae ToxR-regulated porin OmpU confers resistance to antimicrobial peptides. Infect. Immun. 2004, 72, 3577–3583. [Google Scholar] [CrossRef] [PubMed]
- Mathur, J.; Davis, B.M.; Waldor, M.K. Antimicrobial peptides activate the Vibrio cholerae sigmaE regulon through an OmpU-dependent signalling pathway. Mol. Microbiol. 2007, 63, 848–858. [Google Scholar] [CrossRef]
- Mathieu-Denoncourt, A.; Whitfield, G.B.; Vincent, A.T.; Berne, C.; Pauzé-Foixet, J.; Mahieddine, F.C.; Brun, Y.V.; Duperthuy, M. The carRS-ompV-virK operon of Vibrio cholerae senses antimicrobial peptides and activates the expression of multiple resistance systems. Sci. Rep. 2025, 15, 13686. [Google Scholar] [CrossRef]
- Sakharwade, S.C.; Sharma, P.K.; Mukhopadhaya, A. Vibrio cholerae porin OmpU induces pro-inflammatory responses, but down-regulates LPS-mediated effects in RAW 264.7, THP-1 and human PBMCs. PLoS ONE 2013, 8, e76583. [Google Scholar] [CrossRef]
- Xu, D.; Zhang, J.; Liu, J.; Xu, J.; Zhou, H.; Zhang, L.; Zhu, J.; Kan, B. Outer membrane protein OmpW is the receptor for typing phage VP5 in the Vibrio cholerae O1 El Tor biotype. J. Virol. 2014, 88, 7109–7111. [Google Scholar] [CrossRef]
- Fan, F.; Li, X.; Pang, B.; Zhang, C.; Li, Z.; Zhang, L.; Li, J.; Zhang, J.; Yan, M.; Liang, W.; et al. The outer-membrane protein TolC of Vibrio cholerae serves as a second cell-surface receptor for the VP3 phage. J. Biol. Chem. 2018, 293, 4000–4013. [Google Scholar] [CrossRef]
- Lång, H.A.; Palva, E.T. A major outer membrane protein in Vibrio cholerae is maltose-inducible. Microb. Pathog. 1987, 3, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, S.R.; Chaudhuri, K.; Sen, K.; Das, J. Porins of Vibrio cholerae: Purification and characterization of OmpU. J. Bacteriol. 1996, 178, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; McCandlish, A.C.; Gronenberg, L.S.; Chng, S.S.; Silhavy, T.J.; Kahne, D. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. USA 2006, 103, 11754–11759. [Google Scholar] [CrossRef]
- Genevrois, S.; Steeghs, L.; Roholl, P.; Letesson, J.J.; van der Ley, P. The Omp85 protein of Neisseria meningitidis is required for lipid export to the outer membrane. EMBO J. 2003, 22, 1780–1789. [Google Scholar] [CrossRef]
- Pohlner, J.; Meyer, T.F.; Jalajakumari, M.B.; Manning, P.A. Nucleotide sequence of ompV, the gene for a major Vibrio cholerae outer membrane protein. Mol. Gen. Genet. 1986, 205, 494–500. [Google Scholar] [CrossRef]
- Pohlner, J.; Meyer, T.F.; Manning, P.A. Serological properties and processing in Escherichia coli K12 of OmpV fusion proteins of Vibrio cholerae. Mol. Gen. Genet. 1986, 205, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, G.; Leavesley, D.I.; Lagnado, C.A.; Heuzenroeder, M.W.; Manning, P.A. Purification of the 25-kDa Vibrio cholerae major outer-membrane protein and the molecular cloning of its gene: ompV. Eur. J. Biochem. 1985, 148, 385–390. [Google Scholar] [CrossRef]
- Kabir, S. Composition and immunochemical properties of outer membrane proteins of Vibrio cholerae. J. Bacteriol. 1980, 144, 382–389. [Google Scholar] [CrossRef]
- Kelley, J.T.; Parker, C.D. Identification and preliminary characterization of Vibrio cholerae outer membrane proteins. J. Bacteriol. 1981, 145, 1018–1024. [Google Scholar] [CrossRef]
- Sahu, G.K.; Chowdhury, R.; Das, J. Heat shock response and heat shock protein antigens of Vibrio cholerae. Infect. Immun. 1994, 62, 5624–5631. [Google Scholar] [CrossRef]
- Xu, C.; Wang, S.; Ren, H.; Lin, X.; Wu, L.; Peng, X. Proteomic analysis on the expression of outer membrane proteins of Vibrio alginolyticus at different sodium concentrations. Proteomics 2005, 5, 3142–3152. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Ren, H.; Wang, S.; Peng, X. Proteomic analysis of salt-sensitive outer membrane proteins of Vibrio parahaemolyticus. Res. Microbiol. 2004, 155, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Lin, X.; Wang, F.; Ye, D.; Xiao, X.; Wang, S.; Peng, X. OmpW and OmpV are required for NaCl regulation in Photobacterium damsela. J. Proteome Res. 2006, 5, 2250–2257. [Google Scholar] [CrossRef] [PubMed]
- Kaur, D.; Gandhi, S.; Mukhopadhaya, A. Salmonella Typhimurium adhesin OmpV activates host immunity to confer protection against systemic and gastrointestinal infection in mice. Infect. Immun. 2021, 89, e0012121. [Google Scholar] [CrossRef]
- Liu, F.; Tang, X.; Sheng, X.; Xing, J.; Zhan, W. Comparative study of the vaccine potential of six outer membrane proteins of Edwardsiella tarda and the immune responses of flounder (Paralichthys olivaceus) after vaccination. Vet. Immunol. Immunopathol. 2017, 185, 38–47. [Google Scholar] [CrossRef]
- Langlete, P.; Krabberod, A.K.; Winther-Larsen, H.C. Vesicles from Vibrio cholerae contain AT-rich DNA and shorter mRNAs that do not correlate with their protein products. Front. Microbiol. 2019, 10, 2708. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, T.; Wang, Q.; Huang, J.; Zhu, Y.; Liu, X.; Liu, R.; Yang, B.; Zhou, K. Vibrio cholerae senses human enteric alpha-defensin 5 through a CarSR two-component system to promote bacterial pathogenicity. Commun. Biol. 2022, 5, 559. [Google Scholar] [CrossRef]
- Matson, J.S.; Livny, J.; DiRita, V.J. A putative Vibrio cholerae two-component system controls a conserved periplasmic protein in response to the antimicrobial peptide polymyxin B. PLoS ONE 2017, 12, e0186199. [Google Scholar] [CrossRef]
- Bina, J.E.; Provenzano, D.; Wang, C.; Bina, X.R.; Mekalanos, J.J. Characterization of the Vibrio cholerae vexAB and vexCD efflux systems. Arch. Microbiol. 2006, 186, 171–181. [Google Scholar] [CrossRef]
- Bilecen, K.; Fong, J.C.; Cheng, A.; Jones, C.J.; Zamorano-Sanchez, D.; Yildiz, F.H. Polymyxin B resistance and biofilm formation in Vibrio cholerae are controlled by the response regulator CarR. Infect. Immun. 2015, 83, 1199–1209. [Google Scholar] [CrossRef]
- Ko, D.; Sung, D.; Kim, T.Y.; Choi, G.; Bang, Y.J.; Choi, S.H. CarRS two-component system essential for Polymyxin B resistance of Vibrio vulnificus responds to multiple host environmental signals. Microbiol. Spectr. 2023, 11, e0030523. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, W.; von Rechenberg, M.; Holtje, J.V. Demonstration of molecular interactions between the murein polymerase PBP1B, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J. Biol. Chem. 1999, 274, 6726–6734. [Google Scholar] [CrossRef] [PubMed]
- Janet-Maitre, M.; Job, V.; Bour, M.; Robert-Genthon, M.; Brugiere, S.; Triponney, P.; Cobessi, D.; Coute, Y.; Jeannot, K.; Attree, I. Pseudomonas aeruginosa MipA-MipB envelope proteins act as new sensors of polymyxins. mBio 2024, 15, e0221123. [Google Scholar] [CrossRef]
- Meza-Villezcas, A.; Gallego-Hernández, A.L.; Yildiz, F.H.; Jaime-Acuña, O.E.; Raymond-Herrera, O.; Huerta-Saquero, A. Effect of antimicrobial nanocomposites on Vibrio cholerae lifestyles: Pellicle biofilm, planktonic and surface-attached biofilm. PLoS ONE 2019, 14, e0217869. [Google Scholar] [CrossRef]
- Jalajakumari, M.B.; Manning, P.A. Nucleotide sequence of the gene, ompW, encoding a 22kDa immunogenic outer membrane protein of Vibrio cholerae. Nucleic Acids Res. 1990, 18, 2180. [Google Scholar] [CrossRef]
- Manning, P.A.; Bartowsky, E.J.; Leavesly, D.I.; Hackett, J.A.; Heuzenroeder, M.W. Molecular cloning using immune sera of a 22-kDal minor outer membrane protein of Vibrio cholerae. Gene 1985, 34, 95–103. [Google Scholar] [CrossRef]
- Guan, H.; Xue, P.; Zhou, H.; Sha, D.; Wang, D.; Gao, H.; Li, J.; Diao, B.; Zhao, H.; Kan, B.; et al. A multiplex PCR assay for the detection of five human pathogenic Vibrio species and Plesiomonas. Mol. Cell. Probes 2021, 55, 101689. [Google Scholar] [CrossRef]
- Li, Z.; Guan, H.; Wang, W.; Gao, H.; Feng, W.; Li, J.; Diao, B.; Zhao, H.; Kan, B.; Zhang, J. Development of a Rapid and Fully Automated Multiplex Real-Time PCR Assay for Identification and Differentiation of Vibrio cholerae and Vibrio parahaemolyticus on the BD MAX Platform. Front. Cell. Infect. Microbiol. 2021, 11, 639473. [Google Scholar] [CrossRef] [PubMed]
- Soleimani, F.; Taherkhani, R.; Dobaradaran, S.; Spitz, J.; Saeedi, R. Molecular detection of E. coli and Vibrio cholerae in ballast water of commercial ships: A primary study along the Persian Gulf. J. Environ. Health Sci. Eng. 2021, 19, 457–463. [Google Scholar] [CrossRef]
- Das, M.; Chopra, A.K.; Cantu, J.M.; Peterson, J.W. Antisera to selected outer membrane proteins of Vibrio cholerae protect against challenge with homologous and heterologous strains of V. cholerae. FEMS Immunol. Med. Microbiol. 1998, 22, 303–308. [Google Scholar] [CrossRef]
- Zareitaher, T.; Sadat Ahmadi, T.; Latif Mousavi Gargari, S. Immunogenic efficacy of DNA and protein-based vaccine from a chimeric gene consisting OmpW, TcpA and CtxB, of Vibrio cholerae. Immunobiology 2022, 227, 152190. [Google Scholar] [CrossRef] [PubMed]
- Tehrani, S.S.; Jahangiri, A.; Taheri-Anganeh, M.; Maghsoudi, H.; Khalili, S.; Fana, S.E.; Maniati, M.; Amani, J. Designing an Outer Membrane Protein (Omp-W) Based Vaccine for Immunization against Vibrio and Salmonella: An in silico Approach. Recent Pat. Biotechnol. 2020, 14, 312–324. [Google Scholar] [CrossRef]
- Hong, H.; Patel, D.R.; Tamm, L.K.; van den Berg, B. The outer membrane protein OmpW forms an eight-stranded beta-barrel with a hydrophobic channel. J. Biol. Chem. 2006, 281, 7568–7577. [Google Scholar] [CrossRef]
- Thompson Dorothea, K.; Beliaev Alexander, S.; Giometti Carol, S.; Tollaksen Sandra, L.; Khare, T.; Lies Douglas, P.; Nealson Kenneth, H.; Lim, H.; Yates, J.; Brandt Craig, C.; et al. Transcriptional and Proteomic Analysis of a Ferric Uptake Regulator (Fur) Mutant of Shewanella oneidensis: Possible Involvement of Fur in Energy Metabolism, Transcriptional Regulation, and Oxidative Stress. Appl. Environ. Microbiol. 2002, 68, 881–892. [Google Scholar] [CrossRef]
- Gil, F.; Ipinza, F.; Fuentes, J.; Fumeron, R.; Villarreal, J.M.; Aspée, A.; Mora, G.C.; Vásquez, C.C.; Saavedra, C. The ompW (porin) gene mediates methyl viologen (paraquat) efflux in Salmonella enterica serovar Typhimurium. Res. Microbiol. 2007, 158, 529–536. [Google Scholar] [CrossRef]
- Fu, X.; Zhang, J.; Li, T.; Zhang, M.; Li, J.; Kan, B. The outer membrane protein OmpW enhanced V. cholerae growth in hypersaline conditions by transporting carnitine. Front. Microbiol. 2017, 8, 2703. [Google Scholar] [CrossRef] [PubMed]
- Martinac, B.; Buechner, M.; Delcour, A.H.; Adler, J.; Kung, C. Pressure-sensitive ion channel in Escherichia coli. Proc. Natl. Acad. Sci. USA 1987, 84, 2297–2301. [Google Scholar] [CrossRef] [PubMed]
- Shahjee, H.M.; Banerjee, K.; Ahmad, F. Comparative analysis of naturally occurring L-amino acid osmolytes and their D-isomers on protection of Escherichia coli against environmental stresses. J. Biosci. 2002, 27, 515–520. [Google Scholar] [CrossRef]
- Brown, A.D. Microbial water stress. Bacteriol. Rev. 1976, 40, 803–846. [Google Scholar] [CrossRef]
- Fu, X.; Liang, W.; Du, P.; Yan, M.; Kan, B. Transcript changes in Vibrio cholerae in response to salt stress. Gut Pathog. 2014, 6, 47. [Google Scholar] [CrossRef]
- Singleton, F.L.; Attwell, R.; Jangi, S.; Colwell, R.R. Effects of temperature and salinity on Vibrio cholerae growth. Appl. Environ. Microbiol. 1982, 44, 1047–1058. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.F. Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Syst. 2005, 1, 5. [Google Scholar] [CrossRef] [PubMed]
- Pathania, M.; Acosta-Gutierrez, S.; Bhamidimarri, S.P.; Baslé, A.; Winterhalter, M.; Ceccarelli, M.; van den Berg, B. Unusual constriction zones in the major porins OmpU and OmpT from Vibrio cholerae. Structure 2018, 26, 708–721.e4. [Google Scholar] [CrossRef]
- Paauw, A.; Trip, H.; Niemcewicz, M.; Sellek, R.; Heng, J.M.E.; Mars-Groenendijk, R.H.; de Jong, A.L.; Majchrzykiewicz-Koehorst, J.A.; Olsen, J.S.; Tsivtsivadze, E. OmpU as a biomarker for rapid discrimination between toxigenic and epidemic Vibrio cholerae O1/O139 and non-epidemic Vibrio cholerae in a modified MALDI-TOF MS assay. BMC Microbiol. 2014, 14, 158. [Google Scholar] [CrossRef] [PubMed]
- Duret, G.; Delcour, A.H. Size and dynamics of the Vibrio cholerae porins OmpU and OmpT probed by polymer exclusion. Biophys. J. 2010, 98, 1820–1829. [Google Scholar] [CrossRef]
- Benz, R.; Maier, E.; Chakraborty, T. Purification of OmpU from Vibrio cholerae classical strain 569B: Evidence for the formation of large cation-selective ion-permeable channels by OmpU. Microbiologia 1997, 13, 321–330. [Google Scholar]
- Li, H.; Zhang, W.; Dong, C. Crystal structure of the outer membrane protein OmpU from Vibrio cholerae at 2.2 Å resolution. Acta Crystallogr. D Struct. Biol. 2018, 74, 21–29. [Google Scholar] [CrossRef]
- Simonet, V.C.; Basle, A.; Klose, K.E.; Delcour, A.H. The Vibrio cholerae porins OmpU and OmpT have distinct channel properties. J. Biol. Chem. 2003, 278, 17539–17545. [Google Scholar] [CrossRef]
- Yang, J.S.; Jeon, J.H.; Jang, M.S.; Kang, S.-S.; Ahn, K.B.; Song, M.; Yun, C.-H.; Han, S.H. Vibrio cholerae OmpU induces IL-8 expression in human intestinal epithelial cells. Mol. Immunol. 2018, 93, 47–54. [Google Scholar] [CrossRef]
- Fong, J.C.N.; Syed, K.A.; Klose, K.E.; Yildiz, F.H. Role of Vibrio polysaccharide (vps) genes in VPS production, biofilm formation and Vibrio cholerae pathogenesis. Microbiology 2010, 156, 2757–2769. [Google Scholar] [CrossRef]
- Bandyopadhaya, A.; Bhowmick, S.; Chaudhuri, K. Activation of proinflammatory response in human intestinal epithelial cells following Vibrio cholerae infection through PI3K/Akt pathway. Can. J. Microbiol. 2009, 55, 1310–1318. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhaya, A.; Sarkar, M.; Chaudhuri, K. Human intestinal epithelial cell cytokine mRNA responses mediated by NF-κB are modulated by the motility and adhesion process of Vibrio cholerae. Int. J. Biochem. Cell Biol. 2007, 39, 1863–1876. [Google Scholar] [CrossRef]
- Dhar, V.; Gandhi, S.; Sakharwade, S.C.; Chawla, A.; Mukhopadhaya, A. Vibrio cholerae porin OmpU activates dendritic cells via TLR2 and the NLRP3 inflammasome. Infect. Immun. 2023, 91, e0033222. [Google Scholar] [CrossRef] [PubMed]
- Sakharwade, S.C.; Mukhopadhaya, A. Vibrio cholerae porin OmpU induces LPS tolerance by attenuating TLR-mediated signaling. Mol. Immunol. 2015, 68, 312–324. [Google Scholar] [CrossRef]
- Prasad, G.; Dhar, V.; Mukhopadhaya, A. Vibrio cholerae OmpU mediates CD36-dependent reactive oxygen species generation triggering an additional pathway of MAPK activation in macrophages. J. Immunol. 2019, 202, 2431–2450. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Prasad, G.V.; Mukhopadhaya, A. Vibrio cholerae porin OmpU induces Caspase-independent programmed cell death upon translocation to the host cell mitochondria. J. Biol. Chem. 2015, 290, 31051–31068. [Google Scholar] [CrossRef]
- Jiang, H.H.; Zhou, Y.; Liu, M.; Larios-Valencia, J.; Lee, Z.; Wang, H.; Gao, X.H.; Zhu, J. Vibrio cholerae Virulence Activator ToxR Regulates Manganese Transport and Resistance to Reactive Oxygen Species. Infect. Immun. 2020, 88. [Google Scholar] [CrossRef]
- Crawford, J.A.; Kaper, J.B.; DiRita, V.J. Analysis of ToxR-dependent transcription activation of ompU, the gene encoding a major envelope protein in Vibrio cholerae. Mol. Microbiol. 1998, 29, 235–246. [Google Scholar] [CrossRef]
- Li, C.C.; Crawford, J.A.; DiRita, V.J.; Kaper, J.B. Molecular cloning and transcriptional regulation of ompT, a ToxR-repressed gene in Vibrio cholerae. Mol. Microbiol. 2000, 35, 189–203. [Google Scholar] [CrossRef]
- Medrano, A.I.; DiRita, V.J.; Castillo, G.; Sanchez, J. Transient transcriptional activation of the Vibrio cholerae El Tor virulence regulator toxT in response to culture conditions. Infect. Immun. 1999, 67, 2178–2183. [Google Scholar] [CrossRef]
- Provenzano, D.; Schuhmacher, D.A.; Barker, J.L.; Klose, K.E. The virulence regulatory protein ToxR mediates enhanced bile resistance in Vibrio cholerae and other pathogenic Vibrio species. Infect. Immun. 2000, 68, 1491–1497. [Google Scholar] [CrossRef] [PubMed]
- Provenzano, D.; Klose, K.E. Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc. Natl. Acad. Sci. USA 2000, 97, 10220–10224. [Google Scholar] [CrossRef]
- Chomvarin, C.; Jumroenjit, W.; Chaicumpar, K.; Namwat, W. Association of ompU gene in Vibrio cholerae from patients and environment with bile resistance. Southeast Asian J. Trop. Med. Public Health 2008, 39, 876–881. [Google Scholar]
- Provenzano, D.; Lauriano, C.M.; Klose, K.E. Characterization of the role of the ToxR-modulated outer membrane porins OmpU and OmpT in Vibrio cholerae virulence. J. Bacteriol. 2001, 183, 3652–3662. [Google Scholar] [CrossRef] [PubMed]
- Merrell, D.S.; Bailey, C.; Kaper James, B.; Camilli, A. The ToxR-mediated organic acid tolerance response of Vibrio cholerae requires OmpU. J. Bacteriol. 2001, 183, 2746–2754. [Google Scholar] [CrossRef]
- Duperthuy, M.; Binesse, J.; Le Roux, F.; Romestand, B.; Caro, A.; Got, P.; Givaudan, A.; Mazel, D.; Bachere, E.; Destoumieux-Garzon, D. The major outer membrane protein OmpU of Vibrio splendidus contributes to host antimicrobial peptide resistance and is required for virulence in the oyster Crassostrea gigas. Environ. Microbiol. 2010, 12, 951–963. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.M.; Waldor, M.K. High-throughput sequencing reveals suppressors of Vibrio cholerae rpoE mutations: One fewer porin is enough. Nucleic Acids Res. 2009, 37, 5757–5767. [Google Scholar] [CrossRef]
- Missiakas, D.; Mayer, M.P.; Lemaire, M.; Georgopoulos, C.; Raina, S. Modulation of the Escherichia coli sigmaE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol. Microbiol. 1997, 24, 355–371. [Google Scholar] [CrossRef]
- Walsh, N.P.; Alba, B.M.; Bose, B.; Gross, C.A.; Sauer, R.T. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 2003, 113, 61–71. [Google Scholar] [CrossRef]
- Wilken, C.; Kitzing, K.; Kurzbauer, R.; Ehrmann, M.; Clausen, T. Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease. Cell 2004, 117, 483–494. [Google Scholar] [CrossRef]
- Ades, S.E.; Connolly, L.E.; Alba, B.M.; Gross, C.A. The Escherichia coli sigma(E)-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev. 1999, 13, 2449–2461. [Google Scholar] [CrossRef] [PubMed]
- Kanehara, K.; Ito, K.; Akiyama, Y. YaeL (EcfE) activates the sigma(E) pathway of stress response through a site-2 cleavage of anti-sigma(E), RseA. Genes Dev. 2002, 16, 2147–2155. [Google Scholar] [CrossRef]
- Pennetzdorfer, N.; Höfler, T.; Wölflingseder, M.; Tutz, S.; Schild, S.; Reidl, J. σ(E) controlled regulation of porin OmpU in Vibrio cholerae. Mol. Microbiol. 2021, 115, 1244–1261. [Google Scholar] [CrossRef]
- Dartigalongue, C.; Missiakas, D.; Raina, S. Characterization of the Escherichia coli sigma E regulon. J. Biol. Chem. 2001, 276, 20866–20875. [Google Scholar] [CrossRef]
- Kovacikova, G.; Skorupski, K. The alternative sigma factor sigma(E) plays an important role in intestinal survival and virulence in Vibrio cholerae. Infect. Immun. 2002, 70, 5355–5362. [Google Scholar] [CrossRef] [PubMed]
- Grant, T.A.; López-Pérez, M.; Haro-Moreno, J.M.; Almagro-Moreno, S. Allelic diversity uncovers protein domains contributing to the emergence of antimicrobial resistance. PLoS Genet. 2023, 19, e1010490. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, B.J.; Levade, I.; Kovacikova, G.; Taylor, R.K.; Almagro-Moreno, S. Origins of pandemic Vibrio cholerae from environmental gene pools. Nat. Microbiol. 2016, 2, 16240. [Google Scholar] [CrossRef]
- Espinoza-Vergara, G.; Noorian, P.; Silva-Valenzuela, C.A.; Raymond, B.B.A.; Allen, C.; Hoque, M.M.; Sun, S.; Johnson, M.S.; Pernice, M.; Kjelleberg, S.; et al. Vibrio cholerae residing in food vacuoles expelled by protozoa are more infectious in vivo. Nat. Microbiol. 2019, 4, 2466–2474. [Google Scholar] [CrossRef]
- Mitterer, F.; Pombo, J.P.; Schild, S. Vibrio cholerae released by protozoa are hyperinfectious. Trends Microbiol. 2020, 28, 4–6. [Google Scholar] [CrossRef]
- Pennetzdorfer, N.; Lembke, M.; Pressler, K.; Matson, J.S.; Reidl, J.; Schild, S. Regulated proteolysis in Vibrio cholerae allowing rapid adaptation to stress conditions. Front. Cell. Infect. Microbiol. 2019, 9, 214. [Google Scholar] [CrossRef]
- Häse, C.C.; Mekalanos, J.J. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 1998, 95, 730–734. [Google Scholar] [CrossRef] [PubMed]
- Weber, G.G.; Klose, K.E. The complexity of ToxT-dependent transcription in Vibrio cholerae. Indian J. Med. Res. 2011, 133, 201–206. [Google Scholar]
- Ramamurthy, T.; Nandy, R.K.; Mukhopadhyay, A.K.; Dutta, S.; Mutreja, A.; Okamoto, K.; Miyoshi, S.I.; Nair, G.B.; Ghosh, A. Virulence regulation and innate host response in the pathogenicity of Vibrio cholerae. Front. Cell. Infect. Microbiol. 2020, 10, 572096. [Google Scholar] [CrossRef]
- Herzog, R.; Peschek, N.; Fröhlich, K.S.; Schumacher, K.; Papenfort, K. Three autoinducer molecules act in concert to control virulence gene expression in Vibrio cholerae. Nucleic Acids Res. 2019, 47, 3171–3183. [Google Scholar] [CrossRef] [PubMed]
- Waters, C.M.; Lu, W.; Rabinowitz, J.D.; Bassler, B.L. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 2008, 190, 2527–2536. [Google Scholar] [CrossRef]
- Duret, G.; Simonet, V.; Delcour, A.H. Modulation of Vibrio cholerae porin function by acidic pH. Channels 2007, 1, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Mey Alexandra, R.; Craig Stephanie, A.; Payne Shelley, M. Effects of amino acid supplementation on porin expression and ToxR levels in Vibrio cholerae. Infect. Immun. 2012, 80, 518–528. [Google Scholar] [CrossRef]
- Miller, V.L.; Mekalanos, J.J. A novel suicide vector and its use in construction of insertion mutations: Osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 1988, 170, 2575–2583. [Google Scholar] [CrossRef]
- Duret, G.; Delcour, A.H. Deoxycholic acid blocks Vibrio cholerae OmpT but not OmpU porin. J. Biol. Chem. 2006, 281, 19899–19905. [Google Scholar] [CrossRef]
- Li, C.C.; Merrell, D.S.; Camilli, A.; Kaper, J.B. ToxR interferes with CRP-dependent transcriptional activation of ompT in Vibrio cholerae. Mol. Microbiol. 2002, 43, 1577–1589. [Google Scholar] [CrossRef]
- Song, T.; Sabharwal, D.; Wai, S.N. VrrA mediates Hfq-dependent regulation of OmpT synthesis in Vibrio cholerae. J. Mol. Biol. 2010, 400, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Teschler, J.K.; Zamorano-Sanchez, D.; Utada, A.S.; Warner, C.J.; Wong, G.C.; Linington, R.G.; Yildiz, F.H. Living in the matrix: Assembly and control of Vibrio cholerae biofilms. Nat. Rev. Microbiol. 2015, 13, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Bari, W.; Lee, K.-M.; Yoon, S.S. Structural and functional importance of outer membrane proteins in Vibrio cholerae flagellum. J. Microbiol. 2012, 50, 631–637. [Google Scholar] [CrossRef]
- Chatterjee, D.; Chaudhuri, K. Association of cholera toxin with Vibrio cholerae outer membrane vesicles which are internalized by human intestinal epithelial cells. FEBS Lett. 2011, 585, 1357–1362. [Google Scholar] [CrossRef] [PubMed]
- Rasti, E.S.; Brown, A.C. Cholera toxin encapsulated within several Vibrio cholerae O1 Serotype Inaba outer membrane vesicles lacks a functional B-subunit. Toxins 2019, 11, 207. [Google Scholar] [CrossRef]
- Lang, H.; Maki, M.; Rantakari, A.; Korhonen, T.K. Characterization of adhesive epitopes with the OmpS display system. Eur. J. Biochem. 2000, 267, 163–170. [Google Scholar] [CrossRef]
- Lång, H.A.; Jonson, G.; Svennerholm, A.M.; Palva, E.T. The maltose-inducible 43 kDa major outer membrane protein in Vibrio cholerae is immunogenic and common to different isolates. Microb. Pathog. 1988, 5, 169–175. [Google Scholar] [CrossRef]
- Norris, N.; Alcolombri, U.; Keegstra, J.M.; Yawata, Y.; Menolascina, F.; Frazzoli, E.; Levine, N.M.; Fernandez, V.I.; Stocker, R. Bacterial chemotaxis to saccharides is governed by a trade-off between sensing and uptake. Biophys. J. 2022, 121, 2046–2059. [Google Scholar] [CrossRef]
- LaRocque Regina, C.; Krastins, B.; Harris Jason, B.; Lebrun Lauren, M.; Parker Kenneth, C.; Chase, M.; Ryan Edward, T.; Qadri, F.; Sarracino, D.; Calderwood Stephen, B. Proteomic analysis of Vibrio cholerae in human stool. Infect. Immun. 2008, 76, 4145–4151. [Google Scholar] [CrossRef]
- Wimley, W.C. The versatile β-barrel membrane protein. Curr. Opin. Struct. Biol. 2003, 13, 404–411. [Google Scholar] [CrossRef]
- Vogt, J.; Schulz, G.E. The structure of the outer membrane protein OmpX from Escherichia coli reveals possible mechanisms of virulence. Structure 1999, 7, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- Rath, P.; Sharpe, T.; Hiller, S. The electrostatic core of the outer membrane protein X from E. coli. Biochim. Biophys. Acta (BBA)-Biomembr. 2020, 1862, 183031. [Google Scholar] [CrossRef]
- Alm, R.A.; Braun, G.; Morona, R.; Manning, P.A. Detection of an OmpA-like protein in Vibrio cholerae. FEMS Microbiol. Lett. 1986, 37, 99–104. [Google Scholar] [CrossRef]
- Smith, S.G.J.; Mahon, V.; Lambert, M.A.; Fagan, R.P. A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol. Lett. 2007, 273, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Valeru, S.P.; Shanan, S.; Alossimi, H.; Saeed, A.; Sandstrom, G.; Abd, H. Lack of outer membrane protein A enhances the release of outer membrane vesicles and survival of Vibrio cholerae and suppresses viability of Acanthamoeba castellanii. Int. J. Microbiol. 2014, 2014, 610190. [Google Scholar] [CrossRef]
- Song, T.; Wai, S.N. A novel sRNA that modulates virulence and environmental fitness of Vibrio cholerae. RNA Biol. 2009, 6, 254–258. [Google Scholar] [CrossRef]
- Song, T.; Sabharwal, D.; Gurung, J.M.; Cheng, A.T.; Sjöström, A.E.; Yildiz, F.H.; Uhlin, B.E.; Wai, S.N. Vibrio cholerae utilizes direct sRNA regulation in expression of a biofilm matrix protein. PLoS ONE 2014, 9, e101280. [Google Scholar] [CrossRef]
- Saul-McBeth, J.; Matson, J.S. A periplasmic antimicrobial peptide-binding protein is required for stress survival in Vibrio cholerae. Front. Microbiol. 2019, 10, 161. [Google Scholar] [CrossRef]
- Meibom, K.L.; Li, X.B.; Nielsen, A.T.; Wu, C.Y.; Roseman, S.; Schoolnik, G.K. The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. USA 2004, 101, 2524–2529. [Google Scholar] [CrossRef]
- Hayes, C.A.; Dalia, T.N.; Dalia, A.B. Systematic genetic dissection of chitin degradation and uptake in Vibrio cholerae. Environ. Microbiol. 2017, 19, 4154–4163. [Google Scholar] [CrossRef]
- Keyhani, N.O.; Li, X.B.; Roseman, S. Chitin catabolism in the marine bacterium Vibrio furnissii. Identification and molecular cloning of a chitoporin. J. Biol. Chem. 2000, 275, 33068–33076. [Google Scholar] [CrossRef] [PubMed]
- Soysa, H.S.M.; Aunkham, A.; Schulte, A.; Suginta, W. Single-channel properties, sugar specificity, and role of chitoporin in adaptive survival of Vibrio cholerae type strain O1. J. Biol. Chem. 2020, 295, 9421–9432. [Google Scholar] [CrossRef]
- Aunkham, A.; Zahn, M.; Kesireddy, A.; Pothula, K.R.; Schulte, A.; Baslé, A.; Kleinekathöfer, U.; Suginta, W.; van den Berg, B. Structural basis for chitin acquisition by marine Vibrio species. Nat. Commun. 2018, 9, 220. [Google Scholar] [CrossRef] [PubMed]
- Suginta, W.; Chumjan, W.; Mahendran, K.R.; Schulte, A.; Winterhalter, M. Chitoporin from Vibrio harveyi, a channel with exceptional sugar specificity. J. Biol. Chem. 2013, 288, 11038–11046. [Google Scholar] [CrossRef]
- Davis, B.M.; Waldor, M.K. RNase E-dependent processing stabilizes MicX, a Vibrio cholerae sRNA. Mol. Microbiol. 2007, 65, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Kunkle, D.E.; Bina, X.R.; Bina, J.E. Vibrio cholerae OmpR contributes to virulence repression and fitness at alkaline pH. Infect. Immun. 2020, 88. [Google Scholar] [CrossRef]
- Yoon, Y.; Song, S. Structural insights into the lipopolysaccharide transport (Lpt) system as a novel antibiotic target. J. Microbiol. 2024, 62, 261–275. [Google Scholar] [CrossRef]
- Konovalova, A.; Kahne, D.E.; Silhavy, T.J. Outer membrane biogenesis. Annu. Rev. Microbiol. 2017, 71, 539–556. [Google Scholar] [CrossRef]
- Zha, Z.; Li, C.; Li, W.; Ye, Z.; Pan, J. LptD is a promising vaccine antigen and potential immunotherapeutic target for protection against Vibrio species infection. Sci. Rep. 2016, 6, 38577. [Google Scholar] [CrossRef]
- Qiao, S.; Luo, Q.; Zhao, Y.; Zhang, X.C.; Huang, Y. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature 2014, 511, 108–111. [Google Scholar] [CrossRef]
- von Krüger, W.M.A.; Humphreys, S.; Ketley, J.M. A role for the PhoBR regulatory system homologue in the Vibrio cholerae phosphate-limitation response and intestinal colonization. Microbiology 1999, 145 Pt 9, 2463–2475. [Google Scholar] [CrossRef] [PubMed]
- Goulart, C.L.; Lery, L.M.S.; Diniz, M.M.P.; Vianez-Junior, J.L.; Neves-Ferreira, A.G.C.; Perales, J.; Bisch, P.M.; von Krüger, W.M.A. Molecular analysis of VCA1008: A putative phosphoporin of Vibrio cholerae. FEMS Microbiol. Lett. 2009, 298, 241–248. [Google Scholar] [CrossRef]
- von Kruger, W.M.; Lery, L.M.; Soares, M.R.; de Neves-Manta, F.S.; Batista e Silva, C.M.; Neves-Ferreira, A.G.; Perales, J.; Bisch, P.M. The phosphate-starvation response in Vibrio cholerae O1 and phoB mutant under proteomic analysis: Disclosing functions involved in adaptation, survival and virulence. Proteomics 2006, 6, 1495–1511. [Google Scholar] [CrossRef]
- Goulart, C.L.; Bisch, P.M.; von Krüger, W.M.A.; Homblé, F. VCA1008: An anion-selective porin of Vibrio cholerae. Biochim. Biophys. Acta (BBA)-Biomembr. 2015, 1848, 680–687. [Google Scholar] [CrossRef] [PubMed]
- Osorio, C.G.; Martinez-Wilson, H.; Camilli, A. The ompU Paralogue vca1008 is required for virulence of Vibrio cholerae. J. Bacteriol. 2004, 186, 5167–5171. [Google Scholar] [CrossRef]
- Osorio, C.G.; Crawford, J.A.; Michalski, J.; Martinez-Wilson, H.; Kaper, J.B.; Camilli, A. Second-generation recombination-based in vivo expression technology for large-scale screening for Vibrio cholerae genes induced during infection of the mouse small intestine. Infect. Immun. 2005, 73, 972–980. [Google Scholar] [CrossRef]
- Goulart, C.L.; Dos Santos, G.G.; Barbosa, L.C.; Lery, L.M.S.; Bisch, P.M.; von Krüger, W.M.A. A ToxR-dependent role for the putative phosphoporin VCA1008 in bile salt resistance in Vibrio cholerae El Tor N16961. Microbiology 2010, 156, 3011–3020. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, A.; Vibhuti, R.K. Molecular mechanism of iron transport systems in Vibrio. J. Pure Appl. Microbiol. 2022, 16, 116–129. [Google Scholar] [CrossRef]
- Mey, A.R.; Payne, S.M. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol. Microbiol. 2001, 42, 835–849. [Google Scholar] [CrossRef]
- Ferguson, A.D.; Deisenhofer, J. TonB-dependent receptors—Structural perspectives. Biochim. Biophys. Acta (BBA)-Biomembr. 2002, 1565, 318–332. [Google Scholar] [CrossRef]
- Noinaj, N.; Guillier, M.; Barnard, T.J.; Buchanan, S.K. TonB-dependent transporters: Regulation, structure, and function. Annu. Rev. Microbiol. 2010, 64, 43–60. [Google Scholar] [CrossRef]
- Occhino, D.A.; Wyckoff, E.E.; Henderson, D.P.; Wrona, T.J.; Payne, S.M. Vibrio cholerae iron transport: Haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol. Microbiol. 1998, 29, 1493–1507. [Google Scholar] [CrossRef]
- Wyckoff, E.E.; Allred, B.E.; Raymond, K.N.; Payne, S.M. Catechol siderophore transport by Vibrio cholerae. J. Bacteriol. 2015, 197, 2840–2849. [Google Scholar] [CrossRef]
- Mey, A.R.; Wyckoff, E.E.; Oglesby, A.G.; Rab, E.; Taylor, R.K.; Payne, S.M. Identification of the Vibrio cholerae enterobactin receptors VctA and IrgA: IrgA is not required for virulence. Infect. Immun. 2002, 70, 3419–3426. [Google Scholar] [CrossRef] [PubMed]
- Rogers Marc, B.; Sexton Jessica, A.; DeCastro, G.J.; Calderwood Stephen, B. Identification of an operon required for ferrichrome iron utilization in Vibrio cholerae. J. Bacteriol. 2000, 182, 2350–2353. [Google Scholar] [CrossRef] [PubMed]
- Ma, A.T.; Tyrell, B.; Beld, J. Specificity of cobamide remodeling, uptake and utilization in Vibrio cholerae. Mol. Microbiol. 2020, 113, 89–102. [Google Scholar] [CrossRef]
- Han, L.; Zheng, J.; Wang, Y.; Yang, X.; Liu, Y.; Sun, C.; Cao, B.; Zhou, H.; Ni, D.; Lou, J.; et al. Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat. Struct. Mol. Biol. 2016, 23, 192–196. [Google Scholar] [CrossRef]
- Malinverni, J.C.; Werner, J.; Kim, S.; Sklar, J.G.; Kahne, D.; Misra, R.; Silhavy, T.J. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol. 2006, 61, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Noinaj, N.; Kuszak, A.J.; Gumbart, J.C.; Lukacik, P.; Chang, H.; Easley, N.C.; Lithgow, T.; Buchanan, S.K. Structural insight into the biogenesis of β-barrel membrane proteins. Nature 2013, 501, 385–390. [Google Scholar] [CrossRef]
- Sánchez-Pulido, L.; Devos, D.; Genevrois, S.; Vicente, M.; Valencia, A. POTRA: A conserved domain in the FtsQ family and a class of β-barrel outer membrane proteins. Trends Biochem. Sci. 2003, 28, 523–526. [Google Scholar] [CrossRef]
- Gentle, I.E.; Burri, L.; Lithgow, T. Molecular architecture and function of the Omp85 family of proteins. Mol. Microbiol. 2005, 58, 1216–1225. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, R.; Schütz, M.; Oberhettinger, P.; Faulstich, M.; Bermejo, I.; Rudel, T.; Diederichs, K.; Zeth, K. Structure of BamA, an essential factor in outer membrane protein biogenesis. Acta Crystallogr. Sect. D Struct. Biol. 2014, 70, 1779–1789. [Google Scholar] [CrossRef]
- Kim, S.; Malinverni, J.C.; Sliz, P.; Silhavy, T.J.; Harrison, S.C.; Kahne, D. Structure and function of an essential component of the outer membrane protein assembly machine. Science 2007, 317, 961–964. [Google Scholar] [CrossRef] [PubMed]
- Gatzeva-Topalova, P.Z.; Warner, L.R.; Pardi, A.; Sousa, M.C. Structure and flexibility of the complete periplasmic domain of BamA: The protein insertion machine of the outer membrane. Structure 2010, 18, 1492–1501. [Google Scholar] [CrossRef]
- Noinaj, N.; Gumbart, J.C.; Buchanan, S.K. The β-barrel assembly machinery in motion. Nat. Rev. Microbiol. 2017, 15, 197–204. [Google Scholar] [CrossRef]
- Ni, D.; Wang, Y.; Yang, X.; Zhou, H.; Hou, X.; Cao, B.; Lu, Z.; Zhao, X.; Yang, K.; Huang, Y. Structural and functional analysis of the β-barrel domain of BamA from Escherichia coli. FASEB J. 2014, 28, 2677–2685. [Google Scholar] [CrossRef] [PubMed]
- Gatzeva-Topalova, P.Z.; Walton, T.A.; Sousa, M.C. Crystal structure of YaeT: Conformational flexibility and substrate recognition. Structure 2008, 16, 1873–1881. [Google Scholar] [CrossRef]
- Xu, Q.; Guo, M.; Yu, F. β-Barrel assembly machinery (BAM) complex as novel antibacterial drug target. Molecules 2023, 28, 3758. [Google Scholar] [CrossRef]
- Noinaj, N.; Kuszak, A.J.; Balusek, C.; Gumbart, J.C.; Buchanan, S.K. Lateral opening and exit pore formation are required for BamA function. Structure 2014, 22, 1055–1062. [Google Scholar] [CrossRef]
- Shen, C.; Chang, S.; Luo, Q.; Chan, K.C.; Zhang, Z.; Luo, B.; Xie, T.; Lu, G.; Zhu, X.; Wei, X.; et al. Structural basis of BAM-mediated outer membrane β-barrel protein assembly. Nature 2023, 617, 185–193. [Google Scholar] [CrossRef]
- Kim, K.H.; Aulakh, S.; Paetzel, M. The bacterial outer membrane β-barrel assembly machinery. Protein Sci. 2012, 21, 751–768. [Google Scholar] [CrossRef] [PubMed]
- Ebenberger, S.P.; Cakar, F.; Chen, Y.-C.; Pressler, K.; Eberl, L.; Schild, S. The activity of the quorum sensing regulator HapR is modulated by the bacterial extracellular vesicle (BEV)-associated protein ObfA of Vibrio cholerae. J. Extracell. Vesicles 2024, 13, e12507. [Google Scholar] [CrossRef]
- van Kempen, M.; Kim, S.S.; Tumescheit, C.; Mirdita, M.; Lee, J.; Gilchrist, C.L.M.; Soding, J.; Steinegger, M. Fast and accurate protein structure search with Foldseek. Nat. Biotechnol. 2024, 42, 243–246. [Google Scholar] [CrossRef]
- Yildiz, Ö.; Vinothkumar, K.R.; Goswami, P.; Kühlbrandt, W. Structure of the monomeric outer-membrane porin OmpG in the open and closed conformation. EMBO J. 2006, 25, 3702–3713. [Google Scholar] [CrossRef]
- Köster, S.; van Pee, K.; Yildiz, Ö. Chapter Eight—Purification, refolding, and crystallization of the outer membrane protein OmpG from Escherichia coli. In Methods in Enzymology; Shukla, A.K., Ed.; Academic Press: Cambridge, MA, USA, 2015; Volume 557, pp. 149–166. [Google Scholar]
- Misra, R.; Benson, S.A. A novel mutation, cog, which results in production of a new porin protein (OmpG) of Escherichia coli K-12. J. Bacteriol. 1989, 171, 4105–4111. [Google Scholar] [CrossRef] [PubMed]
- Antonova Elena, S.; Hammer Brian, K. Genetics of natural competence in Vibrio cholerae and other Vibrios. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Li, Y.-D.; Ren, H.-L.; Lu, S.-Y.; Zhou, Y.; Han, X.; Gong, B.-B.; Zhang, Y.-Y.; Liu, Z.-S. Cloning, expression, and genus-specificity analysis of 28-kDa OmpK from Vibrio alginolyticus. J. Food Sci. 2010, 75, M198–M203. [Google Scholar] [CrossRef] [PubMed]
- Schwechheimer, C.; Hebert, K.; Tripathi, S.; Singh, P.K.; Floyd, K.A.; Brown, E.R.; Porcella, M.E.; Osorio, J.; Kiblen, J.T.M.; Pagliai, F.A.; et al. A tyrosine phosphoregulatory system controls exopolysaccharide biosynthesis and biofilm formation in Vibrio cholerae. PLoS Pathog. 2020, 16, e1008745. [Google Scholar] [CrossRef]
- Schwabe, J.; Pérez-Burgos, M.; Herfurth, M.; Glatter, T.; Søgaard-Andersen, L. Evidence for a widespread third system for bacterial polysaccharide export across the outer membrane comprising a composite OPX/β-barrel translocon. mBio 2022, 13, e02032-22. [Google Scholar] [CrossRef]
- Guest, T.; Haycocks, J.R.J.; Warren, G.Z.L.; Grainger, D.C. Genome-wide mapping of Vibrio cholerae VpsT binding identifies a mechanism for c-di-GMP homeostasis. Nucleic Acids Res. 2022, 50, 149–159. [Google Scholar] [CrossRef]
- Ayala, J.C.; Wang, H.; Silva, A.J.; Benitez, J.A. Repression by H-NS of genes required for the biosynthesis of the Vibrio cholerae biofilm matrix is modulated by the second messenger cyclic diguanylic acid. Mol. Microbiol. 2015, 97, 630–645. [Google Scholar] [CrossRef] [PubMed]
- Giles, D.K.; Hankins, J.V.; Guan, Z.; Trent, M.S. Remodelling of the Vibrio cholerae membrane by incorporation of exogenous fatty acids from host and aquatic environments. Mol. Microbiol. 2011, 79, 716–728. [Google Scholar] [CrossRef] [PubMed]
- Pride Aaron, C.; Herrera Carmen, M.; Guan, Z.; Giles David, K.; Trent, M.S. The outer surface lipoprotein VolA mediates utilization of exogenous lipids by Vibrio cholerae. mBio 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Pavoncello, V.; Barras, F.; Bouveret, E. Degradation of exogenous fatty acids in Escherichia coli. Biomolecules 2022, 12, 1019. [Google Scholar] [CrossRef]
- Turgeson, A.; Morley, L.; Giles, D.; Harris, B. Simulated docking predicts putative channels for the transport of long-chain fatty acids in Vibrio cholerae. Biomolecules 2022, 12, 1269. [Google Scholar] [CrossRef]
- Heffernan, E.J.; Reed, S.; Hackett, J.; Fierer, J.; Roudier, C.; Guiney, D. Mechanism of resistance to complement-mediated killing of bacteria encoded by the Salmonella Typhimurium virulence plasmid gene rck. J. Clin. Investig. 1992, 90, 953–964. [Google Scholar] [CrossRef]
- Saksena, S.; Forbes, K.; Rajan, N.; Giles, D. Phylogenetic investigation of Gammaproteobacteria proteins involved in exogenous long-chain fatty acid acquisition and assimilation. Biochem. Biophys. Rep. 2023, 35, 101504. [Google Scholar] [CrossRef]
- Rivera-Chávez, F.; Mekalanos, J.J. Cholera toxin promotes pathogen acquisition of host-derived nutrients. Nature 2019, 572, 244–248. [Google Scholar] [CrossRef]
- Chapman Claire, M.L.; Kapinos, A.; Rivera-Chávez, F. Modulation of host-microbe metabolism by cholera toxin. Infect. Immun. 2023, 91, e00435-22. [Google Scholar] [CrossRef]
- Beyhan, S.; Yildiz, F.H. Smooth to rugose phase variation in Vibrio cholerae can be mediated by a single nucleotide change that targets c-di-GMP signalling pathway. Mol. Microbiol. 2007, 63, 995–1007. [Google Scholar] [CrossRef]
- Peterson, K.M.; Mekalanos, J.J. Characterization of the Vibrio cholerae ToxR regulon: Identification of novel genes involved in intestinal colonization. Infect. Immun. 1988, 56, 2822–2829. [Google Scholar] [CrossRef] [PubMed]
- Hughes, K.J.; Everiss, K.D.; Kovach, M.E.; Peterson, K.M. Isolation and characterization of the Vibrio cholerae acfA gene, required for efficient intestinal colonization. Gene 1995, 156, 59–61. [Google Scholar] [CrossRef] [PubMed]
- Parsot, C.; Mekalanos, J.J. Structural analysis of the acfA and acfD genes of Vibrio cholerae: Effects of DNA topology and transcriptional activators on expression. J. Bacteriol. 1992, 174, 5211–5218. [Google Scholar] [CrossRef]
- Withey, J.H.; DiRita, V.J. Activation of both acfA and acfD transcription by Vibrio cholerae ToxT requires binding to two centrally located DNA sites in an inverted repeat conformation. Mol. Microbiol. 2005, 56, 1062–1077. [Google Scholar] [CrossRef]
- Gallego-Hernandez, A.L.; DePas, W.H.; Park, J.H.; Teschler, J.K.; Hartmann, R.; Jeckel, H.; Drescher, K.; Beyhan, S.; Newman, D.K.; Yildiz, F.H. Upregulation of virulence genes promotes Vibrio cholerae biofilm hyperinfectivity. Proc. Natl. Acad. Sci. USA 2020, 117, 11010–11017. [Google Scholar] [CrossRef] [PubMed]
- Marrero, K.; Sánchez, A.; Rodríguez-Ulloa, A.; González, L.J.; Castellanos-Serra, L.; Paz-Lago, D.; Campos, J.; Rodríguez, B.L.; Suzarte, E.; Ledón, T.; et al. Anaerobic growth promotes synthesis of colonization factors encoded at the Vibrio pathogenicity island in Vibrio cholerae El Tor. Res. Microbiol. 2009, 160, 48–56. [Google Scholar] [CrossRef]
- Sharma, M.K.; Jani, D.; Thungapathra, M.; Gautam, J.K.; Meena, L.S.; Singh, Y.; Ghosh, A.; Tyagi, A.K.; Sharma, A.K. Expression of accessory colonization factor subunit A (ACFA) of Vibrio cholerae and ACFA fused to cholera toxin B subunit in transgenic tomato (Solanum lycopersicum). J. Biotechnol. 2008, 135, 22–27. [Google Scholar] [CrossRef]
- Cheng, A.T.; Ottemann, K.M.; Yildiz, F.H. Vibrio cholerae response regulator VxrB controls colonization and regulates the type VI secretion system. PLoS Pathog. 2015, 11, e1004933. [Google Scholar] [CrossRef]
- Beaber John, W.; Hochhut, B.; Waldor Matthew, K. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 2002, 184, 4259–4269. [Google Scholar] [CrossRef]
- Asakura, H.; Ishiwa, A.; Arakawa, E.; Makino, S.i.; Okada, Y.; Yamamoto, S.; Igimi, S. Gene expression profile of Vibrio cholerae in the cold stress-induced viable but non-culturable state. Environ. Microbiol. 2007, 9, 869–879. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schutze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Tsenkov, M.; Velankar, S. Challenges in bridging the gap between protein structure prediction and functional interpretation. Proteins Struct. Funct. Bioinform. 2025, 93, 400–410. [Google Scholar] [CrossRef]
- Rosignoli, S.; Pacelli, M.; Manganiello, F.; Paiardini, A. An outlook on structural biology after AlphaFold: Tools, limits and perspectives. FEBS Open Bio 2025, 15, 202–222. [Google Scholar] [CrossRef]
- Chakravarty, D.; Lee, M.; Porter, L.L. Proteins with alternative folds reveal blind spots in AlphaFold-based protein structure prediction. Curr. Opin. Struct. Biol. 2025, 90, 102973. [Google Scholar] [CrossRef]
- Lin, P.-Y.; Huang, S.-C.; Chen, K.-L.; Huang, Y.-C.; Liao, C.-Y.; Lin, G.-J.; Lee, H.; Chen, P.-Y. Analysing protein complexes in plant science: Insights and limitation with AlphaFold 3. Bot. Stud. 2025, 66, 14. [Google Scholar] [CrossRef]
- Wee, J.; Wei, G.-W. Evaluation of AlphaFold 3’s protein–protein complexes for predicting binding free energy changes upon mutation. J. Chem. Inf. Model. 2024, 64, 6676–6683. [Google Scholar] [CrossRef]
- Singh, B.; Jaiswal, S.; Kodgire, P. Outer membrane proteins and vesicles as promising vaccine candidates against Vibrio spp. infections. Crit. Rev. Microbiol. 2024, 50, 417–433. [Google Scholar] [CrossRef]
- Rashid, M.I.; Rehman, S.; Ali, A.; Andleeb, S. Fishing for vaccines against Vibrio cholerae using in silico pan-proteomic reverse vaccinology approach. PeerJ 2019, 7, e6223. [Google Scholar] [CrossRef]
- Lång, H. Outer membrane proteins as surface display systems. Int. J. Med. Microbiol. 2000, 290, 579–585. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Wang, Q.; Wang, Y.; Wen, X.; Peng, H.; Peng, R.; Shi, Q.; Xie, X.; Li, L. Outer membrane porins contribute to antimicrobial resistance in Gram-negative bacteria. Microorganisms 2023, 11, 1690. [Google Scholar] [CrossRef]
- O’Ryan, M.; Stoddard, J.; Toneatto, D.; Wassil, J.; Dull, P.M. A multi-component meningococcal serogroup B vaccine (4CMenB): The clinical development program. Drugs 2014, 74, 15–30. [Google Scholar] [CrossRef]
- Lun, J.; Zheng, P.; Liang, X.; Hu, Y.; An, L.; Xiao, G.; Chen, X.; Chen, Y.; Gong, H.; Zhong, M.; et al. Identification of a conserved cryptic epitope with cross-immunoreactivity in outer membrane protein K (OmpK) from Vibrio species. Vaccine 2025, 53, 126964. [Google Scholar] [CrossRef]
- Kanoktippornchai, B.; Chomvarin, C.; Engchanil, C.; Wongboot, W. Triplex reverse transcription-PCR for detecting viable toxigenic Vibrio cholerae in water samples in Thailand. Southeast Asian J. Trop. Med. Public Health 2014, 45, 375. [Google Scholar]
- Martinez-Govea, A.; Ambrosio, J.; Gutierrez-Cogco, L.; Flisser, A. Identification and strain differentiation of Vibrio cholerae by using polyclonal antibodies against outer membrane proteins. Clin. Diagn. Lab. Immunol. 2001, 8, 768–771. [Google Scholar] [CrossRef] [PubMed]
- Maiti, B.; Dubey, S.; Munang’andu, H.M.; Karunasagar, I.; Karunasagar, I.; Evensen, Ø. Application of outer membrane protein-based vaccines against major bacterial fish pathogens in India. Front. Immunol. 2020, 11, 1362. [Google Scholar] [CrossRef]
- Dehbanipour, R.; Ghalavand, Z. Anti-virulence therapeutic strategies against bacterial infections: Recent advances. Germs 2022, 12, 262–275. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xiong, K.; Pan, Q.; He, W.; Cong, Y. Application of TonB-dependent transporters in vaccine development of Gram-Negative bacteria. Front. Cell. Infect. Microbiol. 2020, 10, 589115. [Google Scholar] [CrossRef]
- Rice, J.J.; Schohn, A.; Bessette, P.H.; Boulware, K.T.; Daugherty, P.S. Bacterial display using circularly permuted outer membrane protein OmpX yields high affinity peptide ligands. Protein Sci. 2006, 15, 825–836. [Google Scholar] [CrossRef]
- Chen, T.; Wang, K.; Chi, X.; Zhou, L.; Li, J.; Liu, L.; Zheng, Q.; Wang, Y.; Yu, H.; Gu, Y.; et al. Construction of a bacterial surface display system based on outer membrane protein F. Microb. Cell Factories 2019, 18, 70. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Song, M.; Li, K.; Yan, S.; Chen, M.; Geng, J.E. coli outer membrane protein T (OmpT) nanopore for peptide sensing. Biochem. Biophys. Res. Commun. 2023, 677, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Khalid, S.; Sansom, M.S.P.; Bayley, H. Outer membrane protein G: Engineering a quiet pore for biosensing. Proc. Natl. Acad. Sci. USA 2008, 105, 6272–6277. [Google Scholar] [CrossRef] [PubMed]
- Samineni, L.; Acharya, B.; Behera, H.; Oh, H.; Kumar, M.; Chowdhury, R. Protein engineering of pores for separation, sensing, and sequencing. Cell Syst. 2023, 14, 676–691. [Google Scholar] [CrossRef]
- Sanganna Gari, R.R.; Seelheim, P.; Liang, B.; Tamm, L.K. Quiet outer membrane protein G (OmpG) nanopore for biosensing. ACS Sens. 2019, 4, 1230–1235. [Google Scholar] [CrossRef]
| OmpV | OmpW | OmpU | OmpT | OmpS | OmpX | OmpA | LptD | BamA | ChiP | PhoE | TBDTs | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| General porin | x | x | x | |||||||||
| Substrate-specific porin | x | x | x | x | x | |||||||
| Osmoregulation | x | |||||||||||
| Antimicrobial resistance | x | x | x | x | ||||||||
| Bile resistance | x | x | ||||||||||
| Membrane vesicle formation | x | |||||||||||
| Biofilm matrix | x | x | x | |||||||||
| Biofilm formation | x | |||||||||||
| Cell-to-cell adhesion | x | |||||||||||
| Adhesion to surfaces | x | |||||||||||
| Signal transduction | x | x | ||||||||||
| Immunomodulation | x | |||||||||||
| Iron acquisition | ||||||||||||
| Structure/cell envelope | x (flagellum) | x (flagellum) | x | x | x | |||||||
| Virulence | x | x | x | x | x |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mathieu-Denoncourt, A.; Duperthuy, M. Functional Versatility of Vibrio cholerae Outer Membrane Proteins. Appl. Microbiol. 2025, 5, 64. https://doi.org/10.3390/applmicrobiol5030064
Mathieu-Denoncourt A, Duperthuy M. Functional Versatility of Vibrio cholerae Outer Membrane Proteins. Applied Microbiology. 2025; 5(3):64. https://doi.org/10.3390/applmicrobiol5030064
Chicago/Turabian StyleMathieu-Denoncourt, Annabelle, and Marylise Duperthuy. 2025. "Functional Versatility of Vibrio cholerae Outer Membrane Proteins" Applied Microbiology 5, no. 3: 64. https://doi.org/10.3390/applmicrobiol5030064
APA StyleMathieu-Denoncourt, A., & Duperthuy, M. (2025). Functional Versatility of Vibrio cholerae Outer Membrane Proteins. Applied Microbiology, 5(3), 64. https://doi.org/10.3390/applmicrobiol5030064






